Introduction
Renal cell carcinoma (RCC), the most common type of
human kidney cancer, is the most lethal urological malignancy
worldwide (1). In total, 295,000
novel cases of RCC and 134,000 mortalities due to RCC are estimated
globally every year (2). RCC may
be divided into three histological subtypes: Clear cell RCC
(ccRCC), papillary RCC and chromophobe RCC (3); these subtypes exhibit distinct
structural and cytogenetic characteristics (4). ccRCC, the most aggressive and
frequent subtype, accounts for ~80% of all diagnosed RCC cases
(5). Radical nephrectomy is the
primary therapeutic method used in patients with ccRCC who have
been diagnosed at an early stage, whereas, patients with local or
distant metastasis are not suitable for surgery (6,7). In
addition, patients with ccRCC benefit minimally from chemotherapy
and radiotherapy treatments (8).
Despite significant progress in therapeutic strategies, the
long-term prognosis of patients with metastatic ccRCC remains poor,
with a median survival period of 1.5 years (9). Therefore, it is urgent to elucidate
the molecular mechanism underlying ccRCC occurrence and
progression, in order to identify novel therapeutic targets for the
improvement of prognosis in patients with this malignancy.
MicroRNAs (miRNAs) are a group of endogenous, single
stranded and non-coding RNA molecules with a length of 19–24
nucleotides (10). Mature miRNAs
mediate gene expression by inducing mRNA degradation and/or
translation inhibition by binding to the complementary sequences in
the 3′-untranslated regions (3′-UTRs) of their target genes
(11). Over one-half of the
characterized miRNAs are located in genomic regions associated with
oncogenes, suggesting that miRNAs may serve important roles in
carcinogenesis and cancer progression (12). An increasing number of previous
studies have demonstrated that miRNAs are dysregulated in
approximately all types of human malignancy, including ccRCC
(13–15). miRNA (miR)-19a (16), miR-224 (17), miR-1274a (18) and miR-543 (19) were upregulated in ccRCC, whereas,
miR-30a (20), miR-200a (21), miR-384 (22) and miR-411 (23) were downregulated. These aberrantly
expressed miRNAs are implicated in the oncogenesis and progression
of ccRCC, and regulate various crucial biological processes,
including cell proliferation, apoptosis, metastasis and
angiogenesis (13,24). Further investigation on the miRNAs
in ccRCC may provide valuable therapeutic strategies for treating
patients with this aggressive disease.
miR-663 was upregulated in nasopharyngeal carcinoma
(25,26), lung cancer (27) and castration-resistant prostate
cancer (28); however, it was
downregulated in papillary thyroid carcinoma (29), pancreatic cancer (30), glioblastoma (31) and gastric cancer (31). However, little is known regarding
the expression pattern and roles of miR-663 in ccRCC. The present
study investigated miR-663 expression in ccRCC and examined its
effects on ccRCC progression. Additionally, the mechanisms
underlying the action of miR-663 in ccRCC were investigated.
p21 activated kinase 4 (PAK4) is a member of the
serine/threonine protein kinase family. PAK4 is located at the
locus 19q13.2 and it was described as a downstream effector of
Ras-related C3 botulinum toxin substrate 1 and cell division
control protein 42 homolog (32).
PAK4 is upregulated in multiple human cancer types, including
colorectal cancer (33), non-small
cell lung cancer (34),
glioblastoma (35) and gastric
cancer (36). PAK4 was implicated
in the carcinogenesis and cancer progression by affecting numerous
biological processes, including cell proliferation, apoptosis,
metastasis and cytoskeletal organisation (37–39).
Notably, it is highly expressed in ccRCC, and serves crucial roles
in the initiation and development of ccRCC (38,40–42).
Therefore, inhibition of PAK4 may represent a potential therapeutic
approach to treat patients with ccRCC.
Materials and methods
Tissue collection
A total of 31 ccRCC tissues and their matched normal
renal tissues were collected from patients at The China-Japan Union
Hospital of Jilin University (Changchun, China) between June 2015
and March 2017. Patients that had received chemotherapy,
radiotherapy or other treatments prior to surgical resection were
excluded from this research. The patients (18 males and 13 females;
age range, 48–71 years) received surgical resection, and the fresh
tissues were quickly frozen in liquid nitrogen and subsequently
stored at −80°C. The present study was approved by the Research
Ethics Committee of The China-Japan Union Hospital, and was
performed in accordance with the Declaration of Helsinki and the
guidelines of the Ethics Committee of The China-Japan Union
Hospital. Written informed consent was obtained from all the
patients for the use of their clinical tissues.
Cell culture
In total, one normal renal cell line (HK-2) and
three human ccRCC cell lines (Caki-1, 786-O and A498) were
purchased from The American Type Culture Collection (Manassas, VA,
USA). HK-2 cells were maintained in keratinocyte serum-free growth
medium containing bovine pituitary extract (0.05 mg/ml) and human
recombinant epidermal growth factor (5 ng/ml) (all from Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). The ccRCC cell
lines were grown in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% v/v fetal bovine serum (FBS), 100 µl/ml
penicillin and 100 mg/ml streptomycin sulphates (all from Gibco;
Thermo Fisher Scientific, Inc.). All cells were incubated at 37°C
in a humidified incubator containing 5% CO2.
Transfection assay
Chemically synthesized human miR-663 mimics and
negative control miRNA mimics (miR-NC) were obtained from Shanghai
GenePharma Co., Ltd. (Shanghai, China). The PAK4 overexpression
plasmid pcDNA3.1-PAK4 and empty pcDNA3.1 plasmid were provided by
The Chinese Academy of Sciences (Changchun, China). A total of
5×105 786-O and A498 cells were seeded into 6-well
culture plates and transfected with miRNA mimics (100 pmol) or
plasmid (4 µg) employing Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The miR-663 mimics sequence was
5′-AGGCGGGGCGCCGCGGGACCGC-3′ and the miR-NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. The transfection efficiency was
evaluated by reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) and western blot analysis. Transfected cells
were harvested and used for the subsequent functional assays.
RT-qPCR and cell invasion assay was performed at 48 h
post-transfection. Cell Counting Kit-8 (CCK-8) assays and western
blot analysis were performed after 24 and 72 h of incubation,
respectively.
Extraction of total RNA and
RT-qPCR
An RT-qPCR assay was conducted to measure miR-663
and PAK4 mRNA expression levels in ccRCC tissues and cells. Total
RNA was isolated from tissues or cells using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. For miR-663 detection, total RNA was
subjected to RT reactions using a TaqMan miRNA RT kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The temperature
protocol for RT was as follows: 16°C for 30 min, 42°C for 30 min
and 85°C for 5 min. The cDNA was amplified by qPCR using a TaqMan
miRNA PCR kit (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The qPCR was performed using the following thermocycling
conditions: 50°C for 2 min, 95°C for 10 min; 40 cycles of
denaturation at 95°C for 15 sec and annealing/extension at 60°C for
1 min.
To quantify PAK4 mRNA expression, cDNA was
synthesised using the Moloney Murine Leukemia Virus RT system
(Promega Corporation, Madison, WI, USA) according to the
manufacturer's instructions. Subsequently, a SYBR-Green PCR master
mix (Takara Bio, Inc., Otsu, Japan) was used to perform qPCR. The
thermocycling conditions for qPCR were as follows: 95°C for 10 min,
followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. U6
small nuclear RNA and GAPDH were used as internal reference genes
for miR-663 and PAK4 mRNA, respectively. RT-qPCR analysis was
performed in triplicate, and the data were analyzed based on the
2−ΔΔCq method (43).
The primers were: miR-663, 5′-TGCGGAGGCGGGGCGCCGCGGG-3′ (forward)
and 5′-CCAGTGCAGGGTCCGAGGT-3′ (reverse); U6,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and
5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); PAK4,
5′-AGGGAAGGCGGGAGATGAG-3′ (forward) and 5′-TCAGTTGCTTGTTCGTGC-3′
(reverse); and GAPDH, 5′-GAAGGCGGGAGATGAG-3′ (forward) and
5′-TCAGTTGCTTGTTCGTGC-3′ (reverse).
CCK-8 assay
Cells were harvested, resuspended and plated into
96-well plates at a density of 3×103 cells each well, 24
h following transfection. Cells were subsequently incubated at 37°C
in a humidified incubator containing 5% CO2 for 0, 24,
48 and 72 h. At every time point, transfected cells were subjected
to a CCK-8 assay to measure cell proliferation. In total, 10 µl
CCK-8 solution was added into each well, followed by incubation at
37°C for 2 h. The optical density was determined at 450 nm on the
BioTek™ ELx800™ spectrophotometer (BioTek
China, Beijing, China).
Cell invasion assay
Cell invasive ability was assessed using Transwell
chambers (8.0 µm pore size) that were precoated with Matrigel (both
from Becton, Dickinson and Company, Franklin Lakes, NJ, USA). A
total of 5×104 transfected cells suspended in 200 µl
FBS-free DMEM was added to the upper compartments of the Transwell
chambers, 48 h following transfection. The lower compartments of
the chambers were filled with 500 µl DMEM containing 20% FBS.
Following an incubation of 24 h, the non-invasive cells remaining
on the upper membrane were gently removed using a cotton swab. The
invasive cells were fixed with 95% ethanol at room temperature for
30 min and stained with 0.5% crystal violet at room temperature for
30 min. The number of invasive cells was counted in five randomly
chosen visual fields using an inverted light microscope (IX83;
Olympus Corporation, Tokyo, Japan; magnification, ×200).
miR-663 target prediction and
luciferase reporter assay
A total of two publically available databases,
TargetScan (http://www.targetscan.org/vert_71/) and microRNA
(http://www.microrna.org/microrna/home.do), were used
to predict the potential targets of miR-663. The 3′-UTR of PAK4
containing putative wild-type (Wt) miR-663-binding sites or mutant
(Mut) miR-663-binding sites were purchased from Shanghai GenePharma
Co., Ltd. and cloned into pmirGLO luciferase reporter vector
(Promega Corporation). The Wt and Mut luciferase plasmids were
defined as pmirGLO-Wt-PAK4-3′-UTR and pmirGLO-Mut-PAK4-3′-UTR,
respectively. 786-O and A498 cells (1×105) were
inoculated into 24-well plates in triplicate one night prior to
transfection. Cotransfection with pmirGLO-Wt-PAK4-3′-UTR or
pmirGLO-Mut-PAK4-3′-UTR and miR-663 mimics or miR-NC into cells was
conducted using Lipofectamine® 2000, according to the
manufacturer's protocol. Luciferase activity was measured after 48
h of incubation using the Dual-Luciferase® Reporter
assay system (Promega Corporation). Firefly luciferase activity was
normalized to Renilla luciferase activity.
Western blot analysis
Radioimmunoprecipitation Assay Lysis and Extraction
Buffer (Thermo Fisher Scientific, Inc.) was used to extract total
protein from tissues or cells. The concentration of total protein
was measured using a Bicinchoninic Acid Protein Assay kit (Thermo
Fisher Scientific, Inc.). An equal amount of protein (30 µg/lane)
was separated by 10% SDS-PAGE gels and subsequently transferred to
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). Subsequently, the membranes were blocked with 5% skimmed milk
in Tris-buffered saline containing 0.1% Tween-20 (TBST) at room
temperature for 1 h and incubated at 4°C overnight with primary
antibodies against PAK4 (cat. no. ab62509; 1:500; Abcam, Cambridge,
UK) or GAPDH (cat. no. ab181603; 1:500; Abcam). Following three
washes with TBST, the membranes were incubated at room temperature
with goat anti-mouse horseradish peroxidase-conjugated secondary
antibody (cat. no. ab6721; 1:5,000; Abcam) for 2 h. The protein
signals were detected using an enhanced chemiluminescent Substrate
kit (Pierce™; Thermo Fisher Scientific, Inc.). GAPDH
served as the internal control to normalize PAK4 protein
expression. Quantity One software version 4.62 (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) was used for
densitometry.
Statistical analysis
All data are presented as the mean ± standard
deviation from three independent experiments and were analyzed with
SPSS version 19.0 software (IBM Corp., Armonk, NY, USA). Student's
t-test was used to evaluate the differences between two groups.
One-way analysis of variance combined with Student-Newman-Keuls
test as the post hoc test, was used to compare the difference
between multiple groups. The correlation between expression levels
of miR-663 and PAK4 mRNA was determined using Spearman's
correlation analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-663 is downregulated in ccRCC
tissues and cell lines
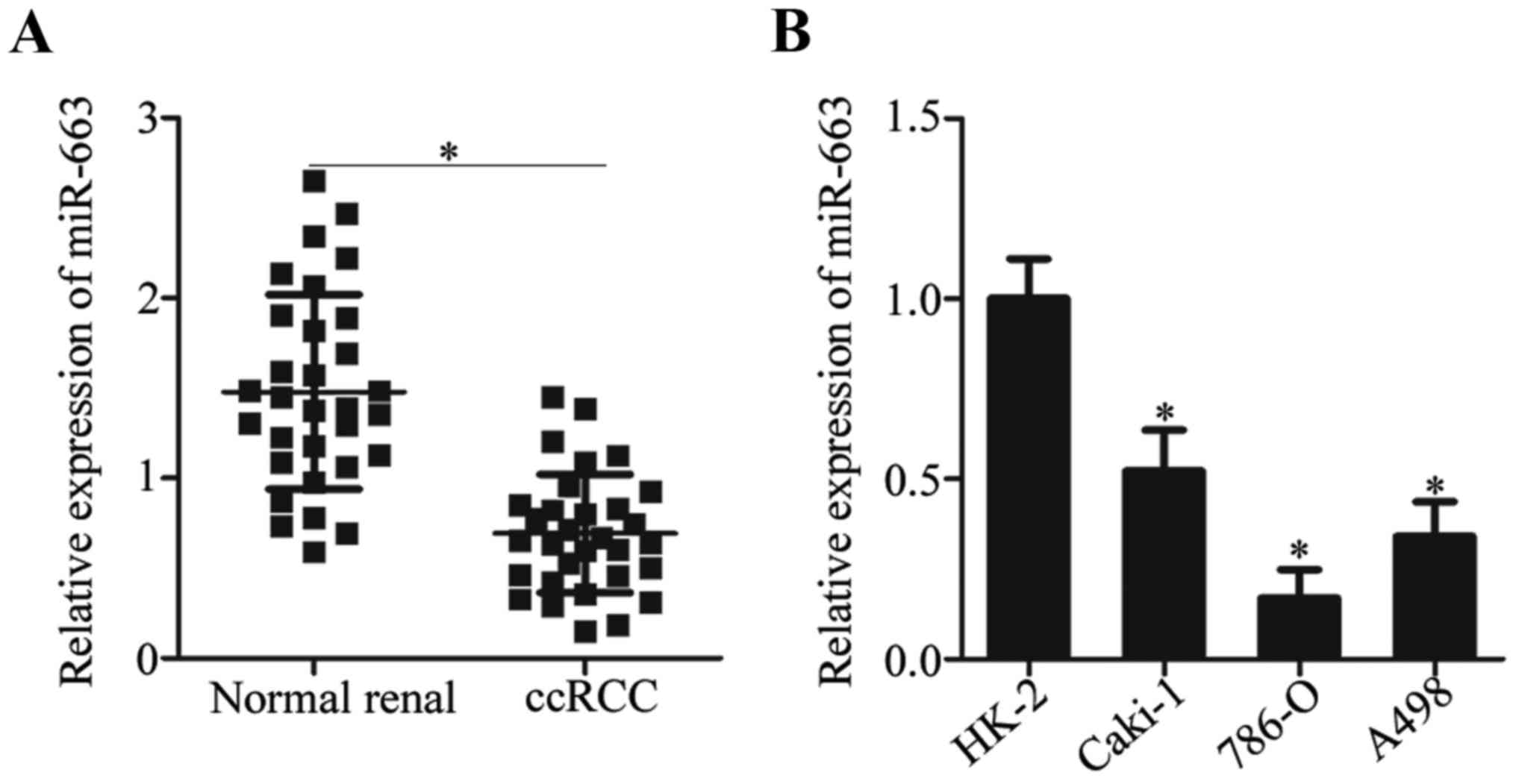
To investigate the expression pattern of miR-663 in
ccRCC, miR-663 expression in 31 paired ccRCC tissues and
corresponding matched normal renal tissues was measured by RT-qPCR.
The miR-663 expression was significantly downregulated in ccRCC
tissues compared with their matched normal renal tissues (Fig. 1A). Subsequently, miR-663 expression
in three ccRCC cell lines (Caki-1, 786-O and A498) and one normal
renal cell line (HK-2) was determined using RT-qPCR. The expression
level of miR-663 was decreased in ccRCC cell lines compared with
the HK-2 cell line (Fig. 1B). The
results suggested that miR-663 expression was downregulated in
ccRCC, and its expression level may be associated with the
development of ccRCC.
miR-663 restoration inhibits the
proliferation and invasion of 786-O and A498 cells
Among the three ccRCC cell lines (Caki-1, 786-O and
A498) investigated, 786-O and A498 cell lines expressed the lowest
level of miR-663 and were selected for the following experiments.
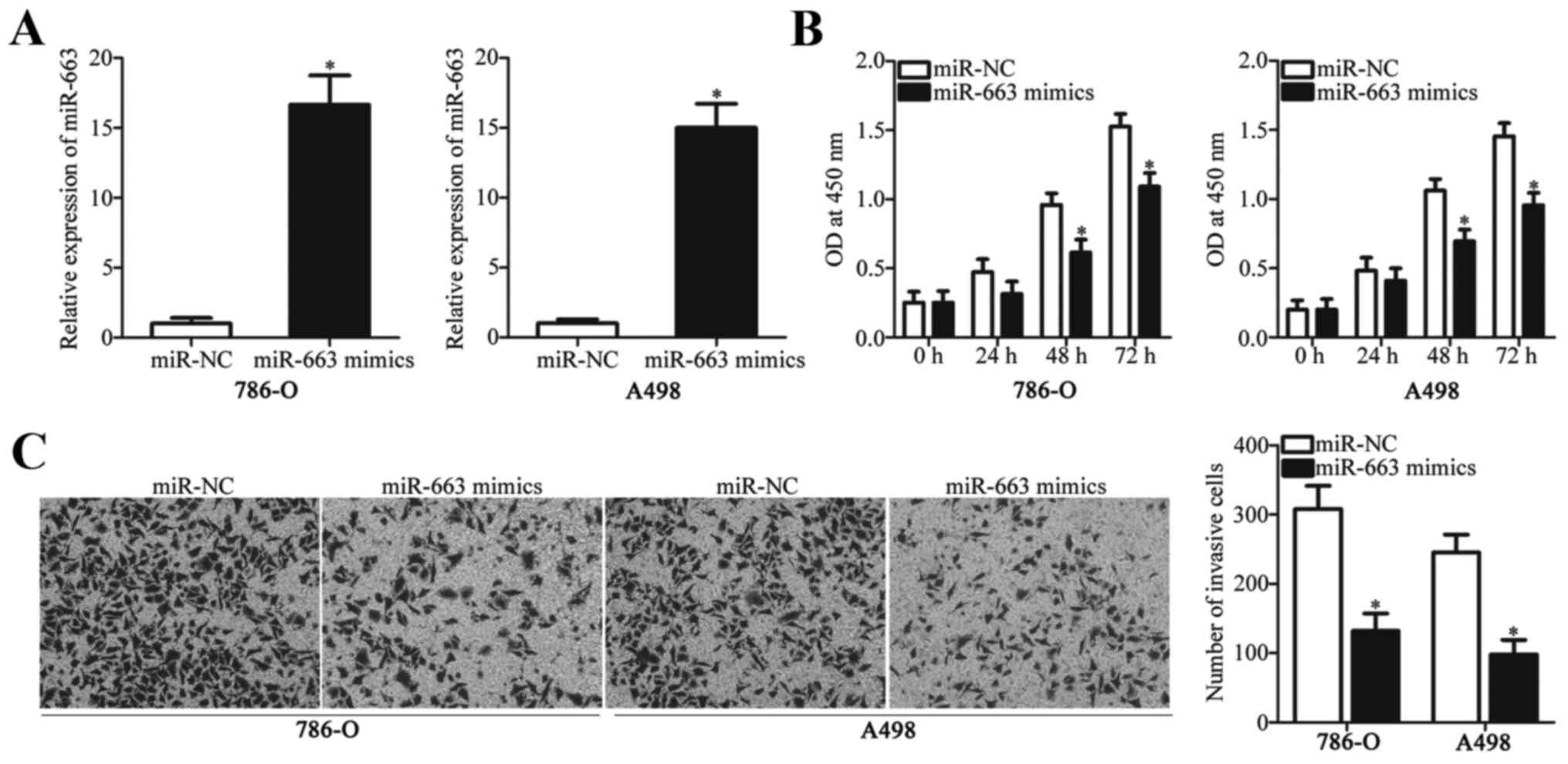
To investigate the roles of miR-663 in ccRCC, miR-663 mimics or
miR-NC was transfected into 786-O and A498 cells. Following
transfection, RT-qPCR analysis was used to evaluate transfection
efficiency. miR-663 expression level was significantly increased in
786-O and A498 cells transfected with miR-663 mimics compared with
the miR-NC (Fig. 2A).
A CCK-8 assay was performed to determine the effect
of miR-663 overexpression on the proliferative ability of ccRCC
cells. Upregulation of miR-663 led to a significant decrease in
cell proliferation in 786-O and A498 cells at 48 and 72 h (Fig. 2B). The invasive ability of 786-O
and A498 cells transfected with miR-663 mimics or miR-NC was
determined by a cell invasion assay. As depicted in Fig. 2C, miR-663 overexpression suppressed
the invasive ability of 786-O and A498 cells compared with cells
transfected with miR-NC. The present results suggested that miR-663
may serve tumor-suppressive roles in ccRCC progression.
PAK4 is a direct target of miR-663 in
ccRCC cells
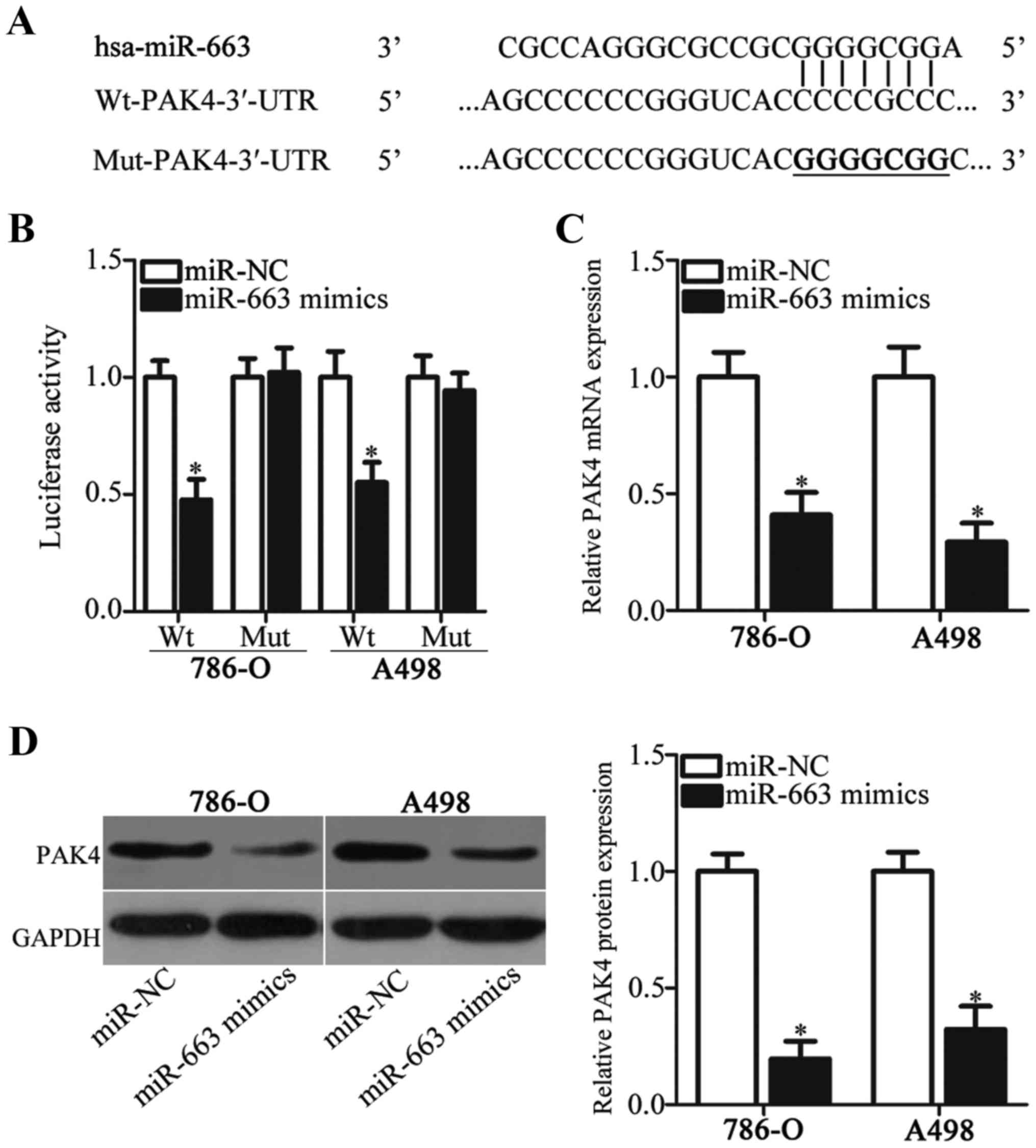
A bioinformatics analysis was performed to predict
the putative targets of miR-663 and to elucidate the mechanisms
underlying the inhibitory effects of miR-663 in ccRCC. Among the
candidates, PAK4 (Fig. 3A) was
selected for further identification since it is known to serve
crucial roles in the tumor formation and progression (38,40–42).
To investigate whether PAK4 is a target of miR-663, a luciferase
reporter assay was performed in 786-O and A498 cells following
cotransfection of miR-663 mimics or miR-NC and
pmirGLO-Wt-PAK4-3′-UTR or pmirGLO-Mut-PAK4-3′-UTR. miR-663
overexpression caused a significant decrease in luciferase activity
in 786-O and A498 cells transfected with the plasmid carrying the
Wt binding sites. However, the luciferase activity in cells
transfected with the plasmid carrying the mutated binding sites was
unaffected (Fig. 3B). To further
investigate this hypothesis, the role of miR-663 in regulating
endogenous PAK4 expression in ccRCC cells was examined. RT-qPCR and
western blot analysis demonstrated that the mRNA (Fig. 3C) and protein (Fig. 3D) expression levels of PAK4 in
786-O and A498 cells transfected with miR-663 mimics were
significantly decreased compared with the respective miR-NC. The
present results demonstrated that PAK4 was a direct target gene of
miR-663 in ccRCC cells.
miR-663 expression is negatively
correlated with PAK4 expression in ccRCC tissues
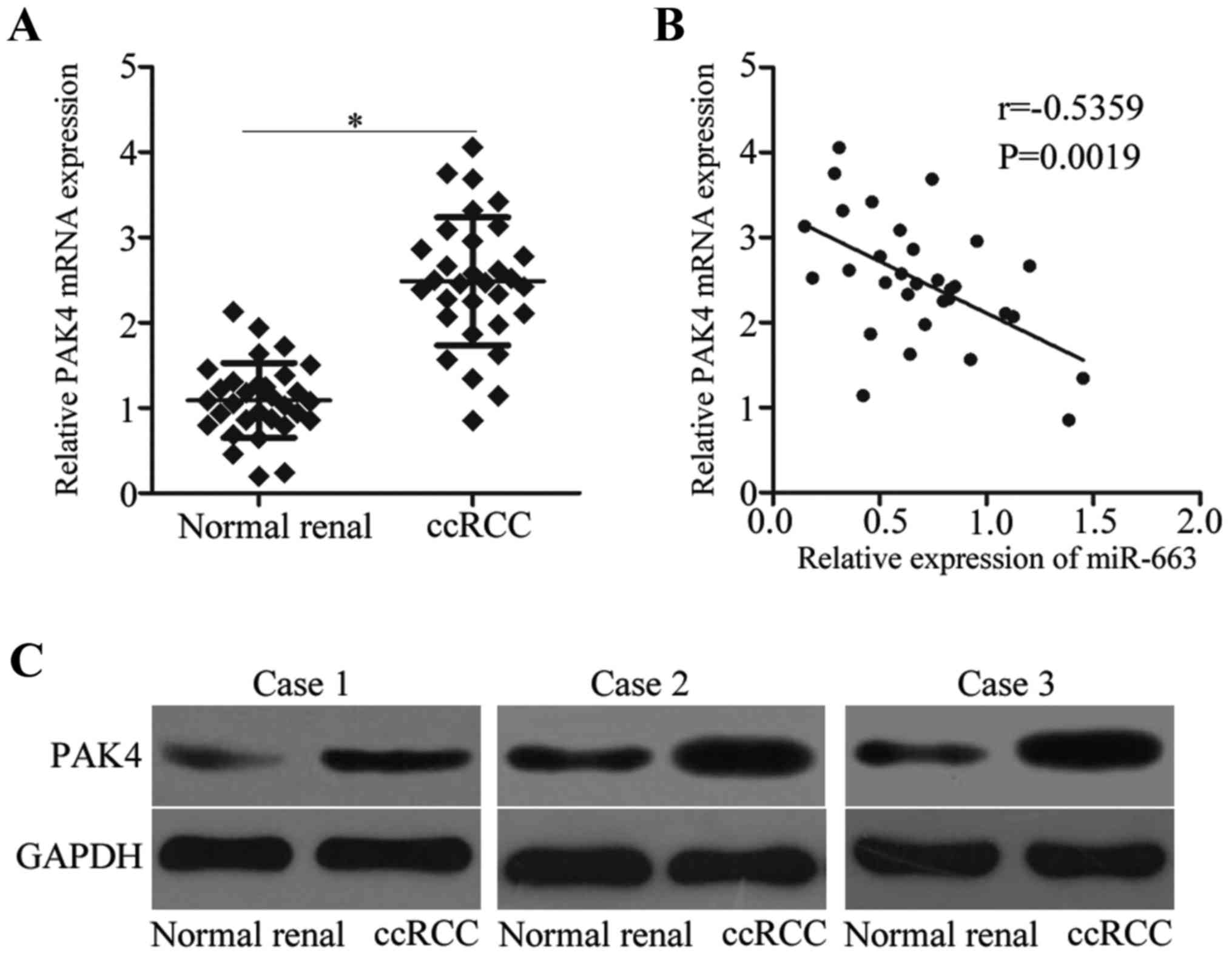
To further test the regulatory effects of miR-663 on
PAK4 in ccRCC, the mRNA expression level of PAK4 in 31 paired ccRCC
tissues and their corresponding matched normal renal tissues was
measured by RT-qPCR. The PAK4 mRNA expression level was
significantly increased in ccRCC tissues compared with matched
normal renal tissues (Fig. 4A).
The correlation analysis for miR-663 and PAK4 demonstrated that the
expression level of miR-663 was inversely correlated with PAK4
expression level in ccRCC tissues (Fig. 4B). Furthermore, the protein
expression level of PAK4 was upregulated in ccRCC tissues compared
with normal renal tissues (Fig.
4C).
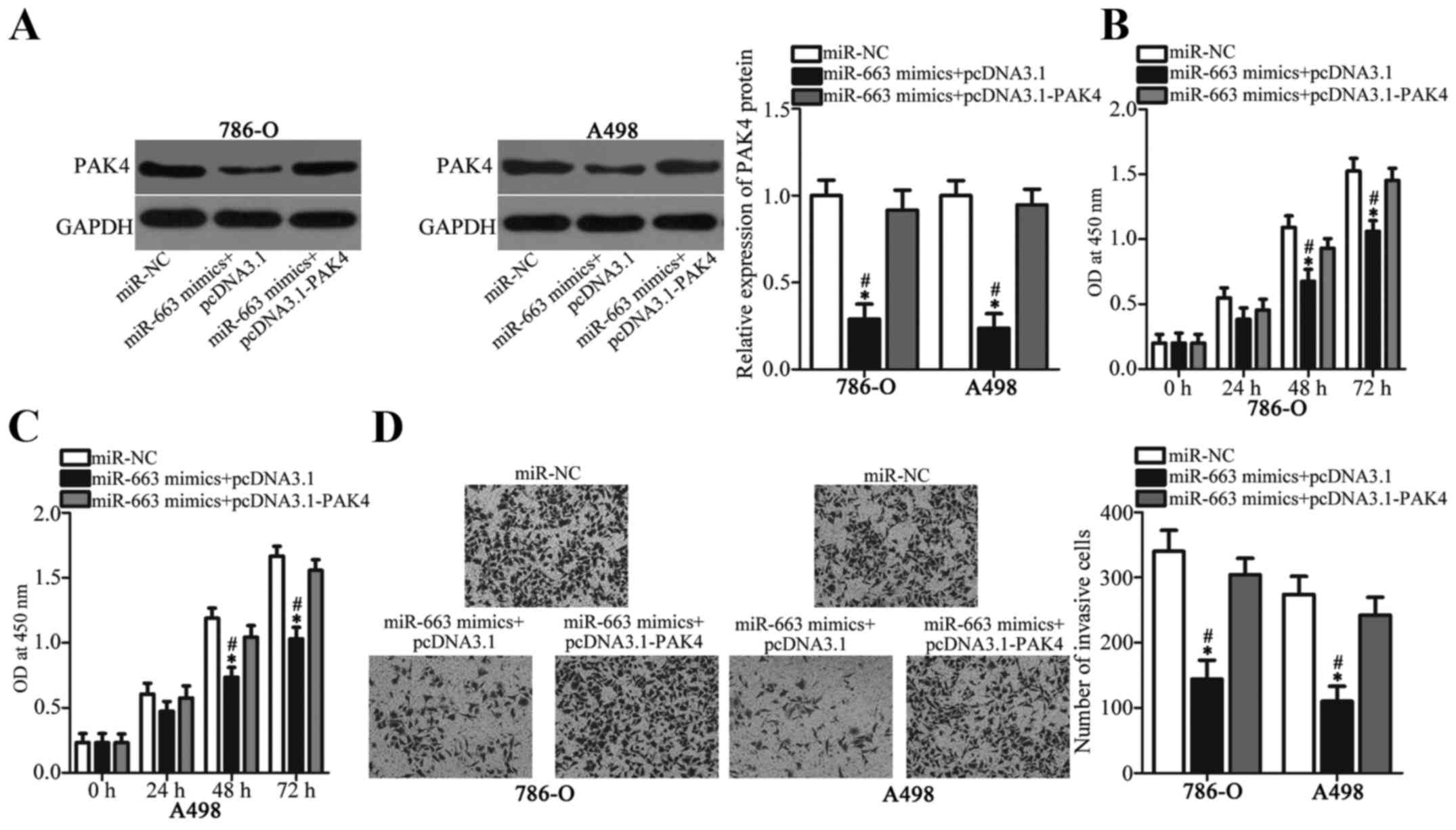
PAK4 upregulation attenuates the
tumor-suppressing roles of miR-663 in ccRCC cells
The PAK4 role in mediating the suppressive effects
of miR-663 in ccRCC cells was investigated by performing rescue
experiments. 786-O and A498 cells were cotransfected with miR-663
mimics and PAK4 overexpression plasmid or empty pcDNA3.1 plasmid,
as the NC. The western blot analysis demonstrated that the PAK4
protein expression level in 786-O and A498 cells, which was
decreased by miR-663 mimics, was re-established by cotransfection
of PAK4 expression plasmid (Fig.
5A). Subsequent functional experiments demonstrated that PAK4
overexpression attenuated the inhibitory effects of miR-663
overexpression on the proliferative (Fig. 5B and C) and invasive ability
(Fig. 5D) of 786-O and A498 cells.
The present results suggested that miR-663 may serve as a tumor
suppressor in ccRCC partly by downregulating PAK4 expression.
Discussion
Accumulating evidence has demonstrated that miRNAs
are key gene regulators and are abnormally expressed in ccRCC
(20,44,45).
The dysregulation of miRNAs has been demonstrated to affect the
initiation and progression of ccRCC (46,47).
Therefore, identification of ccRCC-associated miRNAs may facilitate
the determination of promising therapeutic targets for anti-cancer
treatment. In the present study, the expression levels of miR-663
in ccRCC tissues and cell lines were significantly decreased
compared with matched normal renal tissues and a normal renal cell
line, respectively. In addition, miR-663 inhibited the
proliferation and invasion of ccRCC cells. PAK4 was identified as a
direct target gene of miR-663 in ccRCC cells. PAK4 expression was
upregulated in ccRCC tissues, and its expression level was
inversely correlated with miR-663 expression level. Furthermore,
restoration of PAK4 expression was able to significantly inhibit
the effects of miR-663 overexpression on ccRCC cells. miR-663 may
serve as a novel type of tumor suppressor by directly targeting
PAK4 in ccRCC.
miR-663 has been previously identified to be
upregulated in multiple types of human cancer. miR-663 is
overexpressed in nasopharyngeal carcinoma tissues, cell lines and
serum (25,26,48).
The high expression of miR-663 was correlated with grade, lymph
node metastasis and clinical stage of nasopharyngeal carcinoma
(25,26). An increase in miR-663 expression
correlated with poor five-year survival rate and poor relapse-free
survival of patients with nasopharyngeal carcinoma (25). miR-663 was highly expressed in lung
cancer (27) and
castration-resistant prostate (28) cancer. However, miR-663 expression
is downregulated in papillary thyroid carcinoma tissues and cell
lines (29). Decreased miR-663
expression was associated with age and tumor size of papillary
thyroid carcinoma (29).
Downregulation of miR-663 was observed in pancreatic cancer
(30), glioblastoma (31) and gastric cancer (31). The previous findings suggested that
the expression pattern of miR-663 is tissue specific. Therefore,
this miRNA may be a promising biomarker for the detection and
prediction of prognosis of patients with these specific human
cancer types.
miR-663 serves oncogenic roles in numerous types of
human cancer. miR-663 overexpression was suggested to promote cell
proliferation in nasopharyngeal carcinoma by inducing cell cycle
arrest at the G1 stage in vitro and increasing
tumor growth in vivo (26,48).
Liu et al (27) and Fiori
et al (49) demonstrated
that miR-663 inhibition suppressed cell proliferation and induced
apoptosis in lung cancer. Jiao et al (28) identified that miR-663
overexpression attenuated dihydrotestosterone-induced upregulation
of prostate-specific antigen expression, and promoted
castration-resistant prostate cancer cell proliferation, invasion
and neuroendocrine differentiation. Nevertheless, miR-663 serves as
a tumor suppressor in papillary thyroid carcinoma by inhibiting
cell migration and invasion (29).
Zang et al (30)
demonstrated that restoration of miR-663 expression restricted the
growth and metastasis of pancreatic cancer in vitro and
in vivo. Pan et al (31) demonstrated that ectopic miR-663
expression suppressed the growth and metastasis of glioblastoma
cells. Collectively, the previous findings suggested that the
biological roles of miR-663 are tissue specific, and that miR-663
may be investigated as a therapeutic target for antineoplastic
therapy.
Numerous targets of miR-663 have been previously
identified, including cyclin dependent kinase inhibitor 2A
(26), cyclin dependent kinase
inhibitor 1A (48) in
nasopharyngeal carcinoma, p53 upregulated modulator of apoptosis
(49), BTG anti-proliferation
factor 2 (49) in lung cancer,
transforming growth factor β1 (29) in papillary thyroid carcinoma, and
elongation factor 1-α 2 (30) in
pancreatic cancer. PAK4, a member of the serine/threonine protein
kinase family, was identified as a direct target of miR-663 in
ccRCC. In ccRCC, an increased expression level of PAK4 correlated
with high Fuhrman grade and tumor necrosis (40). Patients with ccRCC with increased
expression levels of PAK4, exhibited poor overall survival and
recurrence-free survival compared with patients with decreased
expression levels of PAK4 (40).
Deregulated PAK4 expression was implicated in tumor oncogenesis and
development, and PAK4 was described to regulate a multitude of
biological processes, including ccRCC occurrence (38,41,42).
Therefore, miR-663-based therapy targeting PAK4 expression may
represent an effective therapeutic strategy for patients with
ccRCC.
In the present study, miR-663 served tumor
suppressive roles in ccRCC partly by targeting PAK4. The present
study provided better understanding of the formation and
progression of ccRCC, contributing to the investigation of novel
therapeutic targets for treating patients with ccRCC in the near
future. However, in the present study, a miR-663 inhibitor was not
tested to knockdown endogenous miR-663 expression, and the effects
of miR-663 knockdown in ccRCC cells was not examined. In addition,
the regulatory roles of miR-663 on ccRCC cell apoptosis were not
tested. These limitations of the present study require further
investigation in future studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZT designed the present study. YL and DJ performed
the functional experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of The China-Japan Union Hospital (Changchun,
China), and was performed in accordance with the Declaration of
Helsinki and the guidelines of the Research Ethics Committee of The
China-Japan Union Hospital. Written informed consent was obtained
from all patients for the use of their clinical tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Linehan WM: The genetic basis of kidney
cancer: Implications for management and use of targeted therapeutic
approaches. Eur Urol. 61:896–898. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nerich V, Hugues M, Paillard MJ, Borowski
L, Nai T, Stein U, Nguyen Tan Hon T, Montcuquet P, Maurina T,
Mouillet G, et al: Clinical impact of targeted therapies in
patients with metastatic clear-cell renal cell carcinoma. Onco
Targets Ther. 7:365–374. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meloni-Ehrig AM: Renal cancer: Cytogenetic
and molecular genetic aspects. Am J Med Genet. 115:164–172. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheville JC, Lohse CM, Zincke H, Weaver AL
and Blute ML: Comparisons of outcome and prognostic features among
histologic subtypes of renal cell carcinoma. Am J Surg Pathol.
27:612–624. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rini BI, Rathmell WK and Godley P: Renal
cell carcinoma. Curr Opin Oncol. 20:300–306. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ljungberg B, Bensalah K, Canfield S,
Dabestani S, Hofmann F, Hora M, Kuczyk MA, Lam T, Marconi L,
Merseburger AS, et al: EAU guidelines on renal cell carcinoma: 2014
update. Eur Urol. 67:913–924. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Albiges L, Choueiri T, Escudier B, Galsky
M, George D, Hofmann F, Lam T, Motzer R, Mulders P, Porta C, et al:
A systematic review of sequencing and combinations of systemic
therapy in metastatic renal cancer. Eur Urol. 67:100–110. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu D, Li M, Wang L, Zhou Y, Zhou J, Pan H
and Qu P: MicroRNA-145 inhibits cell proliferation, migration and
invasion by targeting matrix metallopeptidase-11 in renal cell
carcinoma. Mol Med Rep. 10:393–398. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kurozumi A, Goto Y, Okato A, Ichikawa T
and Seki N: Aberrantly expressed microRNAs in bladder cancer and
renal cell carcinoma. J Hum Genet. 62:49–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vannini I, Fanini F and Fabbri M: Emerging
roles of microRNAs in cancer. Curr Opin Genet Dev. 48:128–133.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lou W, Liu J, Gao Y, Zhong G, Chen D, Shen
J, Bao C, Xu L, Pan J, Cheng J, et al: MicroRNAs in cancer
metastasis and angiogenesis. Oncotarget. 8:115787–115802. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Niu S, Ma X, Zhang Y, Liu YN, Chen X, Gong
H, Yao Y, Liu K and Zhang X: MicroRNA-19a and microRNA-19b promote
the malignancy of clear cell renal cell carcinoma through targeting
the tumor suppressor RhoB. PLoS One. 13:e01927902018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fujii N, Hirata H, Ueno K, Mori J, Oka S,
Shimizu K, Kawai Y, Inoue R, Yamamoto Y, Matsumoto H, et al:
Extracellular miR-224 as a prognostic marker for clear cell renal
cell carcinoma. Oncotarget. 8:109877–109888. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yoshino H, Yonezawa T, Yonemori M,
Miyamoto K, Sakaguchi T, Sugita S, Osako Y, Tatarano S, Nakagawa M
and Enokida H: Downregulation of microRNA-1274a induces cell
apoptosis through regulation of BMPR1B in clear cell renal cell
carcinoma. Oncol Rep. 39:173–181. 2018.PubMed/NCBI
|
|
19
|
Yang F, Ma J, Tang Q, Zhang W, Fu Q, Sun
J, Wang H and Song B: MicroRNA-543 promotes the proliferation and
invasion of clear cell renal cell carcinoma cells by targeting
Krüppel-like factor 6. Biomed Pharmacother. 97:616–623. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang C, Cai L, Liu J, Wang G, Li H, Wang
X, Xu W, Ren M, Feng L, Liu P and Zhang C: MicroRNA-30a-5p inhibits
the growth of renal cell carcinoma by modulating GRP78 expression.
Cell Physiol Biochem. 43:2405–2419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fu H, Song W, Chen X, Guo T, Duan B, Wang
X, Tang Y, Huang L and Zhang C: miRNA-200a induce cell apoptosis in
renal cell carcinoma by directly targeting SIRT1. Mol Cell Biochem.
437:143–152. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song H, Rao Y, Zhang G and Kong X:
MicroRNA-384 inhibits the growth and invasion of renal cell
carcinoma cells by targeting astrocyte elevated gene 1. Oncol Res.
26:457–466. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Zhang M, Cheng J, Lv Z, Wang F
and Cai Z: miR-411 functions as a tumor suppressor in renal cell
cancer. Int J Biol Markers. 32:e454–e460. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He YH, Chen C and Shi Z: The biological
roles and clinical implications of microRNAs in clear cell renal
cell carcinoma. J Cell Physiol. 233:4458–4465. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liang S, Zhang N, Deng Y, Chen L, Zhang Y,
Zheng Z, Luo W, Lv Z, Li S and Xun T: Increased serum level of
MicroRNA-663 is correlated with poor prognosis of patients with
nasopharyngeal carcinoma. Dis Markers. 2016:76482152016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liang S, Zhang N, Deng Y, Chen L, Zhang Y,
Zheng Z, Luo W, Lv Z, Li S and Xu T: miR-663 promotes NPC cell
proliferation by directly targeting CDKN2A. Mol Med Rep.
16:4863–4870. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu ZY, Zhang GL, Wang MM, Xiong YN and
Cui HQ: MicroRNA-663 targets TGFB1 and regulates lung cancer
proliferation. Asian Pac J Cancer Prev. 12:2819–2823.
2011.PubMed/NCBI
|
|
28
|
Jiao L, Deng Z, Xu C, Yu Y, Li Y, Yang C,
Chen J, Liu Z, Huang G, Li LC and Sun Y: miR-663 induces
castration-resistant prostate cancer transformation and predicts
clinical recurrence. J Cell Physiol. 229:834–844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Z, Zhang H, Zhang P, Dong W and He L:
MicroRNA-663 suppresses cell invasion and migration by targeting
transforming growth factor beta 1 in papillary thyroid carcinoma.
Tumour Biol. 37:7633–7644. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zang W, Wang Y, Wang T, Du Y, Chen X, Li M
and Zhao G: miR-663 attenuates tumor growth and invasiveness by
targeting eEF1A2 in pancreatic cancer. Mol Cancer. 14:372015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pan J, Hu H, Zhou Z, Sun L, Peng L, Yu L,
Sun L, Liu J, Yang Z and Ran Y: Tumor-suppressive mir-663 gene
induces mitotic catastrophe growth arrest in human gastric cancer
cells. Oncol Rep. 24:105–112. 2010.PubMed/NCBI
|
|
32
|
Bokoch GM: Biology of the p21-activated
kinases. Annu Rev Biochem. 72:743–781. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song B, Wang W, Zheng Y, Yang J and Xu Z:
P21-activated kinase 1 and 4 were associated with colorectal cancer
metastasis and infiltration. J Surg Res. 196:130–135. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang X, Lu Y, Feng W, Chen Q, Guo H, Sun X
and Bao Y: A two kinase-gene signature model using CDK2 and PAK4
expression predicts poor outcome in non-small cell lung cancers.
Neoplasma. 63:322–329. 2016.PubMed/NCBI
|
|
35
|
Mao K, Lei D, Zhang H and You C:
MicroRNA-485 inhibits malignant biological behaviour of
glioblastoma cells by directly targeting PAK4. Int J Oncol.
51:1521–1532. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kobayashi K, Inokuchi M, Takagi Y, Otsuki
S, Fujimori Y, Sato Y, Yanaka Y, Higuchi K, Aburatani T, Tomii C,
et al: Prognostic significance of PAK4 expression in gastric
cancer. J Clin Pathol. 69:580–585. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gnad F, Young A, Zhou W, Lyle K, Ong CC,
Stokes MP, Silva JC, Belvin M, Friedman LS, Koeppen H, et al:
Systems-wide analysis of K-Ras, Cdc42, and PAK4 signaling by
quantitative phosphoproteomics. Mol Cell Proteomics. 12:2070–2080.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Siu MK, Chan HY, Kong DS, Wong ES, Wong
OG, Ngan HY, Tam KF, Zhang H, Li Z, Chan QK, et al: p21-activated
kinase 4 regulates ovarian cancer cell proliferation, migration,
and invasion and contributes to poor prognosis in patients. Proc
Natl Acad Sci USA. 107:18622–18627. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ahmed T, Shea K, Masters JR, Jones GE and
Wells CM: A PAK4-LIMK1 pathway drives prostate cancer cell
migration downstream of HGF. Cell Signal. 20:1320–1328. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu W, Yang Y, Liu Y, Liu H, Zhang W, Xu
L, Zhu Y and Xu J: p21-Activated kinase 4 predicts early recurrence
and poor survival in patients with nonmetastatic clear cell renal
cell carcinoma. Urol Oncol. 33:205.e13–e21. 2015. View Article : Google Scholar
|
|
41
|
Abu Aboud O, Chen CH, Senapedis W, Baloglu
E, Argueta C and Weiss RH: Dual and specific inhibition of NAMPT
and PAK4 By KPT-9274 decreases kidney cancer growth. Mol Cancer
Ther. 15:2119–2129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kesanakurti D, Chetty C, Rajasekhar
Maddirela D, Gujrati M and Rao JS: Functional cooperativity by
direct interaction between PAK4 and MMP-2 in the regulation of
anoikis resistance, migration and invasion in glioma. Cell Death
Dis. 3:e4452012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pan YJ, Wei LL, Wu XJ, Huo FC, Mou J and
Pei DS: miR-106a-5p inhibits the cell migration and invasion of
renal cell carcinoma through targeting PAK5. Cell Death Dis.
8:e31552017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kovacova J, Poprach A, Buchler T, Cho WC
and Slaby O: MicroRNAs as predictive biomarkers of response to
tyrosine kinase inhibitor therapy in metastatic renal cell
carcinoma. Clin Chem Lab Med. 56:1426–1431. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Redova M, Svoboda M and Slaby O: MicroRNAs
and their target gene networks in renal cell carcinoma. Biochem
Biophys Res Commun. 405:153–156. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Al-Ali BM, Ress AL, Gerger A and Pichler
M: MicroRNAs in renal cell carcinoma: Implications for
pathogenesis, diagnosis, prognosis and therapy. Anticancer Res.
32:3727–3732. 2012.PubMed/NCBI
|
|
48
|
Yi C, Wang Q, Wang L, Huang Y, Li L, Liu
L, Zhou X, Xie G, Kang T, Wang H, et al: miR-663, a microRNA
targeting p21(WAF1/CIP1), promotes the proliferation and
tumorigenesis of nasopharyngeal carcinoma. Oncogene. 31:4421–4433.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fiori ME, Villanova L, Barbini C, De
Angelis ML and De Maria R: miR-663 sustains NSCLC by inhibiting
mitochondrial outer membrane permeabilization (MOMP) through
PUMA/BBC3 and BTG2. Cell Death Dis. 9:492018. View Article : Google Scholar : PubMed/NCBI
|