Introduction
Meniscal injury is a common disorder that may cause
pain, swelling or mechanical symptoms in patients (1). As articular cartilages of the knee
joint, menisci are crucial for normal function of the knee joint
and protection of the articular surfaces (2). Articular cartilages, as specialized
connective tissues of diarthrodial joints, are affected by overall
fitness levels (3,4).
Previously, the effect of factors including aging
and obesity on articular cartilages has been widely studied. With
aging, changes occur in articular cartilages, including
degenerative changes in the morphology, loss of cartilage matrix
proteins, cleavage of type II collagen, worsening of mechanical
properties and dysregulated expression of associated genes
(4–7). An inverse correlation between
cartilage thickness within the knee joint and age has been
demonstrated (8). The mechanisms
of obesity affecting articular cartilages are complex, and include
biomechanical effects via increased loading and meta-inflammation
(9–14). Abnormalities in the articular
cartilage of the knee are also associated with obesity, as revealed
by the investigation of obese adolescent patients suffering from
knee pain (15).
Details of the gene expression profile in human
meniscal injury have been described previously (16–18).
Expression of vascular endothelial growth factor A (VEGFA), tumor
necrosis factor-α and matrix metalloproteinases (MMPs) were
demonstrated to be varied by age in patients with meniscal tears
(16,19). Neuronal apoptosis-inhibitory
protein, apoptosis inhibitor of macrophage, carbohydrate
sulfotransferase 15 and MMP28 were suggested to be associated with
obesity (17). However, gene
expression changes associated with injuries and aging or obesity in
the human meniscus have not been fully investigated.
Microarray analysis of gene expression changes and
functional pathways may be useful to improve understanding of the
associations between aging or obesity and meniscal injuries. In
previous studies, GSE45233 microarray data was used to study
variation in gene expression signatures in human injured meniscus
with age or body mass index (BMI) and the degree of chondrosis in
the knee (16,17). GSE66635 microarray data was used to
compare transcriptome signatures between the meniscus and articular
cartilage from knees undergoing arthroscopic partial meniscectomy
(18). In the present study, the
GSE45233 and GSE66635 datasets were downloaded and analyzed to
identify genes and functional pathways associated with age or BMI
in human injured meniscus. Differentially expressed genes (DEGs)
were identified and feature genes were obtained by comparing the
DEGs from the 2 datasets. Protein-protein interaction (PPI)
networks of these feature genes were constructed to obtain hub
genes. Functional pathway enrichment analysis for feature genes was
also performed to identify key pathways in the development of age
or BMI-associated meniscal injuries. Furthermore, the GSE51588
genome-wide expression profile was downloaded from the Gene
Expression Omnibus (GEO) database to validate the results. The
results concerning gene expression changes in human injured
meniscus may provide an improved understanding of the associations
between meniscal injuries and aging or obesity.
Materials and methods
Affymetrix microarray data
The gene expression profile datasets GSE66635,
deposited by Rai et al (18), and GSE45233, deposited by Rai et
al (16,17), were downloaded from the GEO
(www.ncbi.nlm.nih.gov/geo/) database.
GSE66635 and GSE45233 were based on the GPL16686 Human Gene 2.0 ST
Array (Affymetrix Inc.; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) and GPL10558 HumanHT-12 V4.0 expression Beadchip
(Illumina, Inc., San Diego, CA, USA) platforms, respectively. A
total of 24 samples were available in the GSE66635 dataset,
including 12 injured meniscus and 12 normal articular cartilages
from patients undergoing partial meniscectomy. The second
microarray profile, GSE45233, included 12 isolated injured meniscus
tissues from patients undergoing arthroscopic partial
meniscectomy.
Data preprocessing and feature genes
screening
The probe-level data in the CEL files were converted
to expression measures, and idle probes were deleted. When multiple
probes corresponded to the same gene, the expression values of
those probes were averaged. The 2 datasets were aggregated
according to gene names, followed by background correction and
quartile data normalization by the robust multiarray average
algorithm in R language (20).
Based on the GSE66635 gene expression profile data,
DEGs in injured menisci from patients undergoing partial
meniscectomy were identified using the Linear Models for Microarray
Data (Limma) package (21) in R
language compared with normal articular cartilage. The cut-off
values were P<0.05 and fold change (FC) >1.5.
Samples of GSE45233 were grouped by age and BMI
according to World Health Organization standards (lean, BMI
<18.5; normal, BMI 18.5–25; obese, BMI >25 kg/m2)
(22). The age group included 6
samples of patients >40 years of age (old group) and 6 samples
of patients <40 years of age (young group). DEGs associated with
age were then identified in the old group using the Limma package
and compared with the young group.
The BMI group contained 8 samples of lean patients
(lean group) and 4 samples of obese patients (obese group); no
patients were in the normal category according to this dataset.
DEGs associated with age were identified in the obese group using
the Limma package and compared with the lean group.
Common DEGs between GSE66635 and the age group in
GSE45233 were defined as ‘feature’ genes associated with
age-associated meniscal injuries. Similarly, common DEGs identified
from GSE66635 and the BMI group in GSE45233 were BMI-associated
feature genes.
Integrative analysis of feature gene
expression levels
A total of 9 samples of patients >40 years of age
and 3 samples of patients <40 years of age were selected from
the injured meniscus samples in GSE66635. Combining these samples
with those in the age group from GSE45233, a new age group with 15
samples of patients >40 years of age and 9 samples of patients
<40 years of age was obtained. Similarly, 8 obese samples from
GSE66635 were selected and combined with those in the BMI group
from GSE45233, obtaining a new BMI group with 8 lean samples and 12
obese samples. The significances of average expression level
differences for feature genes extracted from the two new groups
were analyzed by t-test using the Limma package (23). Heat maps were generated using
Pheatmap (24) in R language, to
exhibit the results of bidirectional hierarchical clustering based
on the expression value of the feature genes.
Classification of the sample
classification model
Support vector machines (SVM) (25) are a useful technique for two-group
classification problems. To verify the identifiability and
classifiability of the feature genes extracted from the two new
groups, the SVM classifier model was performed based on the
expression value of these feature genes. SVMs were initially
optimized using the DEGs in the training set and the feature genes
were examined in the test set to separate the samples in the new
age and BMI groups.
PPI network construction
The Human Protein Reference Database (HPRD;
www.hprd.org/) (26) is a protein database that contains
information on human protein functions, including PPIs,
post-translational modifications, and enzyme-substrate and disease
associations. PPI networks for age-associated feature genes and
BMI-associated feature genes were respectively constructed by
mapping the two groups of feature genes to the PPI pairs downloaded
from HPRD, which were then visualized using Cytoscape (27). Finally, hub genes with the highest
node degree in the PPI networks were identified using CytoNCA
(28) plugin (network without
weight) in Cytoscape.
Pathway enrichment analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway database (www.genome.jp/kegg/pathway.html) (29) is used for the systematic analysis
of gene functions and associated genomic information with higher
order functional information. The feature genes were examined using
the KEGG pathway database to identify functional pathways. The
enrichment process was performed by the Fisher algorithm using
KOBAS2.0 (30) as follows:
P=1-∑i=0x-1(Mi)(N-MK-i)(NK)
where N is the number of total genes in the genome,
M is the number of genes in the pathway and K is the number of
DEGs. The Fisher score indicates the probability of X functional
pathway genes among K DEGs.
Data validation
To validate the results, the GSE51588 genome-wide
expression profile was downloaded from the GEO database. This
dataset was obtained from 40 subchondral bone tissue samples from
patients with osteoarthritis and 10 samples from normal subjects
based on the GPL13497 Agilent-026652 Whole Human Genome Microarray
4×44 K v2 platform. The original data in TXT format were
preprocessed using the Limma package (version 3.30.13; www.bioconductor.org/packages/2.9/bioc/html/limma.html)
(21). The preprocessing methods
included background correction and data normalization. DEGs were
identified using the Bayes method in Limma with a threshold
P<0.05.
The overlapping DEGs between GSE66635 and GSE45233
were selected as candidate genes, which were then compared with the
DEGs obtained from GSE51588.
Results
Screening of candidate feature
genes
Following data normalization, genes exceeding the
difference threshold (P<0.05 and FC >1.5) were screened. A
total of 364 DEGs were identified in injured meniscus samples
compared with normal meniscus samples in GSE66635, among which 135
DEGs were down-regulated and 229 DEGs were upregulated. A total of
424 DEGs were screened in the old group compared with the young
group in GSE45233, including 222 downregulated genes and 202
upregulated genes. In the obese group, compared with the lean
group, 540 genes were differentially expressed, including 340
downregulated and 200 upregulated genes. Comparison of the DEGs in
the GSE66635 dataset with the DEGs of the age group in GSE45233
identified 28 age-associated meniscal injury feature genes
(Table I). Of these, 15 feature
genes were downregulated, which included S100 calcium-binding
protein A1 (S100A1) and BARX homeobox 2, and 13 feature genes were
upregulated, which included cluster of differentiation 36 and
endomucin (EMCN). A total of 20 common genes between the GSE45233
dataset and the BMI group in GSE45233 were identified as
BMI-associated meniscal injury feature genes (Table I). Of these, 11 feature genes were
downregulated, which included EMCN and furry, and 9 feature genes
were upregulated, which included IGF binding protein 1 and S100
calcium-binding protein A8 (S100A8).
 | Table I.Feature genes in age and body mass
index groups. |
Table I.
Feature genes in age and body mass
index groups.
| A, Age group |
|---|
|
|---|
| Gene | P-value | log2FC |
|---|
| S100A1 | 0.0004 | −5.64 |
| BARX2 | 0.0096 | −2.95 |
| IGFBP1 | 0.0341 | −2.46 |
| HAPLN1 | 0.0031 | −2.21 |
| DIXDC1 | 0.0123 | −2.01 |
| FGFBP2 | 0.0381 | −1.92 |
| FBLN7 | 0.0076 | −1.81 |
| VEGFA | 0.0173 | −1.66 |
| GREM1 | 0.0171 | −1.56 |
| TF | 0.0281 | −1.40 |
| BCL2 | 0.0345 | −1.30 |
| DCHS1 | 0.0020 | −1.29 |
| SMOC1 | 0.0042 | −1.23 |
| CAPN6 | 0.0167 | −1.20 |
| PTGES | 0.0352 | −1.08 |
| VIT | 0.0459 | 1.00 |
| LYZ | 0.0258 | 1.23 |
| FGL2 | 0.0033 | 1.24 |
| CCNB1 | 0.0030 | 1.31 |
| PBK | 0.0043 | 1.39 |
| BST2 | 0.0102 | 1.46 |
| CTSS | 0.0222 | 1.48 |
| HTRA4 | 0.0068 | 1.51 |
| DDHD1 | 0.0093 | 1.55 |
| TSPAN7 | 0.0007 | 1.57 |
| CALCRL | 0.0198 | 2.61 |
| EMCN | 0.0171 | 6.40 |
| CD36 | 0.0010 | 9.33 |
|
| B, Body mass
index group |
|
| Gene | P-value | log2FC |
|
| EMCN | 0.0169 | −6.40 |
| FRY | 0.0353 | −4.20 |
| EDNRA | 0.0084 | −3.18 |
| MFAP2 | 0.0304 | −2.26 |
| CXCL12 | 0.0113 | −2.19 |
| ABHD2 | 0.0010 | −1.86 |
| EYA4 | 0.0339 | −1.79 |
| ARHGAP11B | 0.0145 | −1.57 |
| MKL2 | 0.0260 | −1.41 |
| MMP14 | 0.0398 | −1.26 |
| ISLR | 0.0121 | −1.18 |
| DIXDC1 | 0.0150 | 1.03 |
| BTC | 0.0212 | 1.14 |
| RCAN1 | 0.0191 | 1.34 |
| TF | 0.0325 | 1.45 |
| CST6 | 0.0397 | 1.48 |
| RAD51AP2 | 0.0007 | 1.58 |
| KAL1 | 0.0098 | 2.40 |
| S100A8 | 0.0042 | 2.74 |
| IGFBP1 | 0.0157 | 2.95 |
Integrative analysis of expression
levels of feature genes
Following integration of the GSE66635 and GSE45233
datasets, a new age-associated meniscal injury group including 24
samples and a new BMI-associated meniscal injury group including 20
samples were obtained. Intra-group differences of the 28
age-associated meniscal injury feature genes and 20 BMI associated
meniscal injury feature genes were examined by t-test. For the age
group, the average expression levels of the young group were
significantly increased compared with those of the old group
(P=0.021); for the BMI group, the average expression levels of the
lean group were increased compared with those of the obese group,
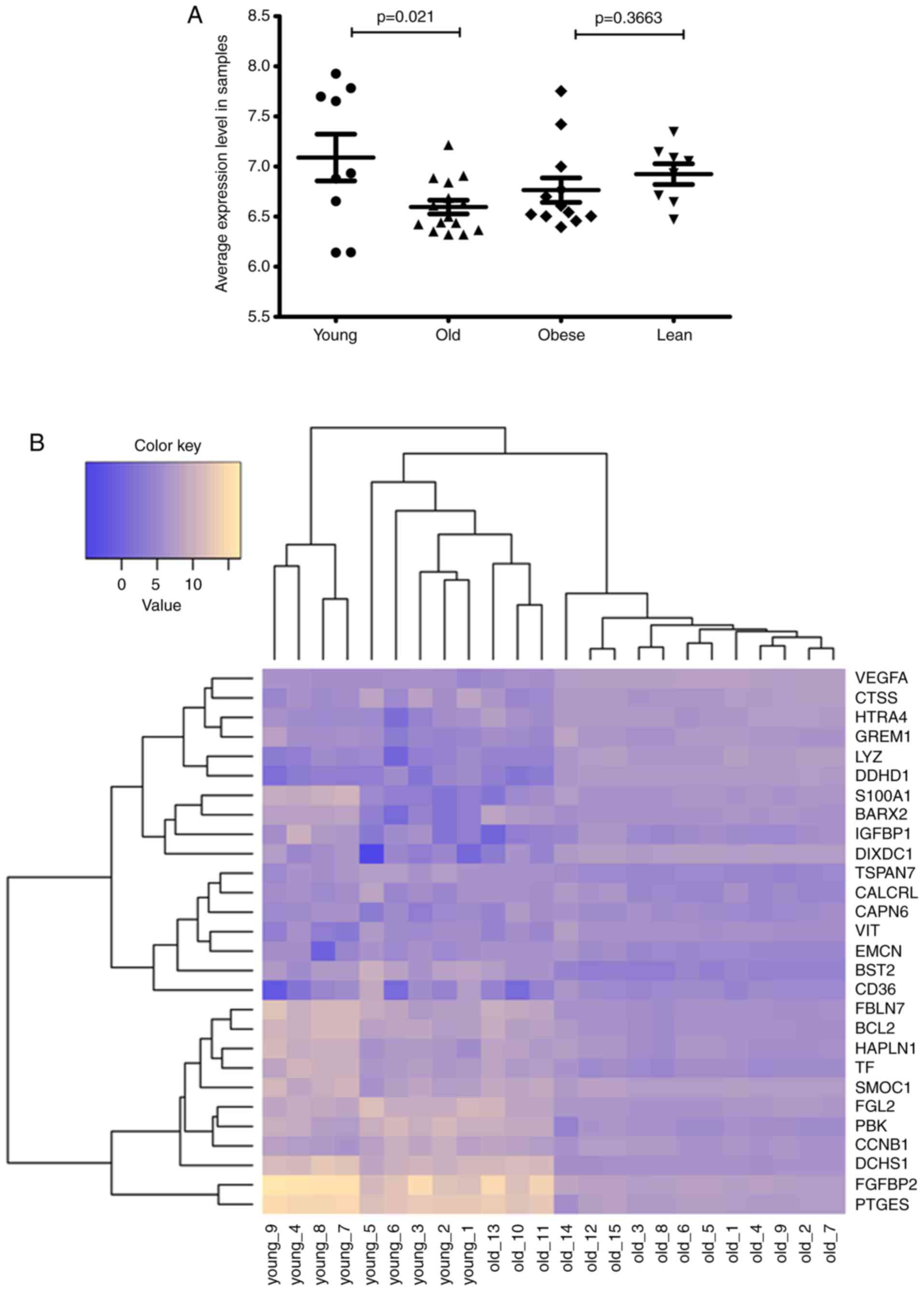
but the difference was not significant (P=0.3663; Fig. 1A). Heat maps (Fig. 1B and C) of bidirectional
hierarchical clustering indicated that the samples within the age
group were completely separated into the young and old groups,
while 1 obese sample (obese-12) in the BMI group was assigned to
the lean group.
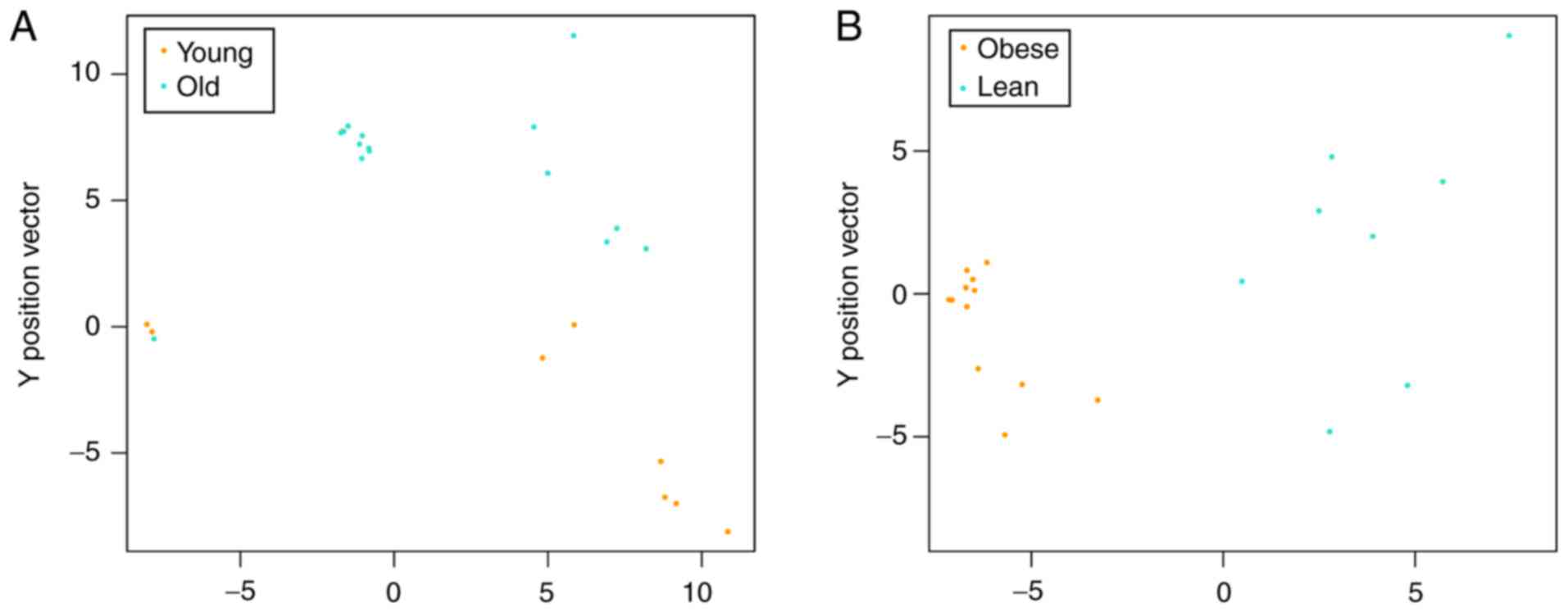
Classification of SVM classifier
model
To observe the classifiability of the feature genes
from the age and BMI classifications, the SVM classifier model was
utilized. Samples in the age group were divided into two groups,
but 1 old sample was included with the young samples (Fig. 2A). The samples in the BMI group
were completely divided into lean and obese groups (Fig. 2B).
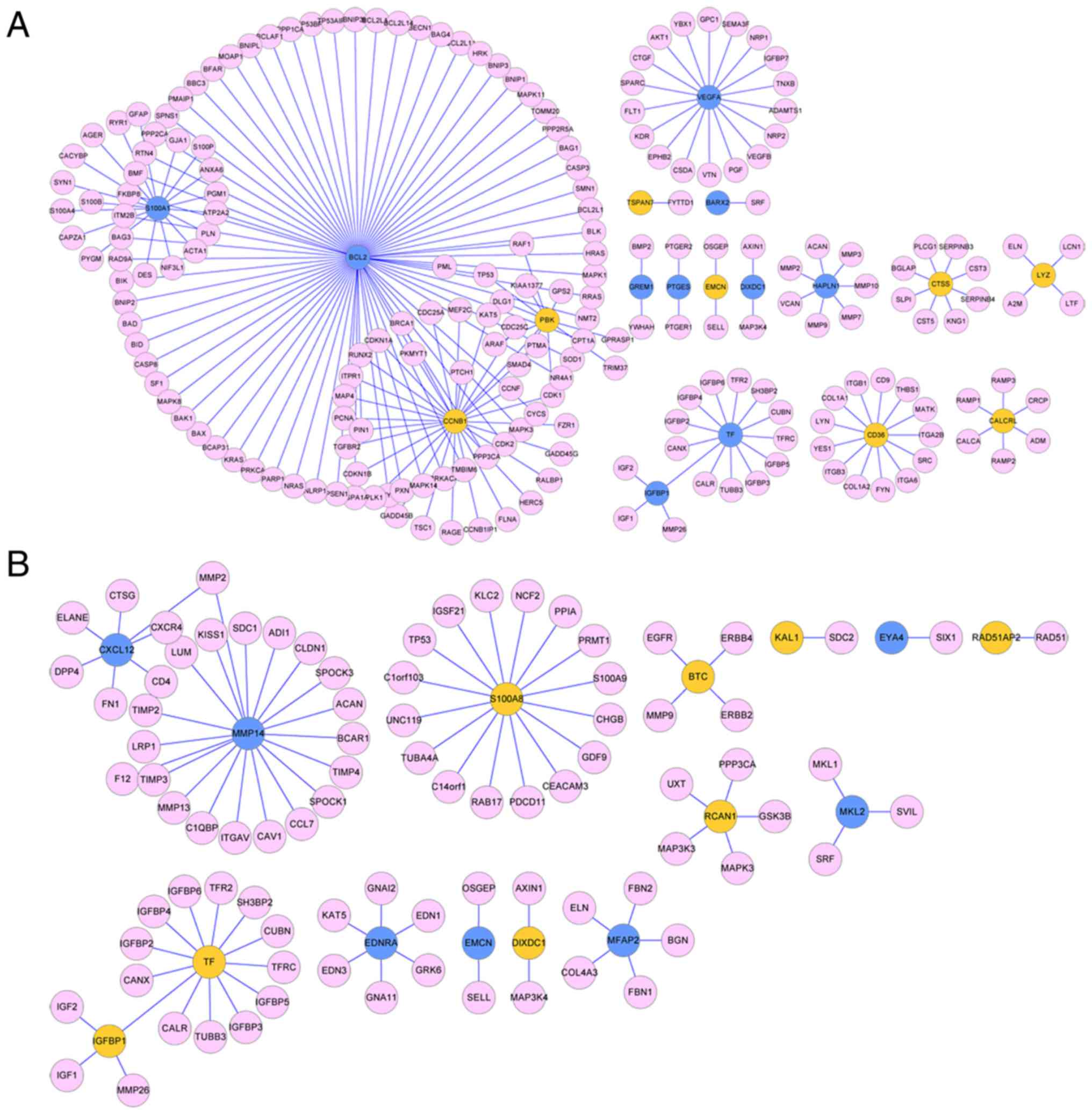
PPI network analysis
Analysis of the HPRD database identified 215 unique
PPI pairs for the age group and 90 unique PPI pairs for the BMI
group. The PPI network for age-associated feature genes contained
222 nodes, including 18 feature genes (Fig. 3A). The PPI network for
BMI-associated feature genes contained 102 nodes, including 15
feature genes (Fig. 3B). Network
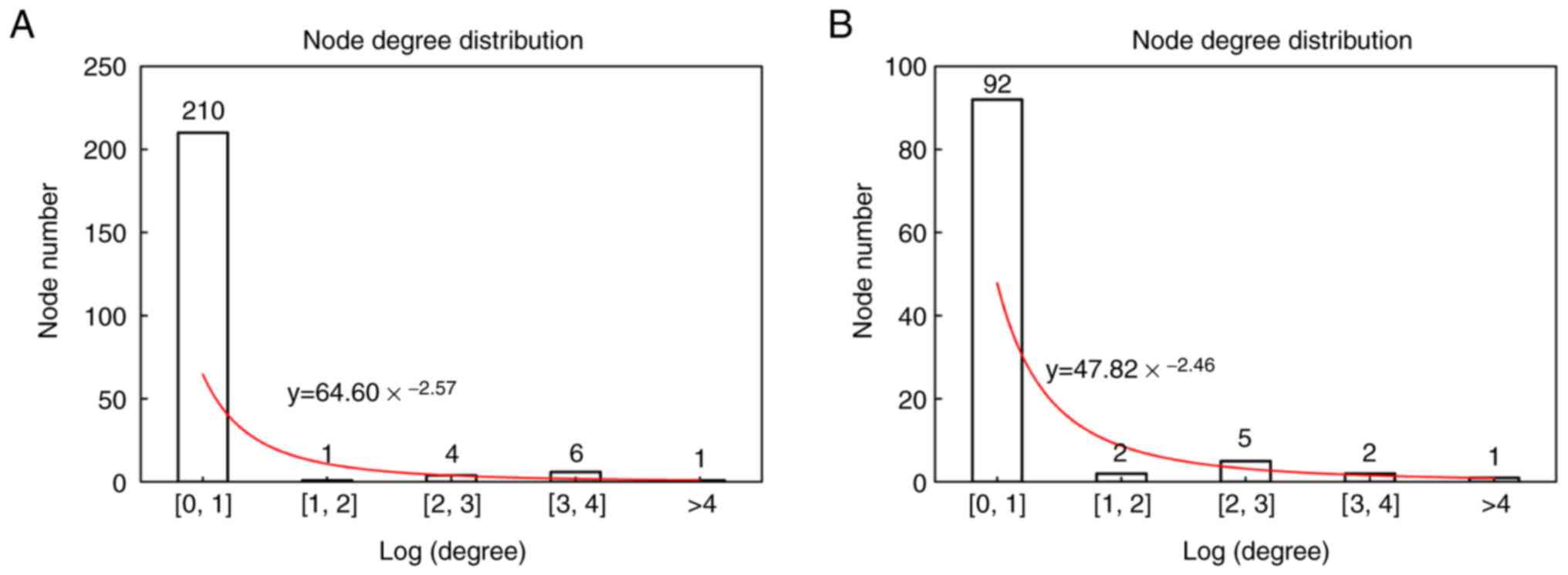
topology analysis indicated that the constructed PPI networks
obeyed scale-free network attributes (Fig. 4) and the node degree of the network
followed the distribution, obtaining y = 64.6 * x−2.57
for the age group and y = 47.82 * x−2.46 for the BMI
group, where × represents node degree. The top 5 nodes with the
higher node degree of the age group were B-cell lymphoma-2 (Bcl-2),
CyclinB1, S100A1, VEGFA and Transferrin (TF) (Table II). The top 5 nodes of the BMI
group were MMP14, S100A8, TF, chemokine (C-X-C motif) ligand 12 and
endothelin receptor type A (Table
II).
 | Table II.List of top 10 highest degree nodes
of the age and body mass index groups. |
Table II.
List of top 10 highest degree nodes
of the age and body mass index groups.
| A, Age group |
|---|
|
|---|
| Gene | P-value | log2FC | Degree |
|---|
| BCL2 | 0.0345 | −1.30 | 74 |
| CCNB1 | 0.0030 | 1.31 | 30 |
| S100A1 | 0.0004 | −5.64 | 18 |
| VEGFA | 0.0173 | −1.66 | 18 |
| TF | 0.0281 | −1.40 | 14 |
| CD36 | 0.0010 | 9.33 | 13 |
| PBK | 0.0043 | 1.39 | 12 |
| CTSS | 0.0222 | 1.48 | 8 |
| HAPLN1 | 0.0031 | −2.21 | 7 |
| CALCRL | 0.0198 | 2.61 | 6 |
|
| B, Body mass
index group |
|
| Gene | P-value | log2FC | Degree |
|
| MMP14 | 0.0398 | −1.26 | 20 |
| S100A8 | 0.0042 | 2.74 | 16 |
| TF | 0.0325 | 1.45 | 14 |
| CXCL12 | 0.0113 | −2.19 | 7 |
| EDNRA | 0.0084 | −3.18 | 6 |
| IGFBP1 | 0.0157 | 2.95 | 5 |
| MFAP2 | 0.0304 | −2.26 | 5 |
| RCAN1 | 0.0191 | 1.34 | 5 |
| BTC | 0.0212 | 1.14 | 4 |
| MKL2 | 0.0260 | −1.41 | 3 |
Pathway enrichment analysis
KEGG pathway enrichment analyses for the top 10
feature genes were performed. Feature genes associated with
age-associated meniscal injury were significantly enriched in the
hypoxia-inducible factor 1 (HIF-1) signaling pathway, including
VEGFA, TF, and Bcl-2 (Table
III). Feature genes associated with BMI-associated meniscal
injury were significantly associated with the mineral absorption
function pathway, in which TF was enriched.
 | Table III.Pathways associated with feature
genes in the age and BMI groups. |
Table III.
Pathways associated with feature
genes in the age and BMI groups.
| Group | Pathway | ID | P-value | Genes |
|---|
| Age | HIF-1 signaling
pathway | hsa04066 | 0.002013 | VEGFA, TF,
BCL2 |
| BMI | Mineral
absorption | hsa04978 | 0.043813 | TF |
Data validation
From GSE51588, a total of 3,403 DEGs (1,853
upregulated and 1,550 downregulated) were identified (data not
shown). Following comparison of GSE66635 and GSE45233, a total of
28 and 20 common DEGs (candidate genes) associated with age and
BMI, respectively, were identified. Finally, 11 overlapping genes
were obtained between candidate genes and DEGs in GSE51588
(Table IV). These included TF and
VEGFA.
 | Table IV.Overlapping genes between candidate
genes and differentially expressed genes in GSE51588. |
Table IV.
Overlapping genes between candidate
genes and differentially expressed genes in GSE51588.
|
| Differentially
expressed genes | Candidate genes in
GSE51588 |
|---|
|
|
|
|
|---|
| Gene name | P-value | logFC | Type | P-value | logFC |
|---|
| TF | 2.81×10-2 | −1.40 | Age | 3.20×10-3 | −0.56 |
| VEGFA | 1.73×10-2 | −1.66 | Age | 7.95×10-3 | −0.62 |
| BST2 | 1.02×10-2 | 1.46 | Age | 6.25×10-3 | 0.64 |
| EMCN | 1.71×10-2 | 6.40 | Age | 1.23×10-5 | 0.65 |
| VIT | 4.59×10-2 | 1.00 | Age | 4.17×10-5 | 0.70 |
| ABHD2 | 9.91×10-4 | −1.86 | BMI | 2.09×10-2 | −0.56 |
| ARHGAP11B | 1.45×10-2 | −1.57 | BMI | 1.19×10-2 | −0.72 |
| BTC | 2.12×10-2 | 1.14 | BMI | 4.12×10-3 | 1.13 |
| DIXDC1 | 1.50×10-2 | 1.03 | BMI | 2.27×10-7 | 0.63 |
| KAL1 | 9.84×10-3 | 2.40 | BMI | 1.39×10-3 | 0.95 |
| RAD51AP2 | 6.60×10-4 | 1.58 | BMI | 4.94×10-2 | 0.37 |
Discussion
Meniscal injury is a common disease caused by
athletic events and activities in daily life. Patient age and BMI
are associated meniscal injuries (17,18).
An understanding of the molecular mechanism of the associations
between meniscal injuries and age or BMI is important. In the
present study, 2 datasets (GSE45233 and GSE66635) associated with
meniscal injury were downloaded from the GEO database and analyzed.
Feature genes associated with age-associated meniscal injury,
including VEGFA, TF and Bcl-2 were involved in the HIF-1 signaling
pathway. In addition, the feature gene TF was also associated with
BMI-associated meniscal injury and was significantly enriched in
the mineral absorption function pathway.
Bcl-2 is a regulator of apoptosis from the B-cell
lymphoma-2 family, and its function is to inhibit cell death,
rather than promote proliferation (18,31–33).
Overexpression of Bcl-2 in mouse bones suppresses apoptosis of bone
cells (34) and suppression of
Bcl-2 increases apoptosis in leukemic cells (35). Meniscal injuries are directly
associated with the development of osteoarthritis (36). Emerging evidence indicates that
apoptosis serves an important role in osteoarthritis pathology
(37). Iwata et al
(38) demonstrated that the levels
of apoptosis in chondrocytes in mice consuming a high-fat diet was
increased. The present study identified that Bcl-2 was
downregulated in the injured meniscus samples compared with the
normal meniscus, meaning that apoptosis may be increased in injured
menisci tissues, which was in accordance with data from Iwata et
al (38). The expression of
Bcl-2 was downregulated in the old group compared with the young
group, in accordance with a previous study (16). Bcl-2 was the hub gene with a node
degree of 74 in the PPI network for the feature genes associated
with age in the injured meniscus. All these data imply that aging
may accelerate apoptosis of the injured menisci of older
patients.
VEGFA is an important angiogenetic protein with a
selective mitogenic effect on vascular endothelial cells (39). As a vascular endothelial growth
factor, VEGFA was demonstrated to be necessary for the survival of
chondrocytes during skeletal development (40) and as a regulator of osteoblast
differentiation during bone development (41). VEGFA may increase the osteogenic
healing capacity by promoting osteogenic and endothelial
differentiation (42). A previous
study revealed that VEGF levels were increased following the
creation of meniscal lesions in rabbits (43). Chen et al (39) recently identified that VEGFA was
downregulated in osteoarthritis chondrocytes compared with normal
chondrocytes. In the present study, the expression of VEGFA was
downregulated in the injured meniscus compared with the normal
meniscus tissues, which is consistent with previous data that the
healing capacity of menisci tissue may be damaged in injured
meniscus, as the angiogenesis function of VEGFA is be inhibited in
the downregulated condition (44,45).
Furthermore, the expression of VEGFA was downregulated in the old
group compared with the young group, consistent with a previous
study (16). Therefore, the
healing capacity for injured meniscus of older patients may be not
as efficient compared with younger patients.
In the present study, TF was identified as a
meniscal injury feature gene associated with age and BMI. TF is an
iron transport protein, which delivers iron from absorption centers
and storage sites to all tissues as it circulates in the plasma
(46). TF serves a major role in
angiogenesis during endochondral bone formation and is produced by
hypertrophic chondrocytes (47).
Hypertrophy and neovascularization are specific signs of healing of
injured menisci (48), through
which TF may be produced and transported. Based on these data, the
upregulation of TF in the injured meniscus indicates that the
healing of the injured meniscus may have occurred, and that TF may
have a role in the healing progress. However, this hypothesis
requires experimental verification. TF was identified to be
downregulated in the old group compared with the young group in the
present study, which is consistent with previous data (16), indicating that recovery of the
injured meniscus in old patients may be slower. Patient age has a
detrimental effect on the healing potential of the injured meniscus
(48). The reason why the
expression of TF was upregulated in the obese group compared with
the lean group is not clear, as a tendency for lower TF saturation
in obesity has been suggested (49). Additional investigation is required
to verify the association between TF and healing of injured
meniscus.
One of the key results of the present study was that
the feature genes in the age group were enriched in the HIF-1
signaling pathway and the feature genes in the BMI group were
enriched in the mineral absorption function pathway. HIF-1 is a
transcriptional activator that targets genes encoding proteins that
increase O2 delivery and mediate adaptive responses to
O2 deprivation (50,51).
HIF-1α may protect articular cartilage by promoting the chondrocyte
phenotype, maintaining chondrocyte viability and supporting
metabolic adaptation to a hypoxic environment (52). Articular cartilage is a hypoxic
tissue, in which HIF-1 is essential for survival, growth, energy
generation and matrix synthesis of chondrocytes (33,44).
HIF-1 is the major regulator of VEGFA in the context of
angiogenesis (53,54). VEGFA may promote angiogenesis
(44,45), which is an indication of the
healing process in injured menisci (48). Expression of oxygen-regulated TF is
positively mediated by HIF-1 (55), which occurs in the hypoxic meniscal
tissues. In addition, Bcl-2 may promote HIF-1-mediated VEGF
expression under hypoxia by increasing the expression level of
HIF-1 (56). In the present study,
VEGFA and Bcl-2 were downregulated in the injured meniscus compared
with the normal meniscus tissue. This indicates that the HIF-1
signaling pathway was inhibited. These results indicate that
apoptosis may be increased, and angiogenesis may be decreased in
meniscal injury tissues. The expression of VEGFA, TF and Bcl-2 was
downregulated in the old group compared with the young group, which
may explain the longer time generally required for older patients
to recover from meniscal injuries compared with younger patients
(57). However, TF was upregulated
in the injured meniscus, which was in contrast to the data that TF
is positively mediated by HIF-1 under hypoxic condition (55). This may be a result of
co-regulation by the HIF-1 signaling pathway and the mineral
absorption function pathway. An additional contradiction is that TF
was upregulated in the obese group and, contrarily, TF saturation
in obesity is usually decreased (49). Experimental verification of these
contradictions is important for additional understanding of the
mechanisms of meniscal injuries.
There are certain limitations in the present study.
The key genes identified were not validated in clinical or in
vivo studies, although data validation was performed.
Additionally, the sample size was small. Therefore, future clinical
and in vivo studies will be performed to validate the
results of the present study, and to investigate the underlying
molecular mechanisms of the effects of aging and obesity on
meniscal injuries.
Aging may affect the development of meniscal
injuries through the HIF-1 signaling pathway, in which VEGFA, TF,
and Bcl-2 are involved. Obesity may affect the mineral absorption
function pathway in injured menisci by regulating the expression of
TF. However, there remains a lack of experimental evidence to
confirm the hypotheses in the present study, and the molecular
mechanisms of the effects of aging and obesity on these pathways
remain unclear. In addition, why and how TF is involved in the
HIF-1 signaling pathway and the mineral absorption function pathway
requires additional study.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Minhang District
Natural Science Project (grant no. 2017MH77).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
PH contributed to data collection, analysis and
interpretation, obtained funding and wrote the manuscript. SW and
YH contributed to the study design. LG performed the experiments
and contributed to the data collection. JG performed data analysis
and interpretation. JW conducted statistical analysis. MW
contributed to data collection and interpretation. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Greis PE, Bardana DD, Holmstrom MC and
Burks RT: Meniscal injury: I. Basic science and evaluation. J Am
Acad Orthop Surg. 10:168–176. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Noble J and Turner PG: The function,
pathology, and surgery of the meniscus. Clin Orthop Relat Res.
62–68. 1986.PubMed/NCBI
|
|
3
|
Anandacoomarasamy A, Leibman S, Smith G,
Caterson I, Giuffre B, Fransen M, Sambrook PN and March L: Weight
loss in obese people has structure-modifying effects on knee
articular cartilage. Ann Rheum Dis. 71:26–32. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Musumeci G, Szychlinska MA and Mobasheri
A: Age-related degeneration of articular cartilage in the
pathogenesis of osteoarthritis: Molecular markers of senescent
chondrocytes. Histol Histopathol. 30:1–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Armstrong CG and Mow VC: Variations in the
intrinsic mechanical properties of human articular cartilage with
age, degeneration and water content. J Bone Joint Surg Am.
64:88–94. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Buckwalter JA, Roughley PJ and Rosenberg
LC: Age-related changes in cartilage proteoglycans: Quantitative
electron microscopic studies. Microsc Res Tech. 28:398–408. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bank RA, Bayliss MT, Lafeber FP, Maroudas
A and Tekoppele JM: Ageing and zonal variation in
post-translational modification of collagen in normal human
articular cartilage. The age-related increase in non-enzymatic
glycation affects biomechanical properties of cartilage. Biochem J.
330:345–351. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dalla PL, Cova M and Pozzi-Mucelli RS: MRI
appearance of the articular cartilage in the knee according to age.
J Belge Radiol. 80:17–20. 1997.PubMed/NCBI
|
|
9
|
Jones DG: Articular cartilage
degeneration: Etiologic association with obesity. Ochsner J.
9:137–139. 2009.PubMed/NCBI
|
|
10
|
Sowers MR and Karvonen-Gutierrez CA: The
evolving role of obesity in knee osteoarthritis. Curr Opin
Rheumatol. 22:533–537. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mezhov V, Ciccutini FM, Hanna FS, Brennan
SL, Wang YY, Urquhart DM and Wluka AE: Does obesity affect knee
cartilage? A systematic review of magnetic resonance imaging data.
Obes Rev. 15:143–157. 2014.
|
|
12
|
Blazek K, Favre J, Asay J, Erharthledik J
and Andriacchi T: Age and obesity alter the relationship between
femoral articular cartilage thickness and ambulatory loads in
individuals without osteoarthritis. J Orthop Res. 32:394–402. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Travascio F, Eltoukhy M, Cami S and Asfour
S: Altered mechano-chemical environment in hip articular cartilage:
Effect of obesity. Biomech Model Mechanobiol. 13:1–15. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Denning WM, Winward JG, Pardo MB, Hopkins
JT and Seeley MK: Body weight independently affects articular
cartilage catabolism. J Sports Sci Med. 14:290–296. 2015.PubMed/NCBI
|
|
15
|
Widhalm HK, Marlovits S, Welsch GH,
Dirisamer A, Neuhold A, van Griensven M, Seemann R, Vécsei V and
Widhalm K: Obesity-related juvenile form of cartilage lesions: A
new affliction in the knees of morbidly obese children and
adolescents. Eur Radiol. 22:672–681. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rai MF, Patra D, Sandell LJ and Brophy RH:
Transcriptome analysis of injured human meniscus reveals a distinct
phenotype of meniscus degeneration with aging. Arthritis Rheum.
65:2090–2101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rai MF, Patra D, Sandell LJ and Brophy RH:
Relationship of gene expression in the injured human meniscus to
body mass index: A biologic connection between obesity and
osteoarthritis. Arthritis Rheumatol. 66:2152–2164. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rai MF, Sandell LJ, Zhang B, Wright RW and
Brophy RH: RNA microarray analysis of macroscopically normal
articular cartilage from knees undergoing partial medial
meniscectomy: Potential prediction of the risk for developing
osteoarthritis. PLoS One. 11:e01553732016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brophy RH, Rai MF, Zhang Z, Torgomyan A
and Sandell LJ: Molecular analysis of age and sex-related gene
expression in meniscal tears with and without a concomitant
anterior cruciate ligament tear. J Bone Joint Surg Am. 94:385–393.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smyth GK: Limma: Linear models for
microarray data. In: Bioinformatics and computational biology
solutions using R and bioconductor. Gentleman R, Carey VJ, Huber W,
Irizarry RA and Dudoit S: Springer; New York, NY: pp. 397–420.
2005
|
|
22
|
Appiah CA: World Health Organisation.
Global Database on Body Mass Index. World Health Organisation:
Geneva 2006. 2014.
|
|
23
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kolde R: Pheatmap. Pretty Heatmaps.
2015.
|
|
25
|
Cortes C and Vapnik V: Support-vector
networks. Mach Learn. 20:273–297. 1995. View Article : Google Scholar
|
|
26
|
Keshava Prasad TS, Goel R, Kandasamy K,
Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R,
Shafreen B, Venugopal A, et al: Human protein reference
database-2009 update. Nucleic Acids Res. 37:D767–D772. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of protein interaction networks. Bio Systems. 127:67–72. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 27:29–34. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:W316–W322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cory S and Adams JM: The Bcl2 family:
Regulators of the cellular life-or-death switch. Nat Rev Cancer.
2:647–656. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vaux DL, Cory S and Adams JM: Bcl-2 gene
promotes haemopoietic cell survival and cooperates with c-myc to
immortalize pre-B cells. Nature. 335:440–442. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fernández-Torres J, Martínez-Nava GA,
Gutiérrez-Ruíz MC, Gómez-Quiroz LE and Gutiérrez M: Role of HIF-1α
signaling pathway in osteoarthritis: A systematic review. Rev Bras
Reumatol Engl Ed. 57:162–173. 2017.(In Portuguese). PubMed/NCBI
|
|
34
|
Yamashita J, Datta NS, Chun YH, Yang DY,
Carey AA, Kreider JM, Goldstein SA and McCauley LK: Role of Bcl2 in
osteoclastogenesis and PTH anabolic actions in bone. J Bone Miner
Res. 23:621–632. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Williams LB and Adesida AB: Angiogenic
approaches to meniscal healing. Injury. 49:467–472. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zamli Z and Sharif M: Chondrocyte
apoptosis: A cause or consequence of osteoarthritis? Int J Rheum
Dis. 14:159–166. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Iwata M, Ochi H, Hara Y, Tagawa M, Koga D,
Okawa A and Asou Y: Initial responses of articular tissues in a
murine high-fat diet-induced osteoarthritis model: Pivotal role of
the IPFP as a cytokine fountain. PLoS One. 8:e607062013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen H and Tian Y: MiR-15a-5p regulates
viability and matrix degradation of human osteoarthritis
chondrocytes via targeting VEGFA. Biosci Trends. 10:482–488. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zelzer E, Mamluk R, Ferrara N, Johnson RS,
Schipani E and Olsen BR: VEGFA is necessary for chondrocyte
survival during bone development. Development. 131:2161–2171. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Duan X, Murata Y, Liu Y, Nicolae C, Olsen
BR and Berendsen AD: Vegfa regulates perichondrial vascularity and
osteoblast differentiation in bone development. Development.
142:1984–1991. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Behr B, Tang C, Germann G, Longaker MT and
Quarto N: Locally applied VEGFA increases the osteogenic healing
capacity of human adipose-derived stem cells by promoting
osteogenic and endothelial differentiation. Stem Cells. 29:286–296.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ruiz Ibán MÁ, Comellas Melero N,
Martinez-Botas J, Ortiz A and Diaz Heredia J: Growth factor
expression after lesion creation in the avascular zone of the
meniscus: A quantitative PCR study in rabbits. Arthroscopy.
30:1131–1138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Deckers MM, van Bezooijen RL, van der
Horst G, Hoogendam J, van Der Bent C, Papapoulos SE and Löwik CW:
Bone morphogenetic proteins stimulate angiogenesis through
osteoblast-derived vascular endothelial growth factor A.
Endocrinology. 143:1545–1553. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Weijts BG, Bakker WJ, Cornelissen PW,
Liang KH, Schaftenaar FH, Westendorp B, de Wolf CA, Paciejewska M,
Scheele CL, Kent L, et al: E2F7 and E2F8 promote angiogenesis
through transcriptional activation of VEGFA in cooperation with
HIF1. EMBO J. 31:3871–3884. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bonkovsky HL: Iron and the liver. Am J Med
Sci. 301:32–43. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Carlevaro MF, Albini A, Ribatti D, Gentili
C, Benelli R, Cermelli S, Cancedda R and Cancedda FD: Transferrin
promotes endothelial cell migration and invasion: Implication in
cartilage neovascularization. J Cell Biol. 136:1375–1384. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Senan V, Sucheendran J and Prasad and
Balagopal K: Histological features of meniscal injury. Kerala J
Orthop. 24:30–36. 2011.
|
|
49
|
Cheng HL, Bryant C, Cook R, O'Connor H,
Rooney K and Steinbeck K: The relationship between obesity and
hypoferraemia in adults: A systematic review. Obes Rev. 13:150–161.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Semenza G: Signal transduction to
hypoxia-inducible factor 1. Biochem Pharmacol. 64:993–998. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Semenza GL: Hypoxia-inducible factor 1:
Oxygen homeostasis and disease pathophysiology. Trends Mol Med.
7:345–350. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang FJ, Luo W and Lei GH: Role of HIF-1α
and HIF-2α in osteoarthritis. Joint Bone Spine. 82:144–147. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pagès G and Pouysségur J: Transcriptional
regulation of the vascular endothelial growth factor gene-a concert
of activating factors. Cardiovasc Res. 65:564–573. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liao D and Johnson RS: Hypoxia: A key
regulator of angiogenesis in cancer. Cancer Metastasis Rev.
26:281–290. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Rolfs A, Kvietikova I, Gassmann M and
Wenger RH: Oxygen-regulated transferrin expression is mediated by
hypoxia-inducible factor-1. J Biol Chem. 272:20055–20062. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Trisciuoglio D, Gabellini C, Desideri M,
Ragazzoni Y, De Luca T, Ziparo E and Del Bufalo D: Involvement of
BH4 domain of bcl-2 in the regulation of HIF-1-mediated VEGF
expression in hypoxic tumor cells. Cell Death Differ. 18:1024–1035.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Roos H, Adalberth T, Dahlberg L and
Lohmander LS: Osteoarthritis of the knee after injury to the
anterior cruciate ligament or meniscus: The influence of time and
age. Osteoarthritis Cartilage. 3:261–267. 1995. View Article : Google Scholar : PubMed/NCBI
|