Introduction
Lung cancer is the leading cause of
cancer-associated mortality, with non-small cell lung cancer
(NSCLC) accounting for ~85% of lung cancer cases worldwide
(1). Despite recent developments
in cancer treatment, the prognosis for patients with lung cancer
has seen limited improvement, with the 5-year survival rate for
NSCLC remaining at 9–14% since 2010 (2). Due to the discovery of epidermal
growth factor receptor tyrosine kinase receptor inhibitors
(EGFR-TKIs), such as gefitinib and erlotinib, a new generation of
agents has been made available for the treatment of patients with
NSCLC, whose tumor harbors activating mutations in the EGFR gene
(3–5). However, the majority of patients who
initially respond to treatment with TKIs, later develop acquired
resistance, which limits the treatment options (6). Several mechanisms of acquired
resistance to TKIs have been identified, including secondary EGFR
T790 M mutation, MET amplification, HER2 amplification, KRAS
mutation and loss of PTEN (7,8).
Further elucidation of the mechanisms underlying gefitinib
resistance is imperative for the development of therapeutic
strategies.
MicroRNAs (miRNAs) are small non-coding endogenous
RNAs that mainly bind to the 3′-untranslated region (3′-UTR) of
target mRNAs, resulting in the degradation of mRNA or the blockade
of protein translation (9). miRNAs
exert a wide range of biological functions, including
proliferation, differentiation, metabolism and apoptosis (9,10).
The involvement of miRNAs in gefitinib resistance in NSCLC has been
previously reported (11). A study
demonstrated that miR-138-5p was downregulated in
gefitinib-resistant NSCLC cell models, and that the overexpression
of miR-138-5p enhanced gefitinib sensitivity via the suppression of
G protein-coupled receptor 124 (12). In another previous study, the
elevation of miR-127 promoted a shift from the epithelial to the
mesenchymal phenotype and increased gefitinib resistance in lung
cancer cells (13). The aim of the
present study was to identify and investigate novel miRNAs that are
potentially involved in gefitinib resistance in NSCLC.
Materials and methods
Clinical specimens and cell
culture
A total of 25 pairs of lung adenocarcinoma and
adjacent non-cancerous specimens were resected from patients (14
male and 11 female) aged 56–67 years old between 2015 and 2017
diagnosed by clinicians at Taizhou Central Hospital (Taizhou,
China). There were 13 patients at stage I, 4 patients at stage II,
6 patients at stage III and 2 patients at stage IV, classified by
TNM staging (14). All samples
were frozen immediately after surgery and stored at −80°C. Written
informed consent was obtained from each patient. The study was
approved by the Ethics Committee of Taizhou Central Hospital.
The 293 cell line, human normal epithelial cell line
BEAS-2B and the NSCLC cell line PC-9 were obtained from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China). PC-9 and
293 cells were cultured in RPMI 1640, supplemented with 10% fetal
bovine serum (HyClone; GE Healthcare Life Sciences, Logan, UT, USA)
in an incubator with 5% CO2 at 37°C. The 293 cells were
applied in the experiments of luciferase assays, BEAS-2B cells were
used for the reconstruction of pcDNA3-AP1 and PC-9 cells were used
in all other experiments.
Establishment of the
gefitinib-resistant cell line
Gefitinib-resistant cells were obtained by
continuously exposing PC-9 cells to gefitinib, as previously
described (12). Briefly, PC-9
cells were first exposed to 0.2 µmol/l gefitinib (LC Laboratories,
Woburn, MA, USA) for 48 h, washed and cultured in drug-free medium
until 80% confluence was reached, followed by re-exposure to
increasing concentrations of gefitinib. Gefitinib-resistant PC-9
(PC-9GR) cells were obtained after 6 months of continuous exposure.
The resistant cells were able to grow in 10 µmol/l gefitinib and
were maintained in 2 µmol/l gefitinib-containing medium. In order
to eliminate gefitinib, PC-9GR cells were cultured in drug-free
medium for 1 week prior to all experiments.
Cell growth inhibition assay
The effects of gefitinib on cells were determined
using the MTT assay. Briefly, exponentially growing cells were
seeded in 96-well plates at the density of 3×103/well
with 100 µl in each well, and cultured in 37°C overnight. On the
following day, gefitinib was dissolved in dimethyl sulfoxide (DMSO)
and added to each well at different concentrations (1, 2, 4, 8 and
16 µM), while the cells in the control groups were treated with the
equivalent volume of DMSO. Cells in each group were incubated for
48 h. Following incubation, 10 µl MTT was added to each well, and
cells were incubated for a further 3 h. The optical density at 492
nm was measured using a microplate reader (Beckman Coulter, Inc.,
Brea, CA, USA).
Cell apoptosis
Cells (2×105 cells/well) were seeded into
12-well plates and cultured for 48 h at 37°C. Subsequently, flow
cytometry was used for the detection of the cell apoptosis by
Annexin V-fluorescein isothiocyanate (FITC) /propidium iodide (PI)
staining. Cells were washed with cold PBS and fixed in ice-cold 70%
ethanol overnight at −20°C. The next day, cells were stained by
Annexin V-FITC and PI (Roche Diagnostics, Basel, Switzerland) at
room temperature for 15 min in the dark. After incubation for 1 h
at 37°C, cell apoptosis was measured using a flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA). The cell number at each
phase was analyzed by FlowJo software version 7.6.3 (FlowJo LLC,
Ashland, OR, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cells using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), while miRNeasy Mini Kit (Qiagen, Inc., Valencia, CA, USA) was
applied for the extraction of miRNA from tissues and cells. RNA
concentration was tested using NanoDrop 2000 (Thermo Fisher
Scientific, Inc., Wilmington, DE, USA). RNA was reverse transcribed
into cDNA using the First Strand cDNA Synthesis kit (Thermo Fisher
Scientific, Inc.). mirVanaqRT-PCR miRNA Detection kit and SYBR
Green I (Applied Biosystems; Thermo Fisher Scientific, Inc.) were
conducted using an ABI 7500 Real Time PCR System (Thermo Fisher
Scientific, Inc.). The primers were as follows: YAP, forward
5′-TAGCCCTGCGTAGCCAGTTA-3′, reverse 5′-TCATGCTTAGTCCACTGTCTGT-3′;
GAPDH, forward 5′-GGAGCGAGATCCCTCCAAAAT-3′, reverse
5′-GGCTGTTGTCATACTTCTCATGG-3′; miR-506-3p, forward
5′-TGCGGTAAGGCACCCTTCTGAGTA-3′, reverse 5′-CCAGTGCAGGGTCCGAGGT-3′;
U6, forward 5′-CTCGCTTCGGCAGCACA-3′, reverse
5′-AACGCTTCACGAATTTGCGT-3′. The thermocycling conditions were as
follows: 95°C for 3 min, 40 cycles of 95°C for 10 sec, 60°C for 15
sec and 72°C for 31 sec. The relative expression level was
determined using the 2−ΔΔCq method (15). GAPDH and U6 were used as internal
controls for Yes-associated protein 1 (YAP1) and miR-506-3p,
respectively.
Transfection with miR-506-3p mimic and
inhibitor
miR-506-3p mimics (50 nM), miR-506-3p inhibitor (100
nM) or their corresponding miR-negative control (miR-NC; Guangzhou
RiboBio Co., Ltd., Guangzhou, China) were transfected into the
cells using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Transfection with pcDNA3-YAP1
YAP1 was amplified by PCR using cDNA obtained from
BEAS-2B cells, and cloned into pcDNA3 (Thermo Fisher Scientific,
Inc.) to generate pcDNA3-YAP1. pcDNA3-YAP1 and negative control
which were used for gain-of-function experiments were transfected
into cells (1×105) using Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol.
Western blot analysis
Briefly, cells were lysed in
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.), and protein concentration was determined using the BCA
Protein Assay kit (Thermo Fisher Scientific, Inc.). Protein lysate
(15 µg) was separated on 10% SDS-PAGE and transferred to
polyvinylidene difluoride (PVDF) membranes (Thermo Fisher
Scientific, Inc.). Next, the PVDF membranes were blocked in 5%
non-fat milk (Yili, Beijing, China) for 2 h at room temperature,
and then incubated with monoclonal rabbit anti-YAP1 (ab5277;
1:1,000; Abcam, Cambridge, MA, USA), B-cell lymphoma 2 (Bcl-2;
ab32142; 1:1,000; Abcam), Bcl-2-associated X protein (Bax; ab32503;
1:1,000; Abcam) or monoclonal mouse anti-GAPDH (ab9485; 1:1,000;
Abcam) antibody for 3 h at room temperature. Subsequently,
membranes were incubated with goat anti-rabbit secondary antibody
(ab97080; 1:20,000; Abcam) for 1 h at room temperature. Following
washing with Tris-buffered saline, the bands were detected using
the ECL Western Blotting kit (Thermo Fisher Scientific, Inc.).
Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.,
Rockville, MD, USA) was used to analyze the relative protein
expression, represented as the density ratio vs. GAPDH.
Plasmid construction and
dual-luciferase reporter assay
YAP1 3′-UTR sequences
(5′-ACUUUUCUAAAUGUAGUGCCUUU-3′) were amplified by PCR using cDNA
from 293 cells, and cloned into pGL3 basic vectors (Promega
Corporation, Madison, WI, USA) between KpnI and Xhol
restriction enzyme sites. The Quick Change site-directed
mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA, USA)
was used for the mutation of the putative binding site of
miR-506-3p in the YAP1 3′-UTR-containing vector, with the YAP1
3′UTR mutant sequence as follows: 5′-ACUUUUCUAAAUGUAGUGCGAUU-3′.
For dual-luciferase reporter assay, PC-9GR cells were seeded in
24-well plates at the density of 1×105/well and cultured
at 37°C overnight. On the following day, cells transfected with
YAP1 3′UTR WT plasmids or YAP1 3′UTR MUT plasmids were also
transfected with miR-NC mimics or miR-506-3p mimic, and luciferase
reporter plasmids containing Renilla vector (pRL-TK; Promega
Corporation). At 48 h post-transfection, the luciferase activity
was determined using the Dual-Luciferase Reporter Assay System
(Promega Corporation). Luciferase activity was normalized to the
Renilla luciferase activity to determine the transfection
efficiency.
Statistical analysis
All data were analyzed using Graphpad Prism software
(version 6; Graphpad Software, Inc., La Jolla, CA, USA), and are
presented as the mean ± standard deviation. Differences between two
groups were evaluated using the Student's t-test. Differences among
three or more groups were compared using one-way analysis of
variance, followed by the Newman-Keuls test. The association
between miR-506-3p and YAP1 was analyzed by the Pearson correlation
analysis. A value of P<0.05 was considered to indicate a
statistically significant difference. All experiments were
performed in triplicate.
Results
PC-9GR cells are relatively
insensitive to gefitinib treatment, compared with PC-9 cells
To study the underlying molecular mechanism of
gefitinib resistance, a PC-9GR cell model was established by
long-term exposure of PC-9 cells to gefitinib. PC-9 and PC-9GR
cells were treated with increasing concentrations of gefitinib for
48 and 96 h, in order to test their sensitivity to gefitinib.
Compared with parental PC-9 cells, gefitinib exhibited a markedly
weaker cytotoxic efficacy on PC-9GR cells following exposure to
doses of 2, 4, 8 and 16 µM for 48 h (Fig. 1A). In addition, the difference in
the response to gefitinib treatment became more evident after 96-h
treatment, as a low dose of 1 µM gefitinib was able to induce
marked viability reduction in PC-9 cells, but not in PC-9GR cells
(Fig. 1B). In order to investigate
the role of miR-506-3p in mediating gefitinib resistance,
miR-506-3p levels were compared between PC-9 and PC-9GR cells.
Using RT-qPCR, miR-506-3p was found to be significantly
downregulated in PC-9GR cells (Fig.
1C), suggesting that miR-506 may be involved in gefitinib
resistance.
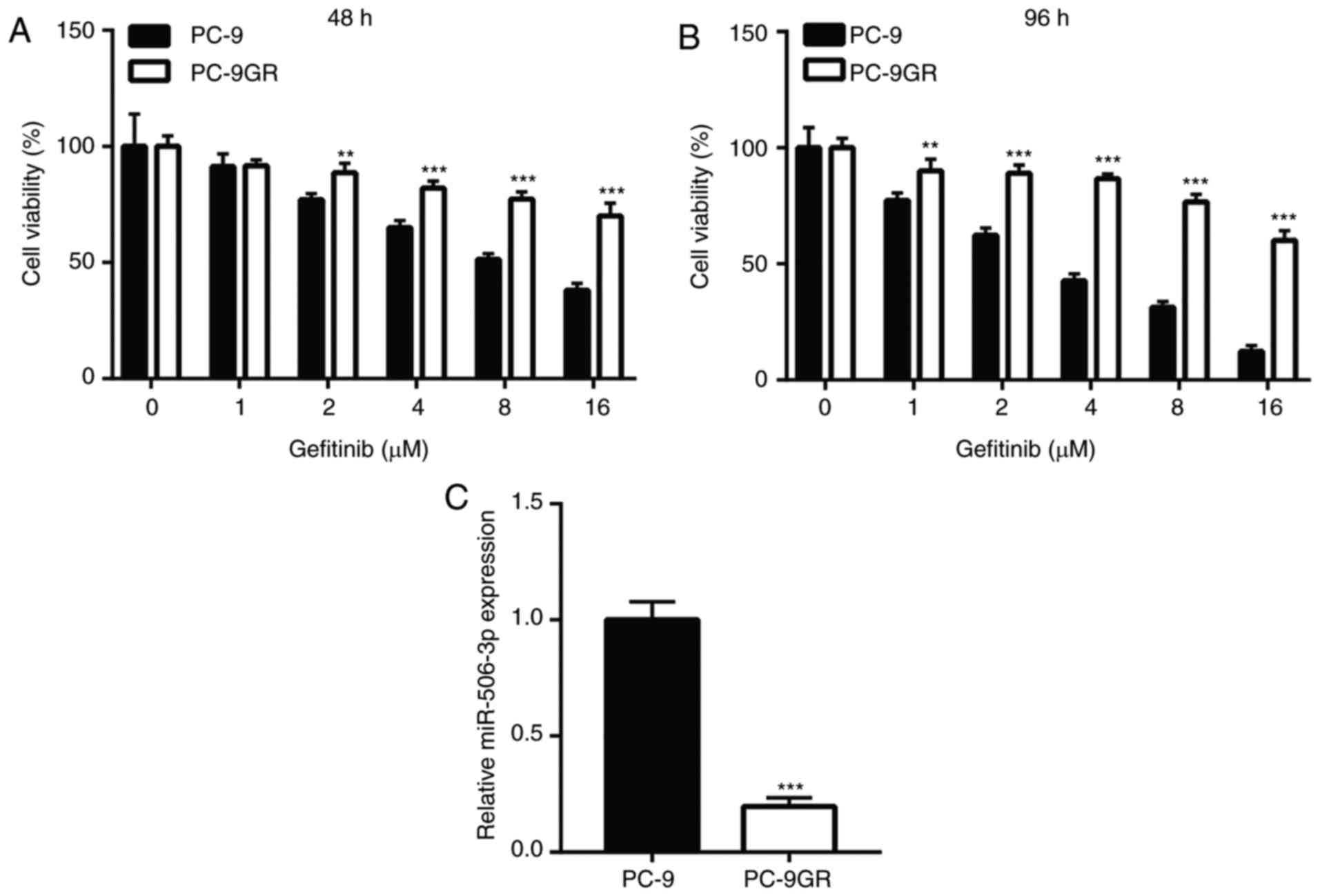 | Figure 1.miR-506-3p was downregulated in PC-9GR
cells, which were relatively insensitive to gefitinib, when
compared with PC-9 cells. The viability reduction was significantly
higher in PC-9 cells following treatment with increasing
concentrations of gefitinib (1, 2, 4, 8 and 16 µM) for (A) 48 h and
(B) 96 h, as compared with that in PC-9GR cells. (C) In PC-9GR
cells, the expression level of miR-506-3p was decreased compared
with that in PC-9 cells. **P<0.01 and ***P<0.001, vs. PC-9
cells at the same concentration. miR, microRNA; PC-9GR,
gefitinib-resistant PC-9. |
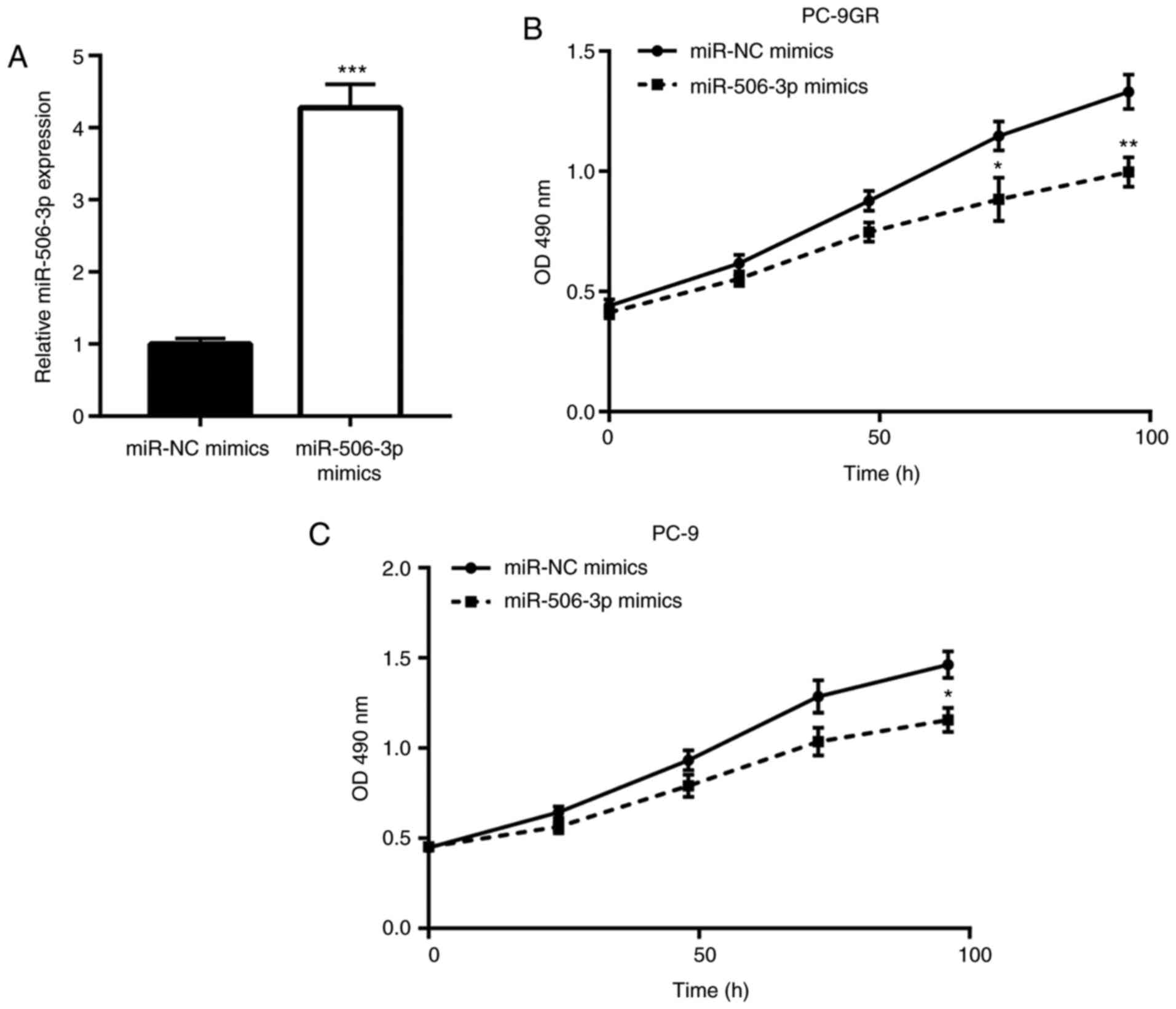
miR-506-3p inhibits the proliferation
of PC-9GR cells
Next, the function of miR-506-3p in controlling the
growth of PC-9GR cells was explored using mimic transfection to
elevate the miR-506-3p levels. As expected, transfection with
miR-506-3p mimics elevated the expression of miR-506-3p in PC-9GR
cells, as compared with that in cells transfected with miR-NC
mimics (Fig. 2A). The
overexpression of miR-506-3p was found to induce significant growth
arrest in PC-9GR cells cultured in gefitinib-free medium in a
time-dependent manner (Fig. 2B).
Notably, miR-506-3p elevation also markedly inhibited the growth of
PC-9 cells, but to a smaller extent (Fig. 2C). These observations indicated
that miR-506-3p is pivotal for the proliferation of PC-9GR
cells.
Downregulation of miR-506-3p drives
gefitinib resistance in PC-9 cells
In order to investigate the role of miR-506-3p in
the development of gefitinib resistance, miR-506-3p expression
levels were elevated or inhibited by the transfection of PC-9GR
cells with miR-506-3p mimic or antagonist, respectively. The
response of these cells to gefitinib treatment was then detected.
Transfection with miR-506-3p antagonist significantly reduced
miR-506-3p expression in PC-9GR cells, as compared with that
observed in miR-NC-transfected cells (Fig. 3A). The overexpression of miR-506-3p
was demonstrated to enhance gefitinib-induced cytotoxicity, whereas
transfection with miR-506-3p antagonist inhibited this cytotoxicity
in PC-9GR cells (Fig. 3B). Flow
cytometric analysis further revealed that compared with gefitinib
group, the overexpression of miR-506-3p significantly increased
cell apoptosis in response to gefitinib treatment (8 µM), while the
inhibition of miR-506-3p reduced the cell apoptosis rate (Fig. 3C and D). Western blot analysis
indicated that Bcl-2 protein expression levels were decreased
following miR-506-3p overexpression and increased following
miR-506-3p inhibition. By contrast, the Bax expression levels were
increased following miR-506-3p overexpression and decreased
following miR-506-3p inhibition (Fig.
3E). In addition, treatment with a low concentration of
gefitinib (1 µM) was able to inhibit cell proliferation in PC-9GR
cells with miR-506-3p overexpression (Fig. 3F), suggesting that miR-506-3p
sensitized PC-9 cells to gefitinib.
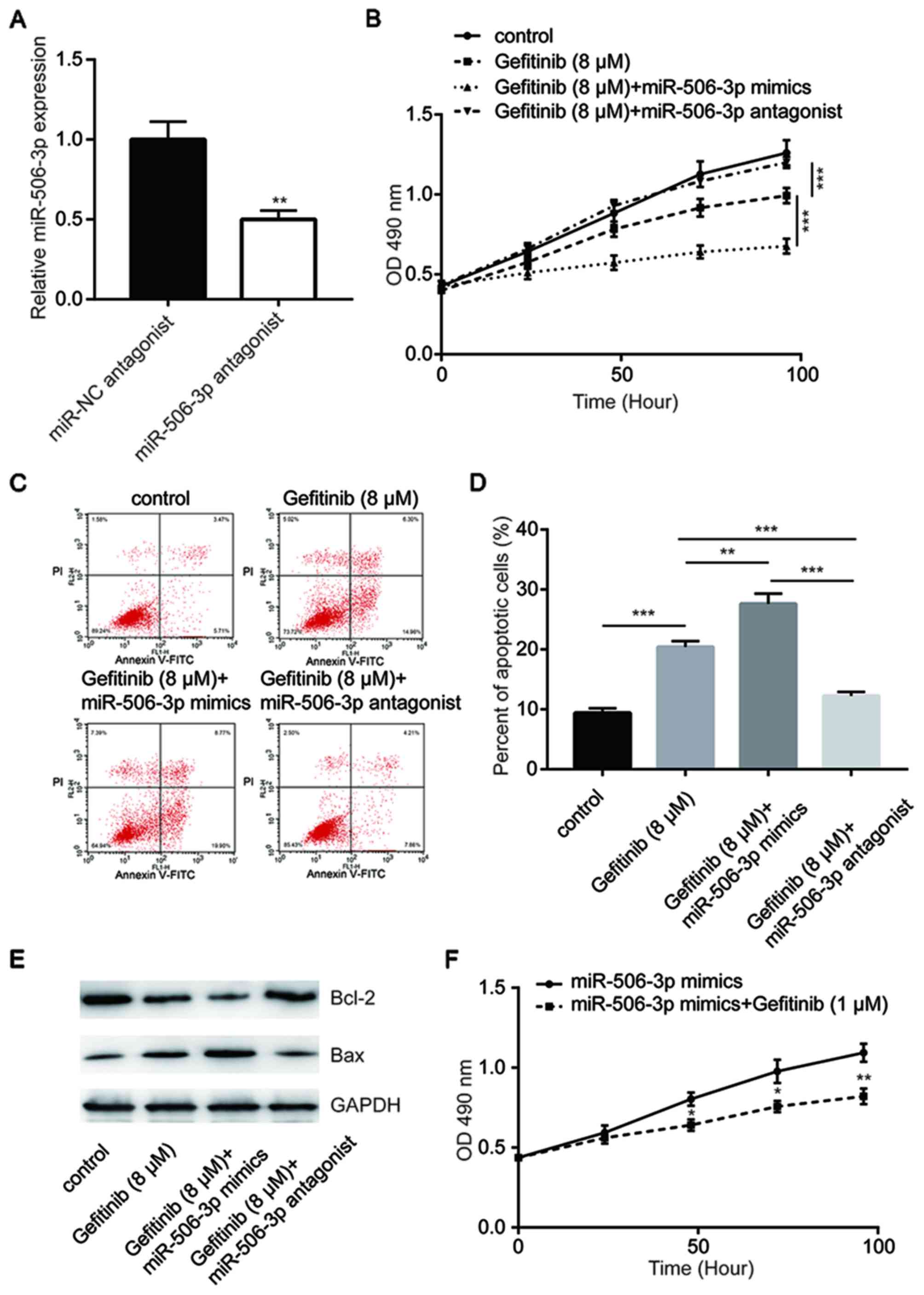 | Figure 3.miR-506-3p-mediated gefitinib
sensitivity in PC-9GR cells. (A) Transfection with miR-506-3p
antagonist decreased the miR-506-3p expression levels in PC-9GR
cells. (B) miR-506-3p mimics enhanced gefitinib (8 µM for 48 h)
treatment-induced reduction in the viability of PC-9GR cells, while
the miR-506-3p antagonist reversed the cell viability reduction.
(C) Flow cytometric assay and (D) quantitative analysis of cell
apoptosis, demonstrating that miR-506-3p mimics enhanced gefitinib
(8 µM for 48 h) treatment-induced apoptosis in PC-9GR cells, while
the miR-506-3p antagonist reversed cell apoptosis. (E) miR-506-3p
mimics enhanced gefitinib (8 µM for 48 h) treatment-induced Bcl-2
reduction and Bax elevation in PC-9GR cells, while miR-506-3p
antagonist reversed the expression alteration of Bcl-2 and Bax. (F)
After treatment for 48 h, 1 µM gefitinib induced growth arrest in
PC-9GR cells transfected with miR-506-3p mimics, although this
concentration did not alter the viability of untransfected PC-9GR
cells. *P<0.05, **P<0.01 and ***P<0.001, vs. corresponding
control group. miR, microRNA; PC-9GR, gefitinib-resistant PC-9; NC,
negative control; Bax, Bcl-2-associated X protein. |
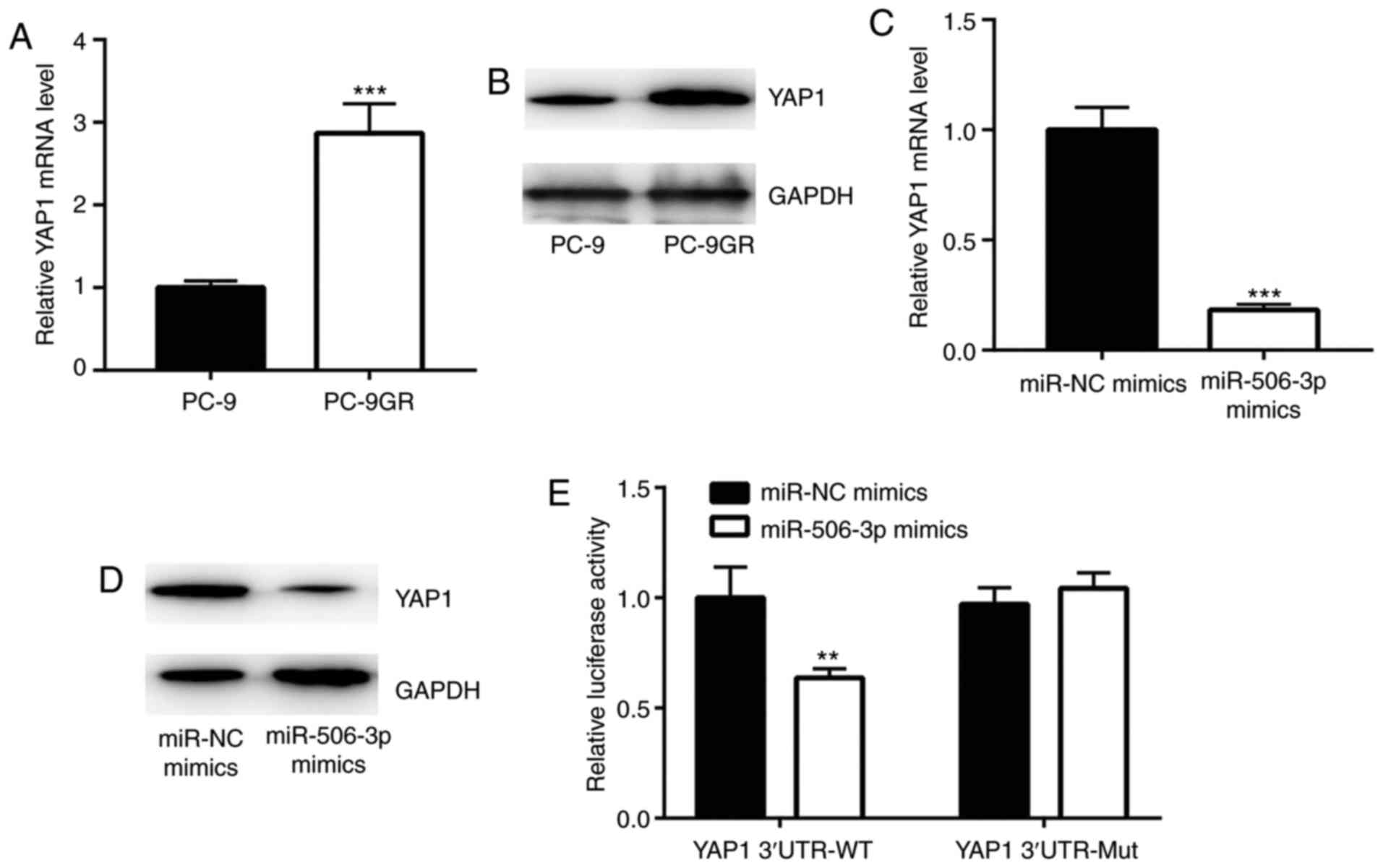
YAP1 is a target gene of miR-506-3p in
PC-9 cells
Previous studies have demonstrated that YAP1 was
directly suppressed by miR-506-3p in breast and liver cancer
(16,17). In the current study, YAP1 mRNA and
protein expression levels were found to be elevated in PC-9GR
cells, as compared with those in PC-9 cells (Fig. 4A and B). However, overexpression of
miR-506-3p significantly reduced YAP1 expression in PC-9GR cells
(Fig. 4C and D). The results of
dual-luciferase reporter assay revealed that miR-506-3p mimic
transfection markedly decreased the luciferase activity of PC-9
cells transfected with YAP1 3′UTR-wild type (Fig. 4E), suggesting that YAP1 was a
target gene of miR-506-3p in NSCLC cells.
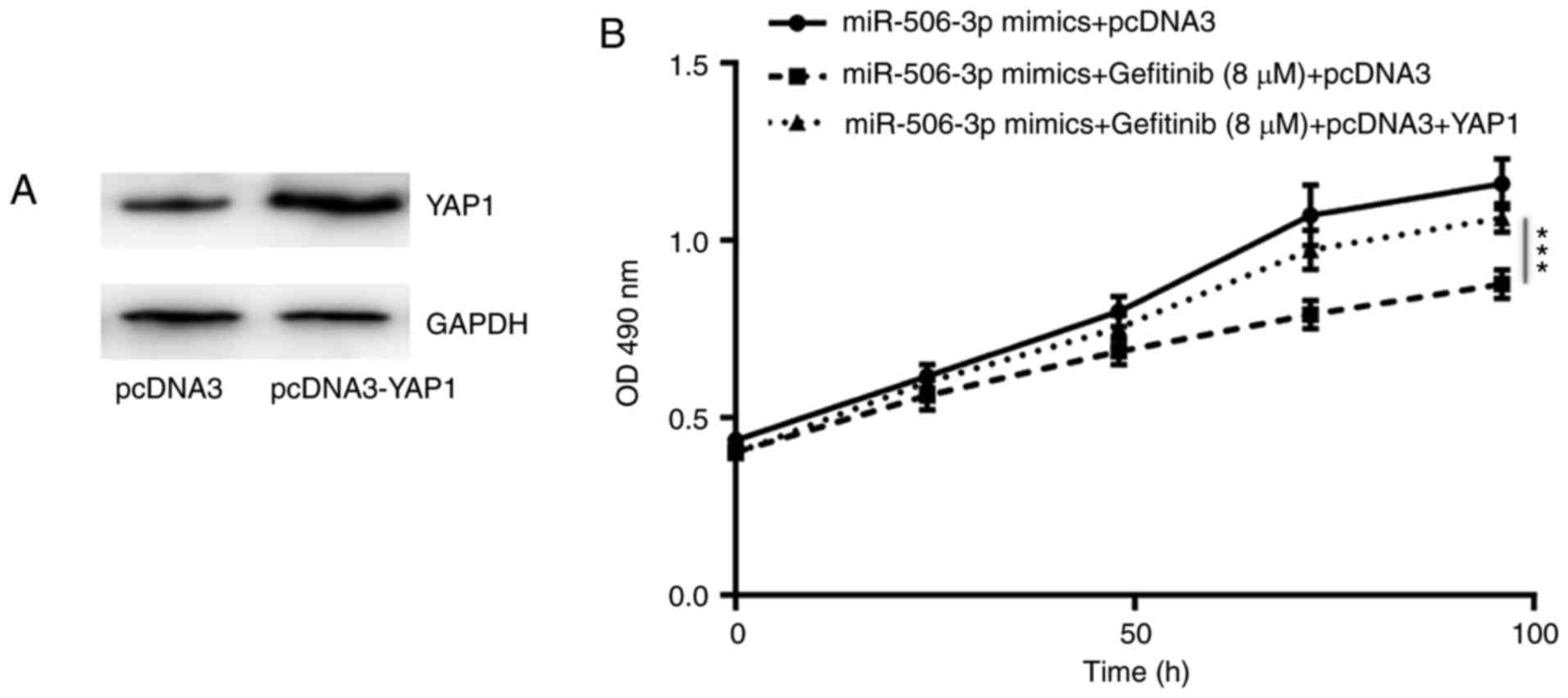
YAP1 is involved in
miR-506-3p-mediated gefitinib sensitivity in PC-9 cells
In order to investigate the association between YAP1
expression and miR-506-3p-associated gefitinib sensitivity in NSCLC
cells, YAP1 overexpression was induced in PC-9GR cells by the
transfection with pcDNA3-YAP1 plasmid (Fig. 5A). As shown in Fig. 5B, the overexpression of YAP1 was
able to partially reverse the elevated sensitivity to gefitinib in
PC-9GR cells transfected with miR-506-3p mimics (Fig. 5B), indicating that YAP1 was pivotal
in the regulatory role of miR-506-3p in gefitinib sensitivity.
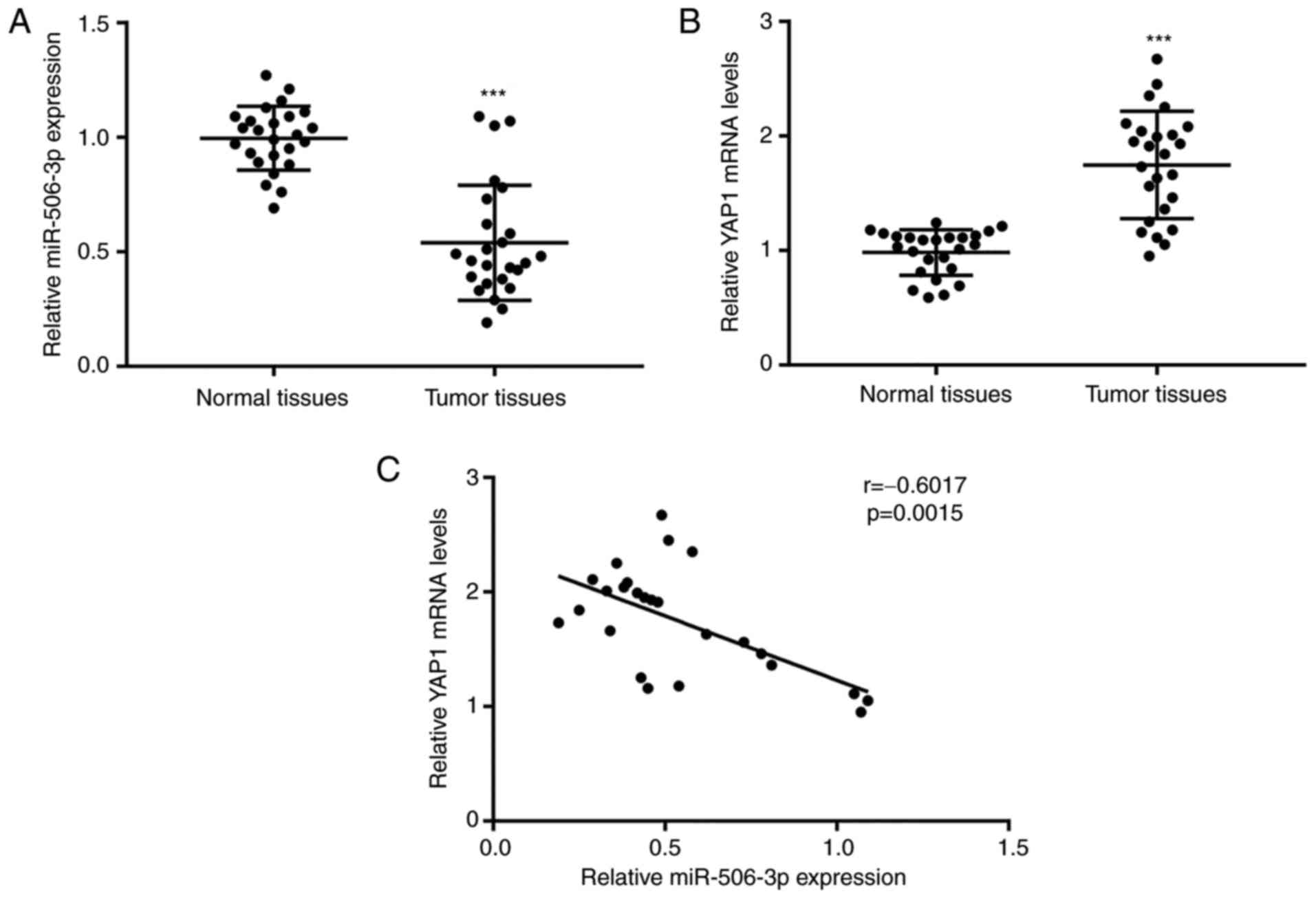
miR-506-3p expression is downregulated
in tumor tissues of patients with NSCLC and correlated with YAP1
mRNA levels
To evaluate the role of miR-506-3p and YAP1
expression levels in NSCLC cells, RT-qPCR was used to detect the
mRNA levels of miR-506-3p and YAP1 in 26 pairs of tumor tissues and
matched normal tissues from patients with NSCLC. As compared with
the normal tissues, a significant decrease in miR-506-3p expression
was observed in tumor tissues, while YAP1 expression was found to
be elevated (Fig. 6A and B).
Pearson correlation analysis revealed a strong negative correlation
(r=−0.6017, P=0.0015) between the miR-506-3p expression and YAP1
mRNA levels in tumor tissues (Fig.
6C). These results further validated the regulatory association
between miR-506-3p and YAP1 in NSCLC tumor tissues.
Discussion
Numerous studies have reported aberrant expression
of miR-506-3p in multiple cancer types, including breast,
pancreatic and colorectal cancer (18–20).
In lung cancer, research on the expression of miR-506-3p has
provided conflicting results. Yin et al (21) reported that miR-506-3p was
upregulated in lung cancer tissue in 83% of patients with lung
cancer; however, the elevation in miR-506-3p expression inhibited
tumor growth in lung cancer cells in vivo (17). Another recent study detected
miR-506-3p downregulation in NSCLC and revealed that miR-506-3p
inhibited NSCLC progression (22).
In the present study, the downregulation of miR-506-3p expression
in NSCLC was confirmed, and the role of miR-506-3p in mediating
gefitinib sensitivity in NSCLC cells was identified.
Various mechanisms for the development of gefitinib
resistance have been identified (23–25).
Altered expression of several miRNAs has also been associated with
gefitinib sensitivity, via the regulation of their target genes in
NSCLC cells (12,26,27).
The expression of miR-506-3p was identified to be a predictor of
the response to chemotherapy in multiple cancer types (19,28,29).
However, the function of miR-506-3p in gefitinib sensitivity
remains unknown. The results of the present study demonstrated a
decrease in the miR-506-3p expression levels in PC-9GR cells, as
compared with those in the parental cells. The cell proliferation
assay demonstrated that miR-506-3p mimics inhibited the growth of
both PC-9GR and PC-9 cells compared with miR-NC mimics, which is
consistent with the findings of a previous study on the
anti-proliferative function of miR-506-3p in NSCLC cells (22). Notably, the overexpression of
miR-506-3p markedly increased the cell viability reduction and
apoptosis following gefitinib treatment in PC-9GR cells, while the
inhibition of miR-506-3p was demonstrated to have the opposing
effect. Bcl-2 and Bax are known regulators of cell apoptosis.
miR-506-3p overexpression increased the gefitinib-induced Bcl-2
reduction and Bax elevation in PC-9GR cells, suggesting enhanced
cytotoxicity. By contrast, the inhibition of miR-506-3p decreased
the Bcl-2 expression and increased the Bax expression following
gefitinib treatment. In addition, in PC-9GR cells transfected with
the miR-506-3p antagonist, treatment with 1 µM gefitinib induced
growth arrest in PC-9GR cells, although this concentration did not
inhibit cell growth in untransfected PC-9GR cells. These data
collectively revealed a gefitinib-sensitizing role of miR-506-3p in
NSCLC cells.
The Hippo/YAP pathway is a well-defined signaling
pathway in mediating cell growth and controlling organ size
(30). In cancer cells, YAP1 is
often elevated, contributing to drug resistance through the
maintenance of stemness (31,32).
Specifically, the elevation of YAP1 has also been found to be
involved in the development of gefitinib resistance in NSCLC cells
(33,34). In the PC-9GR cell line established
in the present study, a significant elevation of mRNA and protein
YAP1 levels was observed, as compared with the same levels in PC-9
cells, suggesting that YAP1 may also be implicated in gefitinib
resistance. Furthermore, YAP1 has been identified as a target gene
of miR-506-3p in breast and liver cancer (35,36).
In the present study, it was observed that the overexpression of
miR-506-3p decreased YAP1 expression in PC-9GR cells. In addition,
dual-luciferase reporter assay confirmed YAP1 as a target gene of
miR-506-3p in PC-9GR cells. Notably, YAP1 elevation partially
reversed miR-506-3p mimic-induced hypersensitivity of PC-9GR cells
to gefitinib treatment, suggesting that YAP1 served a pivotal role
in the mediation of miR-506-3p in gefitinib sensitivity in NSCLC
cells. Furthermore, the analysis of miR-506-3p and YAP1 expression
levels in specimens collected from NSCLC patients indicated a
negative correlation between these levels, providing evidence of
the regulatory association of miR-506-3p and YAP1 in NSCLC.
In conclusion, the results of the present study
demonstrated that the downregulation of miR-506-3p contributes to
gefitinib resistance in NSCLC cells via the regulation of YAP1.
This newly identified miR-506-3p/YAP1 axis in NSCLC provided
insights for the development of a miR-506-based therapeutic
approach to overcome gefitinib resistance in patients with NSCLC.
However, the lack of reasonable correlation between miR-506-3p/YAP1
and EGFR-TKIs is a limitation of the present study, which will be
investigated in our future research.
Acknowledgements
The authors appreciate the support of Taizhou
Central Hospital (Taizhou, China) in the present study.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JZ and LT performed the experiments, and analyzed
the data. LJ conceived the study, analyzed the data and prepared
the manuscript.
Ethics approval and consent to
participate
The current study was approved by the ethics
committee of Taizhou Central Hospital. Each patient was informed
and consented to participate.
Patient consent for publication
Each patient consented to the publication of the
clinical data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeng H, Zheng R, Guo Y, Zhang S, Zou X,
Wang N, Zhang L, Tang J, Chen J, Wei K, et al: Cancer survival in
China, 2003–2005: A population-based study. Int J Cancer.
136:1921–1930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lung cancer (OPTIMAL,
CTONG-0802): A multicentre, open-label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: Gefitinib or chemotherapy for non-small-cell lung cancer
with mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han JY, Park K, Kim SW, Lee DH, Kim HY,
Kim HT, Ahn MJ, Yun T, Ahn JS, Suh C, et al: First-SIGNAL:
First-line single-agent iressa versus gemcitabine and cisplatin
trial in never-smokers with adenocarcinoma of the lung. J Clin
Oncol. 30:1122–1128. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takezawa K, Pirazzoli V, Arcila ME, Nebhan
CA, Song X, de Stanchina E, Ohashi K, Janjigian YY, Spitzler PJ,
Melnick MA, et al: HER2 amplification: A potential mechanism of
acquired resistance to EGFR inhibition in EGFR-mutant lung cancers
that lack the second-site EGFRT790M mutation. Cancer Discov.
2:922–933. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pao W, Miller VA, Politi KA, Riely GJ,
Somwar R, Zakowski MF, Kris MG and Varmus H: Acquired resistance of
lung adenocarcinomas to gefitinib or erlotinib is associated with a
second mutation in the EGFR kinase domain. PLoS Med. 2:e732005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Garofalo M, Romano G, Di Leva G, Nuovo G,
Jeon YJ, Ngankeu A, Sun J, Lovat F, Alder H, Condorelli G, et al:
EGFR and MET receptor tyrosine kinase-altered microRNA expression
induces tumorigenesis and gefitinib resistance in lung cancers. Nat
Med. 18:74–82. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao Y, Fan X, Li W, Ping W, Deng Y and Fu
X: miR-138-5p reverses gefitinib resistance in non-small cell lung
cancer cells via negatively regulating G protein-coupled receptor
124. Biochem Biophys Res Commun. 446:179–186. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi L, Wang Y, Lu Z, Zhang H, Zhuang N,
Wang B, Song Z, Chen G, Huang C, Xu D, et al: miR-127 promotes EMT
and stem-like traits in lung cancer through a feed-forward
regulatory loop. Oncogene. 36:1631–1643. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Horn L, Lovly CM and Johnson DH: Chapter
107: Neoplasms of the lung. Harrison's Principles of Internal
Medicine. Kasper DL, Hauser SL, Jameson JL, Fauci AS, Longo DL and
Loscalzo J: 19th. McGraw-Hill; New York, NY: 2015
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hua K, Yang W, Song H, Song J, Wei C, Li D
and Fang L: Up-regulation of miR-506 inhibits cell growth and
disrupt the cell cycle by targeting YAP in breast cancer cells. Int
J Clin Exp Med. 8:12018–12027. 2015.PubMed/NCBI
|
|
17
|
Wang Y, Cui M, Sun BD, Liu FB, Zhang XD
and Ye LH: MiR-506 suppresses proliferation of hepatoma cells
through targeting YAP mRNA 3′UTR. Acta Pharmacol Sin. 35:1207–1214.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo X, Xiao H, Guo S, Dong L and Chen J:
Identification of breast cancer mechanism based on weighted gene
coexpression network analysis. Cancer Gene Ther. 24:333–341. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Wu H, Li W, Yin L, Guo S, Xu X,
Ouyang Y, Zhao Z, Liu S, Tian Y, et al: Downregulated miR-506
expression facilitates pancreatic cancer progression and
chemoresistance via SPHK1/Akt/NF-κB signaling. Oncogene.
35:5501–5514. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Krawczyk P, Powrozek T, Olesinski T,
Powrózek T, Olesiński T, Dmitruk A, Dziwota J, Kowalski D and
Milanowski J: Evaluation of miR-506 and miR-4316 expression in
early and non-invasive diagnosis of colorectal cancer. Int J
Colorectal Dis. 32:1057–1060. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yin M, Ren X, Zhang X, Luo Y, Wang G,
Huang K, Feng S, Bao X, Huang K, He X, et al: Selective killing of
lung cancer cells by miRNA-506 molecule through inhibiting NF-κB
p65 to evoke reactive oxygen species generation and p53 activation.
Oncogene. 34:691–703. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo S, Yang P, Jiang X, Li X, Wang Y,
Zhang X, Sun B, Zhang Y and Jia Y: Genetic and epigenetic silencing
of mircoRNA-506-3p enhances COTL1 oncogene expression to foster
non-small lung cancer progression. Oncotarget. 8:644–657.
2017.PubMed/NCBI
|
|
23
|
Yu HA, Arcila ME, Rekhtman N, Sima CS,
Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M and Riely GJ:
Analysis of tumor specimens at the time of acquired resistance to
EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers.
Clin Cancer Res. 19:2240–2247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Engelman JA, Zejnullahu K, Mitsudomi T,
Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen
J, et al: MET amplification leads to gefitinib resistance in lung
cancer by activating ERBB3 signaling. Science. 316:1039–1043. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yano S, Yamada T, Takeuchi S, Tachibana K,
Minami Y, Yatabe Y, Mitsudomi T, Tanaka H, Kimura T, Kudoh S, et
al: Hepatocyte growth factor expression in EGFR mutant lung cancer
with intrinsic and acquired resistance to tyrosine kinase
inhibitors in a Japanese cohort. J Thorac Oncol. 6:2011–2017. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu Q, Liu Y, Wen C, Zhao Y, Jin S, Hu Y,
Wang F, Chen L, Zhang B, Wang W, et al: MicroRNA-1 inhibits
tumorigenicity of esophageal squamous cell carcinoma and enhances
sensitivity to gefitinib. Oncol Lett. 15:963–971. 2018.PubMed/NCBI
|
|
27
|
Ping W, Gao Y, Fan X, Li W, Deng Y and Fu
X: MiR-181a contributes gefitinib resistance in non-small cell lung
cancer cells by targeting GAS7. Biochem Biophys Res Commun.
495:2482–2489. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou H, Lin C, Zhang Y, Zhang X, Zhang C,
Zhang P, Xie X and Ren Z: miR-506 enhances the sensitivity of human
colorectal cancer cells to oxaliplatin by suppressing MDR1/P-gp
expression. Cell Prolif. 50:2017. View Article : Google Scholar :
|
|
29
|
Liu G, Xue F and Zhang W: miR-506: A
regulator of chemo-sensitivity through suppression of the
RAD51-homologous recombination axis. Chin J Cancer. 34:485–487.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pan D: The hippo signaling pathway in
development and cancer. Dev Cell. 19:491–505. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Harvey KF, Zhang X and Thomas DM: The
hippo pathway and human cancer. Nat Rev Cancer. 13:246–257. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu S, Cai X, Wu C, Wu L, Wang Y, Liu Y, Yu
Z, Qin S, Ma F, Thiery JP and Chen L: Adhesion glycoprotein CD44
functions as an upstream regulator of a network connecting ERK, AKT
and Hippo-YAP pathways in cancer progression. Oncotarget.
6:2951–2965. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ghiso E, Migliore C, Ciciriello V, Morando
E, Petrelli A, Corso S, De Luca E, Gatti G, Volante M and Giordano
S: YAP-dependent AXL overexpression mediates resistance to EGFR
inhibitors in NSCLC. Neoplasia. 19:1012–1021. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
McGowan M, Kleinberg L, Halvorsen AR,
Helland Å and Brustugun OT: NSCLC depend upon YAP expression and
nuclear localization after acquiring resistance to EGFR inhibitors.
Genes Cancer. 8:497–504. 2017.PubMed/NCBI
|
|
35
|
Hua K, Yang W, Song H, Song J, Wei C, Li D
and Fang L: Up-regulation of miR-506 inhibits cell growth and
disrupt the cell cycle by targeting YAP in breast cancer cells. Int
J Clin Exp Med. 8:12018–12027. 2015.PubMed/NCBI
|
|
36
|
Wang Y, Cui M, Sun BD, Liu FB, Zhang XD
and Ye LH: MiR-506 suppresses proliferation of hepatoma cells
through targeting YAP mRNA 3′UTR. Acta Pharmacol Sin. 35:1207–1214.
2014. View Article : Google Scholar : PubMed/NCBI
|