Introduction
Colorectal cancer (CRC) is now commonly diagnosed
worldwide, particularly in patients who are in favor of a ‘Western
diet’. Statistically, >1,400,000 new cases of CRC were diagnosed
and a cancer-associated mortality rate of 5% among patients with
CRC was reported in 2012. It is estimated that there are likely to
be >2,200,000 new cases diagnosed and 1,100,000 cases of
cancer-associated mortality by the year 2030 (1). CRC normally begins with noncancerous
adenomas and gradually develops into CEC over a period of 10–20
years (2). The presence of
inherited genetic abnormalities, (i.e. family history)
significantly increases the risk of developing CRC compared with
the absence of such an abnormality (3). Studies have identified a substantial
number of genes linked to hereditary CRC syndromes (4). In addition, epigenetic aberrations
have been found to be implicated in the initiation and development
of a variety of tumors, including CRC (5,6).
Given the fact that epigenetic modifications are reversible, a
number of studies have focused on the potential applications of
epigenetic aberration as a therapeutic target.
MicroRNAs (miRNAs), one of the epigenetic regulatory
mechanisms, are important in the downregulation of gene expression
by complementarily binding to the target mRNA (7,8).
miRNAs are small fragments of non-coding RNA with a size of 20–24
nucleotides and are generated through multiple enzymatic excisions
of miRNA precursor pri-miRNA. The RNA-induced silencing complex,
which is composed of mature miRNAs including dicer and other
associated factors, is formed and leads to enzymatic cleavage of
the target mRNA, resulting in substantially decreased levels of
protein translation (7,9). miR-675 is derived from exon 1 of long
noncoding RNA H19 (10). Mice with
deficiency of the H19 transcript grow normally. Hao et al
(11), showed that H19 exhibits
tumor suppression activity, and its associated miR-675 has been
shown to be oncogenic in gastric (12), liver (13) and lung cancer (14). Therefore, the dysregulation of
miR-675 may be used as a potential biomarker for detecting
carcinogenesis in multiple types of cancer.
Cyclin D binding myb like transcription factor 1
(DMTF1) is induced by oncogenic Ras-Raf signaling and functions as
a tumor suppressor (15).
DMTF1-heterozygous and -null mice exhibit accelerated formation of
spontaneously-developed or oncogene-induced tumors (16). Of all types of human non-small lung
cancer, ~40% have been found to have DMTF1 gene deletion (15). In addition, the expression level of
DMTF1 is higher in the colon relative to that in the lung,
according to the proteome database (17); this indicates its potential role in
CRC. In the present study, it was demonstrated that miR-675-3p
directly suppressed DMTF1, which further contributed to the
proliferation of CRC cells.
Materials and methods
Human patients and CRC tissues
CRC tissues and adjacent non-carcinogenic tissues
were collected from patients who underwent surgery between 2012 and
2017 at the Affiliated Hospital of Beihua University (Jilin City,
China). All patients with CRC were diagnosed by colonoscopy
pathology. The total number of patients was 60 with age ranging
between 45 and 81 years. The gender ratio was 1.4:1.0
(male:female). All procedures were conducted under the approval of
the Ethics Committee of the Affiliated Hospital of Beihua
University. The tissues were collected with patients' informed
consent. The collected tissues were immediately stored at −80°C for
future use.
Cell culture and transfection
The SW480 and HT29 CRC cell lines (American Type
Culture Collection, Manassas, VA, USA) were cultured in Dulbecco's
modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (Sigma;
Merck KGaA, Darmstadt, Germany). The cell cultures were maintained
at 37°C under a humidified atmosphere containing 5% CO2.
Transfections were conducted with either Lipofectamine 3000 (Thermo
Fisher Scientific, Inc.) for the overexpression (plasmid) or
RNAiMax for the miRNA mimics and inhibitors.
pCDNA3 was used as a vector to construct the
full-length DMTF1 overexpression plasmid. An empty pCDNA3 vector
was used as the negative control. The miR-675-3p mimics, inhibitors
and the corresponding controls were purchased from Sigma; Merck
KGaA for transfection. The cells were seeded in antibiotic-free
medium prior to transfection to increase the transfection
efficiency. The growth medium was replaced 12 h following
transfection.
RNA extraction, cDNA synthesis and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis
The cultured cells were washed with cold PBS and
then treated with TRIzol (Thermo Fisher Scientific, Inc.). Total
RNA was isolated from the TRIzol-lysed cells following the
manufacturer's protocol. Following isolation, 500 ng of total RNA
was used with 10 µl cDNA synthesis system, including 2 µl of 10X RT
buffer, 0.8 µl of 100 mM NTP mix, 2 µl of 10X RT Random Primers, 1
µl of reverse transcriptase and 3.2 µl of nuclease-free water
(High-Capacity cDNA Reverse Transcript kit; Thermo Fisher
Scientific, Inc.). The temperature protocol for RT-PCR was as
follows: 25°C for 10 min, 37°C for 120 min and 85°C for 5 min. The
qPCR system included: 5 µl of SYBR master mix (Thermo Fisher
Scientific, Inc.), 0.5 µl of synthesized cDNA, 2 µl of primer mix
(5 µM each) and 2.5 µl of water. Specific primers were used for
SYBR green RT-qPCR analysis, as previously described (18): DMTF1, forward
5′-TGTAGCTGATCCATCCGTTGT-3′ and reverse
5′-GGGGTTGCTCCTATTTCTTTG-3′; GAPDH, forward
5′-GCGAGATCGCACTCATCATCT-3′ and reverse
5′-TCAGTGGTGGACCTGACC-3′.
The PCR conditions were as follows: 95°C for 3 min,
40 cycles at 95°C for 10 sec and then at 60°C for 30 sec, followed
by 72°C for 3 min. The total RNA was isolated with TRIzol, and the
qScript microRNA cDNA Synthesis kit (Quantabio) was used for miRNA
according to the manufacturer's protocol. RT-qPCR analysis was
performed using the miScript SYBR Green PCR kit (Qiagen, Inc.,
Valencia, CA, USA) according to the manufacturer's protocol. The
miScript Primer Assay (Qiagen, Inc.) was used for quantification of
miR-675-3p and the U6 small RNA Assay (Qiagen, Inc.) was used for
the housekeeping control (primer sequences are commercially
unavailable). The RT-qPCR was performed as follows: 95°C for 2 min,
40 cycles at 95°C for 5 sec and then at 60°C for 30 sec. Data were
quantified using the 2−∆∆Cq method, as previously
described (19).
MTS assay
The cells were seeded in 96-well plates
(105 cells/well) and were maintained in the growth
medium (100 µl) at 37°C prior to the addition of sterile MTS dye
(0.33 mg/ml, 20 µl). The absorbance was obtained using an enzyme
immunoassay analyzer (BioTek Instruments, Inc., Winooski, VT, USA)
at a wavelength of 490 nm upon a further incubation period of 2
h.
Crystal violet staining
The cells were washed with cold PBS and fixed with
ice-cold methanol for 10 min, followed by staining with 0.5%
crystal violet solution for 10 min. The cells were washed with
deionized water prior to being examined using the Olympus CX21
light microscope (Olympus Corporation, Tokyo, Japan).
Flow cytometry
The cells were washed with cold PBS and fixed in
cold 75% ethanol at 4°C for 30 min. The fixed cells were washed
three times and treated with ribonuclease. The resulting cells were
stained with propidium iodide (50 µg/ml) at 4°C overnight (Abcam,
Cambridge, UK) prior to flow cytometric analysis with excitation at
488 nm (BD Biosciences, Franklin Lakes, NJ, USA).
Vector construction and luciferase
assay
The TargetScan database (targetscan.org) was used to predict potential targets
of miR-675-3p (20). The
3′untranslated region (UTR) of the DMTF1 sequence was amplified and
cloned into a pGL4.10-report vector (Promega Corporation, Madison,
WI, USA). Equal quantities of the pGL4.10-3UTR-DMTF1 and
Renilla expression vector pRL-TK (Promega Corporation) were
co-transfected into the SW480 cells. The luciferase activity was
measured 48 h post-transfection using the Dual-Glo Luciferase Assay
system (Promega Corporation).
Western blot analysis
The cells were lysed with proteinase
inhibitors-containing RIPA buffer to extract total protein. The
protein concentration was measured using the Bradford assay (Sigma;
Merck KGaA). Subsequently, 20 µg of total protein was added to each
well of 10% SDS-PAGE. Then proteins were transferred to PVDF
membranes and incubated in transferring buffer with 5% non-fat milk
at room temperature for 90 min. Membranes were incubated with a
1:2,000 dilution of the primary antibody against human DMTF1 (cat.
no. PA5-29466; Thermo Fisher Scientific, Inc.) or Tubulin (cat. no.
T9026; Sigma-Aldrich; Merck KGaA) at 4°C overnight and then with
secondary antibody (goat anti-rabbit; cat. no. 31460; Thermo Fisher
Scientific, Inc.) at room temperature for 60 min.
Statistical analysis
Data were analyzed using unpaired t-tests or two-way
analysis of variance with Tukey's post hoc test for multiple
comparisons. Statistical analysis was performed using GraphPad
Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-675-3p promotes proliferation in
CRC cells
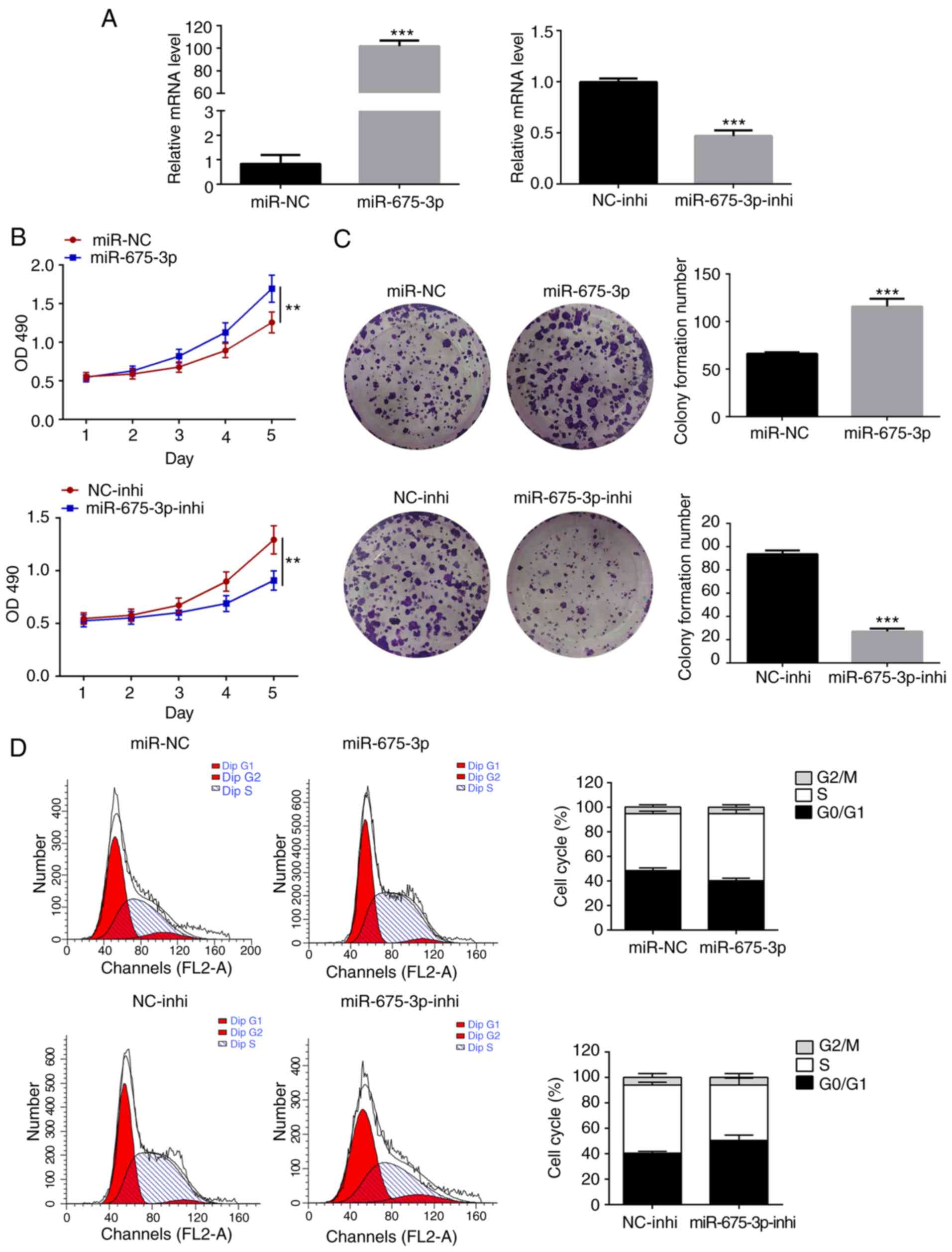
To demonstrate the effect of miR-675-3p on CRC cell
growth, overexpression of miR-675-3p was induced by transfecting
miR-675-3p mimics into the human SW480 CRC cells, whereas the
inhibition assay was performed by transfecting miR-675-3p
inhibitors. The expression level of miR-675-3p was found to be
significantly increased or halved upon transfection with the
miR-675-3p mimic or miR-675-3p inhibitor, respectively, compared
with the corresponding controls (Fig.
1A). Proliferation of the aforementioned cells was evaluated
using an MTS assay. Cells transfected with the miR-675-3p mimics
showed higher values of OD490, indicating the presence
of more viable cells compared with the miR-NC control (Fig. 1B), whereas the inhibition of
miR-675-3p notably reduced the number of viable cells compared with
its control. To confirm the changes observed were caused by
miR-675-3p, crystal violet staining was used to evaluate the
formation of cell colonies. Consistent with the results of the MTS
assay, a significant increase in the number of cell colonies was
observed for the miR-675-3p-overexpressed cells and a decrease was
observed for the miR-675-3p-inhibited cells (Fig. 1C). The increase in the number of
cell colonies induced by miR-675-3p was considered to be due to
enhanced proliferation. Flow cytometry was employed to analyze the
cell cycle: For the miR-675-3p-overexpressed cells, the number of
cells in the S phase increased but those in the
G0/G1 phases decreased, whereas the reverse
was observed for the miR-675-3p-inhibited cells (Fig. 1D).
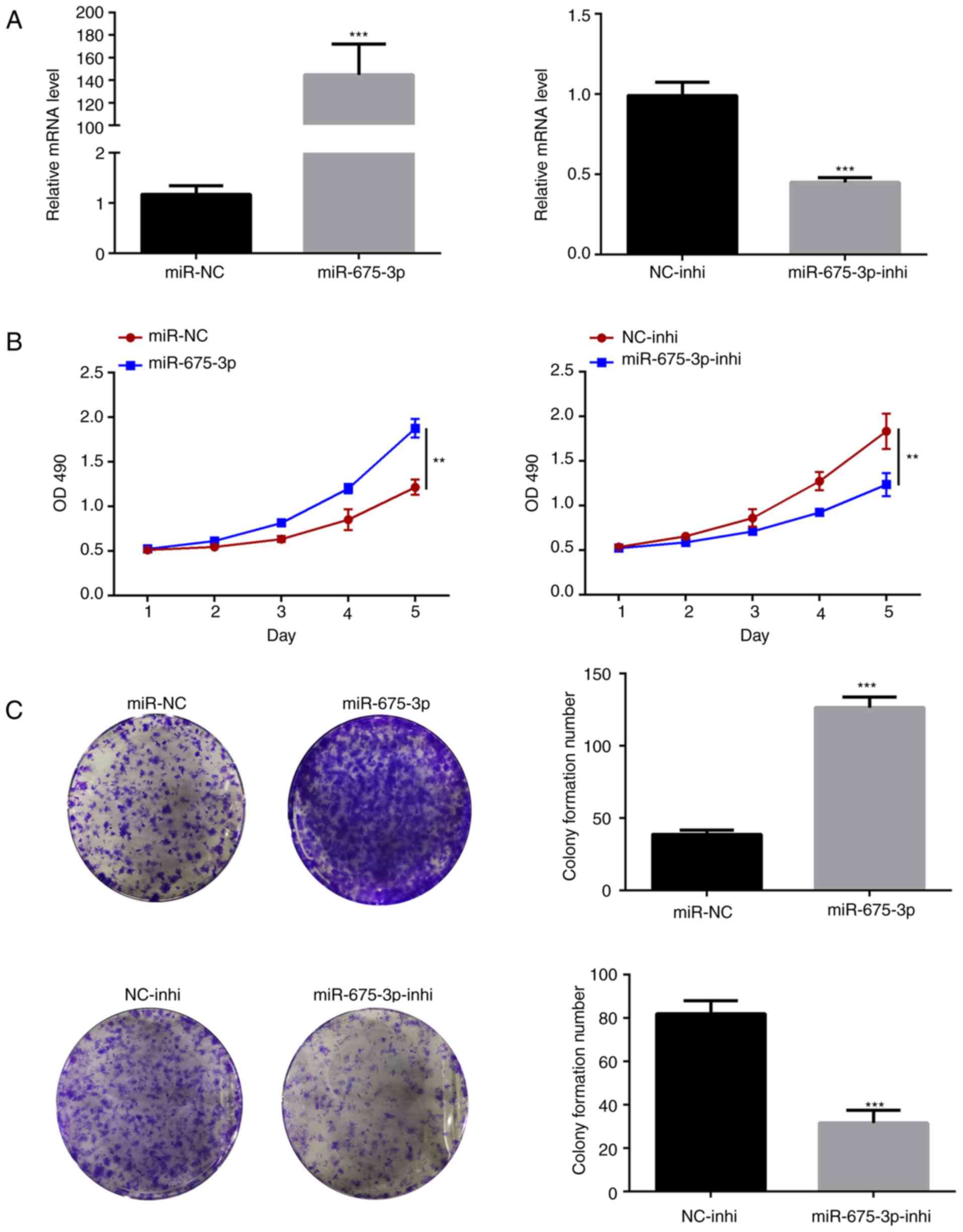
To confirm the concept that miR-675-3p promotes cell
proliferation, the expression of miR675-3p was manipulated in the
HT29 CRC cells. The HT29 cells overexpressing miR675-3p and HT29
cells with suppressed expression of miR675-3p were generated
(Fig. 2A). In these two cell
lines, consistent data were obtained that the overexpression of
miR675-3p significantly increased cell proliferation whereas the
inhibition of miR675-3p lowered proliferation of the HT29 cells,
determined by the OD490 value for viable cells and MTS
assay for cell colony (Fig. 2B and
C).
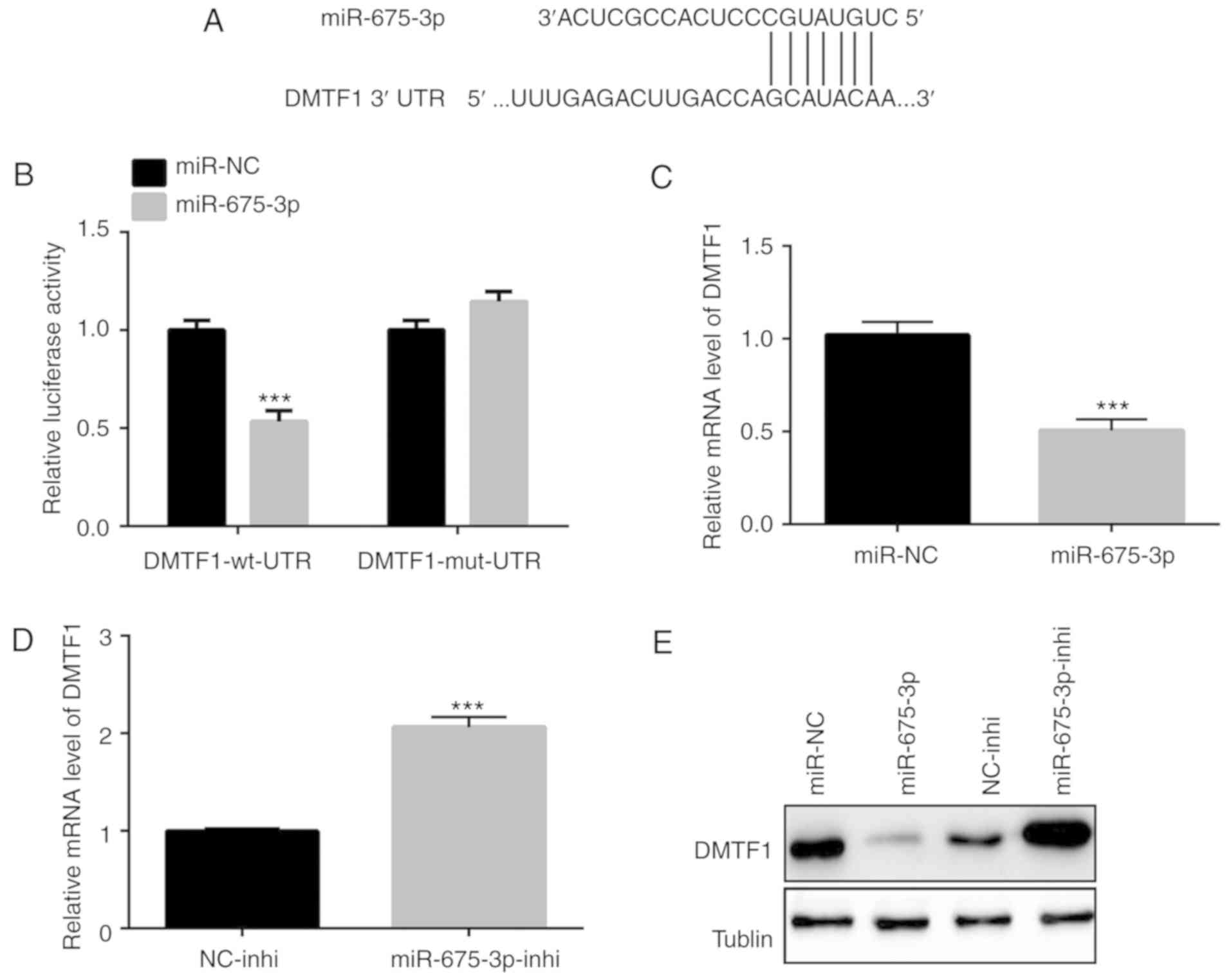
miR-675-3p directly modulates the
expression of DMTF1
The TargetScan database was used to predict
potential targets of miR-675-3p (20). The binding site at the 3′UTR region
of DMTF1, a known tumor suppressor gene (15,21,22),
was found to be a potential target of miR-675-3p (Fig. 3A). A luciferase assay was conducted
to examine the regulation of DMTF1 by miR-675-3p. The
overexpression of miR-675-3p in the SW480 cells significantly
reduced the relative luciferase activity compared with that in the
cells with the mutated DMTF1-3′UTR region, indicating a direct
negative regulation of the expression of DMTF1 by miR-675-3p
through the DMTF1 3′UTR region (Fig.
3B). Consistently, a decrease in the expression of DMTF1 was
found in the SW480 cells with overexpressed miR-675-3p at the mRNA
level (Fig. 3C). Such a decrease
in the expression of DMTF1 was also confirmed by inhibiting the
expression of miR-675-3p in the SW480 cells (Fig. 3D). In addition to the changes of
DMTF1 observed at the mRNA level, western blotting was performed to
examine the protein level changes of DMTF1 in the
miR-675-3p-overexpressed and -inhibited SW480 cells. The results
showed that the overexpression of miR-675-3p suppressed the protein
level of DMTF1, whereas the inhibition of miR-675-3p increased the
level of DMTF1 (Fig. 3E).
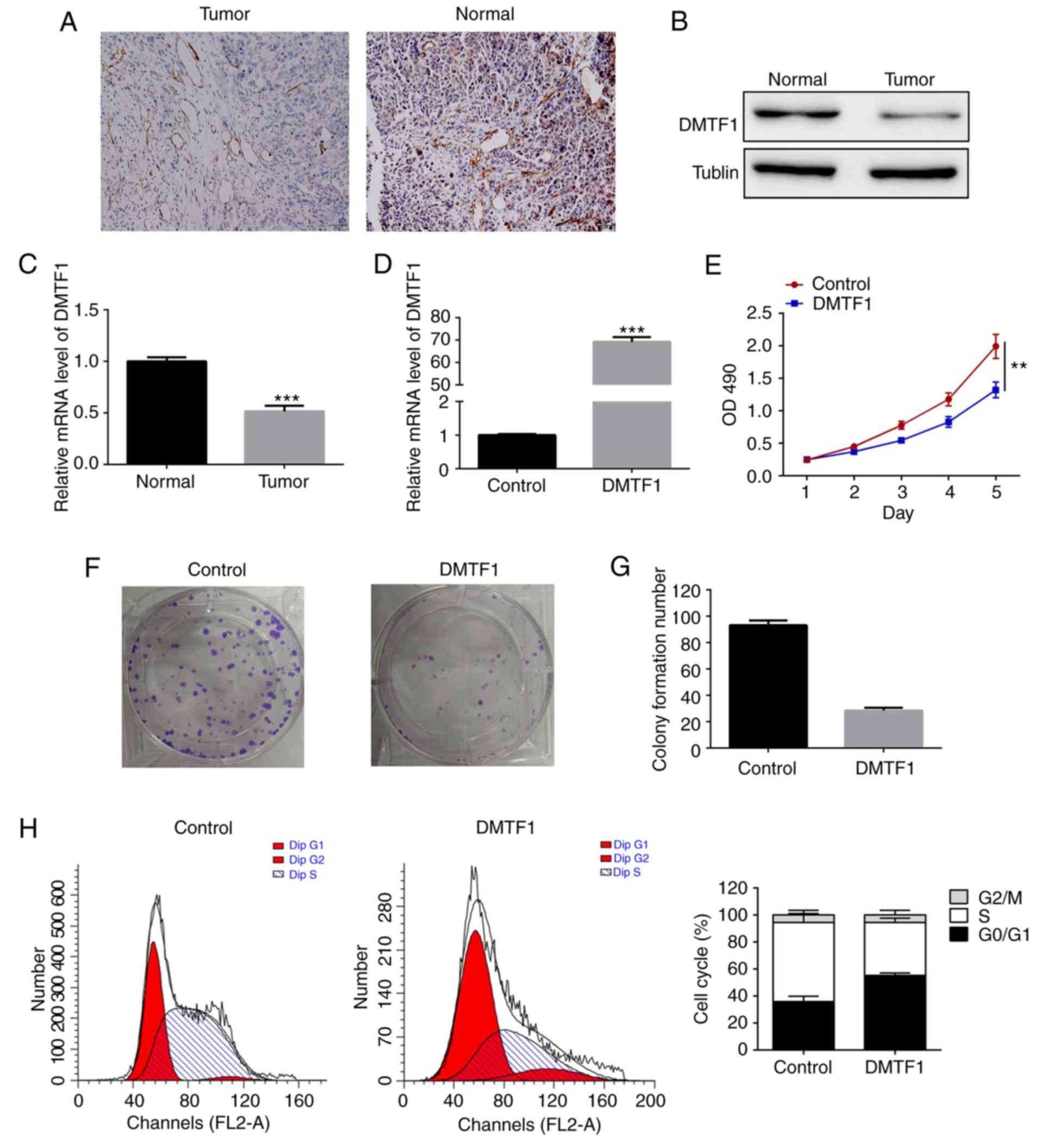
Significant reduction in the
expression of DMTF1 in human CRC tumors
As miR-675-3p promoted CRC cell proliferation and
regulated the downstream expression of DMTF1, it was hypothesized
that DMTF1 was also involved in the development of CRC. The
expression of DMTF1 was examined and was found to be markedly
decreased in the human CRC tumors using immunohistochemistry
(Fig. 4A) and western blot
analysis (Fig. 4B) compared with
that in its adjacent non-carcinogenic tissue. As miR-675-3p was
effective in regulating DMTF1 mRNA, the relative mRNA level of
DMTF1 in tumors was assessed and was also found to be reduced,
consistent with the protein pattern (Fig. 4C).
Overexpression of DMTF1 suppresses
cell proliferation in CRC cells
As DMTF1 was downregulated in CRC tumors, the SW480
cells were transfected to overexpress DMTF1 to illustrate its
function in carcinogenesis. As shown in Fig. 4D, there was an increase of DMTF1 in
the transfected SW480 cells compared with that in the control
group. Cell proliferation (Fig.
4E), cell colony formation (Fig.
4F and G) and cell cycle (Fig.
4H) were also examined using this cell line. The results
demonstrated significant decreases in all three aforementioned
parameters in the DMTF1-overexpressed SW480 cells, indicating the
negatively regulatory role of DMTF1 in cell proliferation.
miR675-3p promotes CRC proliferation
in a DMTF1-dependent manner
It has been shown that miR675-3p and DMTF1 regulated
CRC cell proliferation and that miR675-3p modulated the level of
DMTF1 through binding to its 3′UTR region. A rescue assay was
performed to further demonstrate the correlation between miR-675-3p
and DMTF1 on CRC growth. The SW480 cells were used to overexpress
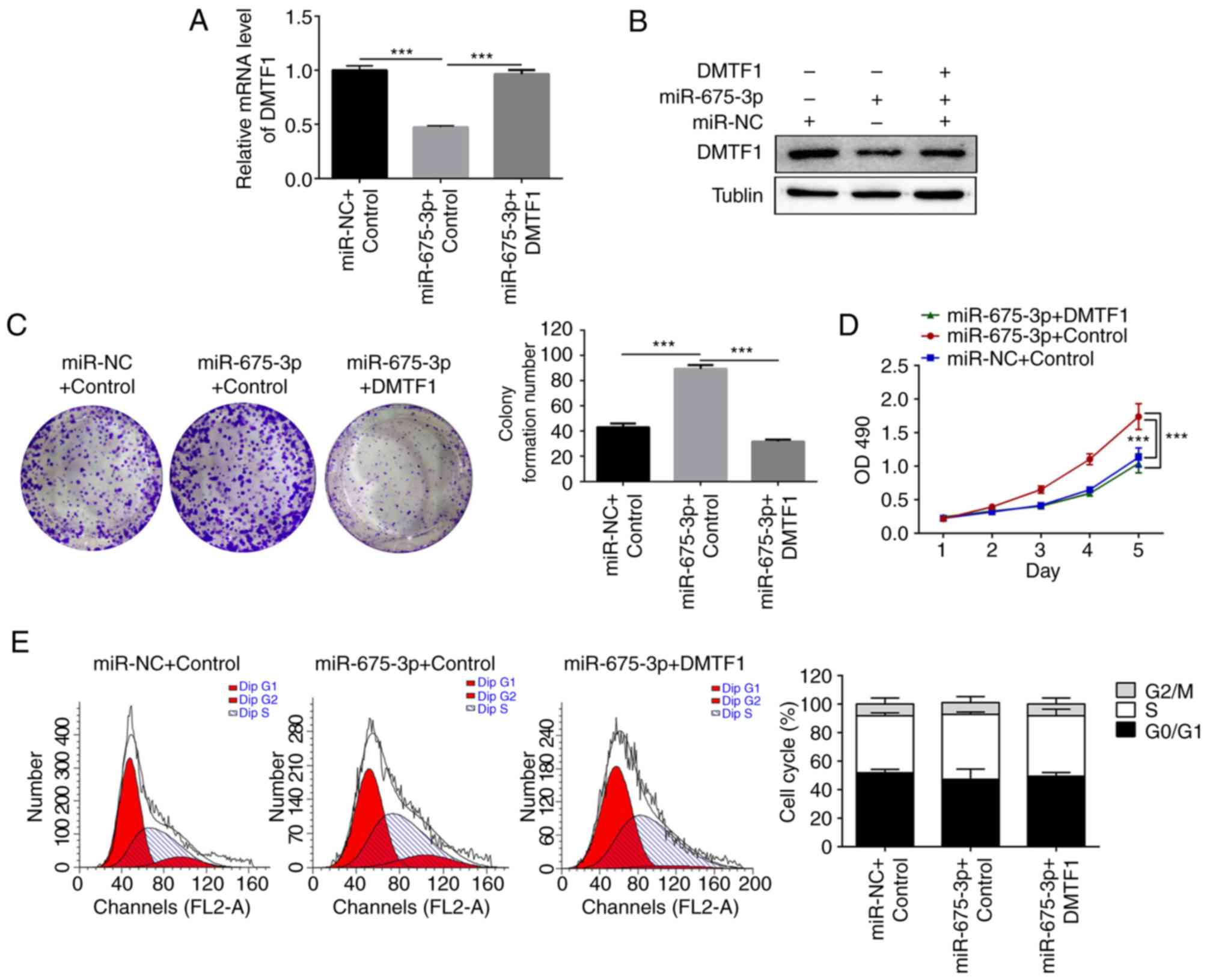
miR-NC, miR-675-3p and miR675-3p + DMTF1, respectively. Consistent
with the previous results, the mRNA level of DMTF1 reduced when
miR-675-3p was overexpressed (Fig.
5A) and co-transfection of the plasmid expressing DMTF1
normalized its expression (Fig.
5A). In addition, the protein level of DMTF1 was examined by
western blotting (Fig. 5B); the
expression level of DMTF1 decreased in the presence of exogenous
miR-675-3p but normalized with additional expressed DMTF1.
Subsequently, crystal violet staining was conducted to evaluate the
single cell colony formation. For the miR-675-3p-overexpressed
SW480 cells, co-transfection of DMTF1 reduced the number of single
colonies (Fig. 5C). It was found
that the increased cell proliferation induced by miR675-3p was
rescued by the expression of DMTF1, based on the MTS results
(Fig. 5D). miR-675-3p
significantly amplified the proportion of cells in the S phase in
the cell cycle assay. Upon co-expression of DMTF1, the S phase
population decreased back to a level comparable to that of the
control group (Fig. 5E).
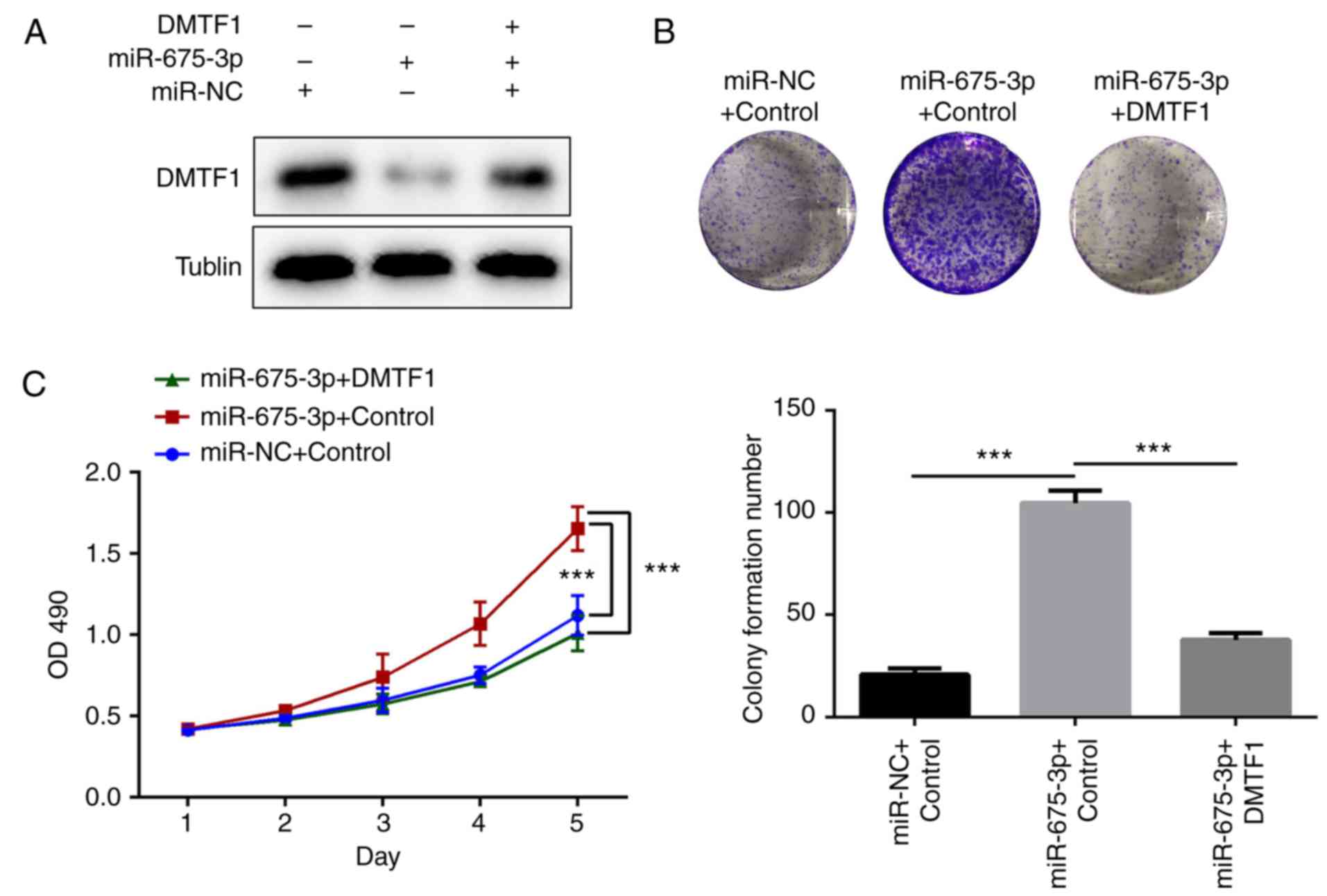
In addition, the DMTF1 rescue experiment was
performed in another CRC cell line (HT29). Consistent with the
SW480 cells, the overexpression of miR675-3p inhibited the
expression of DMTF1 (Fig. 6A). To
elucidate the effect of DMTF1, DMTF1 was overexpressed in HT29
cells overexpressing miR675-3p (Fig.
6A). It was found that DMTF1 negatively regulated the
miR675-3p-induced overgrowth of colonic cells, as determined using
the MTS assay (Fig. 6B) and via
OD490 measurement (Fig.
6C).
Discussion
The initiation and progression of CRC are driven by
multiple factors, including distinct genetic and epigenetic
alterations (6). The therapeutic
manipulation of aberrations in the epigenome, which include DNA
methylation and histone acetylation, has been implicated in the
control of cancer progression. In current clinical settings, two
classes of epigenetic drugs are mainly used: DNA methyltransferase
inhibitors (DNMTis) and histone deacetylase inhibitors (HDACis).
These are designed to restore the tumor suppressor gene expression
by downregulating promoter methylation and upregulating histone
acetylation of the target gene, respectively (23). These two classes of drugs have been
approved and used in the treatment of lymphoma and myelodysplastic
syndrome (24). However, HDACi
failed to generate promising results, based on the clinical phase
study evaluation (25–27). Although the combination of DNMTi
and HDACi treatment in metastatic CRC in phase II was found to
reverse hypermethylation and improve progression-free survival rate
(28), the side-effects of DNMTi
and HDACi were of concern, due to their relatively high in
vivo non-specificity. Alternatively, the modulation of miRNA in
cancer provides a novel therapeutic method by introducing specific
miRNA mimics to restore tumor-suppressor miRNA or inhibitors to
suppress onco-miRNA expression (29).
miR-675-3p has been found to be upregulated in
several types of cancer, however, its role in CRC remains to be
fully elucidated. The present study focused on miR-675-3p and its
downstream factor DMTF1 in their ability to induce CRC cell
proliferation. Firstly, the effect of miR675-3p on CRC cell
proliferation was examined by either overexpressing or inhibiting
its expression in the SW480 cell line. It was found miR-675-3p was
able to promote cell growth (Fig.
1); this was consistent with previous studies (12–14).
An online resource was then used to predict a potential target of
miR-675-3p, DMTF1. It was shown that miR-675-3p regulated the
expression of DMTF1 through its 3′UTR binding site (Fig. 2). DMTF1 is induced by activation of
the Ras-Raf pathway and functions as a tumor suppressor in a
negative feedback-loop (30). The
significant reduction of DMTF1 observed in the human CRC tissue,
compared with its adjacent normal tissue (Fig. 3C), indicated that the dysregulation
of DMTF1 may be responsible for the development of CRC. By
overexpressing DMTF1 in the SW480 cells, the suppressive effect on
CRC cell growth confirmed. As DMTF1 was able to induce cell cycle
arrest at entry to the S phase (31), a cell cycle assay was performed in
the DMTF1-overexpressed SW480 cells. An increased cell population
in the G0/G1 phase at the expense of a
reduced population in the S phase was observed (Fig. 3G). Finally, miR-675-3p and DMTF1
were co-expressed in the SW480 cells to examine their synergistic
effect on cell proliferation. The results showed that increased
expression of DMTF1 in the miR-675-3p-overexpressed SW480 cells
normalized cell growth (Fig.
4C-E). In summary, it was demonstrated that the oncogenic
property of miR-675-3p was dependent on its negative regulation of
tumor suppressor DMTF1. miR-675-3p together with DMTF1 were
important in CRC proliferation.
miRNA-based biomarkers have been implicated in the
prognosis and diagnosis of tumors (32). In particular, miR-675 was one of
the miRNAs differentially expressed in benign and malignant
adrenocortical tumors (33).
miR-675 was also reported to be dysregulated in lung cancer
(14) and significantly correlated
with the overall decreased survival rate of patients with
pancreatic cancer (34). The
significant increase in expression of miR-675-3p and subsequent
decrease in DMTF1 in CRC cells suggests that the comprehensive
comparison of expression levels of miR-675-3p together with DMTF1
can be useful for developing a biomarker for CRC diagnosis.
Accordingly, the introduction of miR-675-3p inhibitor may reduce
cell growth rate in CRC, minimizing the side-effects compared with
those of other non-specific epigenetic drugs.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
WH and XY designed all of the experiments and
revised the manuscript. YLo and MW performed the experiments, and
CL and YLi analyzed and interpreted the data.
Ethics approval and consent to
participate
All procedures were conducted under the approval of
the Ethics Committee of the Affiliated Hospital of Beihua
University (Jilin, China). The study conforms with The Code of
Ethics of the World Medical Association (Declaration of Helsinki),
printed in the British Medical Journal (18 July 1964). Informed
consent was obtained from all individual participants included in
the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arnold M, Sierra MS, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global patterns and trends in
colorectal cancer incidence and mortality. Gut. 66:683–691. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stryker SJ, Wolff BG, Culp CE, Libbe SD,
Ilstrup DM and MacCarty RL: Natural history of untreated colonic
polyps. Gastroenterology. 93:1009–1013. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Butterworth AS, Higgins JP and Pharoah P:
Relative and absolute risk of colorectal cancer for individuals
with a family history: A meta-analysis. Eur J Cancer. 42:216–227.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Valle L: Genetic predisposition to
colorectal cancer: Where we stand and future perspectives. World J
Gastroenterol. 20:9828–9849. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Danese E and Montagnana M: Epigenetics of
colorectal cancer: Emerging circulating diagnostic and prognostic
biomarkers. Ann Transl Med. 5:2792017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pancione M, Remo A and Colantuoni V:
Genetic and epigenetic events generate multiple pathways in
colorectal cancer progression. Patholog Res Int 2012.
5093482012.
|
|
7
|
Jafri MA, Al-Qahtani MH and Shay JW: Role
of miRNAs in human cancer metastasis: Implications for therapeutic
intervention. Semin Cancer Biol. 44:117–131. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peng Y and Croce CM: The role of MicroRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pratt AJ and MacRae IJ: The RNA-induced
silencing complex: A versatile gene-silencing machine. J Biol Chem.
284:17897–17901. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cai X and Cullen BR: The imprinted H19
noncoding RNA is a primary microRNA precursor. RNA. 13:313–316.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hao Y, Crenshaw T, Moulton T, Newcomb E
and Tycko B: Tumour-suppressor activity of H19 RNA. Nature.
365:764–767. 1993. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan J, Zhang Y, She Q, Li X, Peng L, Wang
X, Liu S, Shen X, Zhang W, Dong Y, et al: Long noncoding RNA
H19/miR-675 axis promotes gastric cancer via FADD/caspase 8/caspase
3 signaling pathway. Cell Physiol Biochem. 42:2364–2376. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li H, Li J, Jia S, Wu M, An J, Zheng Q,
Zhang W and Lu D: miR675 upregulates long noncoding RNA H19 through
activating EGR1 in human liver cancer. Oncotarget. 6:31958–31984.
2015.PubMed/NCBI
|
|
14
|
Wang J, Zhao YC, Lu YD and Ma CP:
Integrated bioinformatics analyses identify dysregulated miRNAs in
lung cancer. Eur Rev Med Pharmacol Sci. 18:2270–2274.
2014.PubMed/NCBI
|
|
15
|
Sugiyama T, Frazier DP, Taneja P, Morgan
RL, Willingham MC and Inoue K: Role of DMP1 and its future in lung
cancer diagnostics. Expert Rev Mol Diagn. 8:435–447. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Inoue K, Zindy F, Randle DH, Rehg JE and
Sherr CJ: Dmp1 is haplo-insufficient for tumor suppression and
modifies the frequencies of Arf and p53 mutations in Myc-induced
lymphomas. Genes Dev. 15:2934–2939. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, Olsson I, et al: Proteomics. Tissue-based
map of the human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kobayashi M and Srour EF: Regulation of
murine hematopoietic stem cell quiescence by Dmtf1. Blood.
118:6562–6571. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
21
|
Peng Y, Dong W, Lin TX, Zhong GZ, Liao B,
Wang B, Gu P, Huang L, Xie Y, Lu FD, et al: MicroRNA-155 promotes
bladder cancer growth by repressing the tumor suppressor DMTF1.
Oncotarget. 6:16043–16058. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu S, Mott RT, Fry EA, Taneja P, Kulik G,
Sui G and Inoue K: Cooperation between Dmp1 loss and cyclin D1
overexpression in breast cancer. Am J Pathol. 183:1339–1350. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nervi C, De Marinis E and
Codacci-Pisanelli G: Epigenetic treatment of solid tumours: A
review of clinical trials. Clin Epigenetics. 7:1272015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Santini V, Melnick A, Maciejewski JP,
Duprez E, Nervi C, Cocco L, Ford KG and Mufti G: Epigenetics in
focus: Pathogenesis of myelodysplastic syndromes and the role of
hypomethylating agents. Crit Rev Oncol Hematol. 88:231–245. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fakih MG, Pendyala L, Fetterly G, Toth K,
Zwiebel JA, Espinoza-Delgado I, Litwin A, Rustum YM, Ross ME,
Holleran JL and Egorin MJ: A phase I, pharmacokinetic and
pharmacodynamic study on vorinostat in combination with
5-fluorouracil, leucovorin, and oxaliplatin in patients with
refractory colorectal cancer. Clin Cancer Res. 15:3189–3195. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fakih MG, Fetterly G, Egorin MJ, Muindi
JR, Espinoza-Delgado I, Zwiebel JA, Litwin A, Holleran JL, Wang K
and Diasio RB: A phase I, pharmacokinetic, and pharmacodynamic
study of two schedules of vorinostat in combination with
5-fluorouracil and leucovorin in patients with refractory solid
tumors. Clin Cancer Res. 16:3786–3794. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wilson PM, El-Khoueiry A, Iqbal S, Fazzone
W, LaBonte MJ, Groshen S, Yang D, Danenberg KD, Cole S, Kornacki M,
et al: A phase I/II trial of vorinostat in combination with
5-fluorouracil in patients with metastatic colorectal cancer who
previously failed 5-FU-based chemotherapy. Cancer Chemother
Pharmacol. 65:979–988. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Azad NS, El-Khoueiry A, Yin J, Oberg AL,
Flynn P, Adkins D, Sharma A, Weisenberger DJ, Brown T, Medvari P,
et al: Combination epigenetic therapy in metastatic colorectal
cancer (mCRC) with subcutaneous 5-azacitidine and entinostat: A
phase 2 consortium/stand up 2 cancer study. Oncotarget.
8:35326–35338. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sreeramaneni R, Chaudhry A, McMahon M,
Sherr CJ and Inoue K: Ras-Raf-Arf signaling critically depends on
the Dmp1 transcription factor. Mol Cell Biol. 25:220–232. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Inoue K and Sherr CJ: Gene expression and
cell cycle arrest mediated by transcription factor DMP1 is
antagonized by D-type cyclins through a
cyclin-dependent-kinase-independent mechanism. Mol Cell Biol.
18:1590–1600. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Macha MA, Seshacharyulu P, Krishn SR, Pai
P, Rachagani S, Jain M and Batra SK: MicroRNAs (miRNAs) as
biomarker(s) for prognosis and diagnosis of gastrointestinal (GI)
cancers. Curr Pharm Des. 20:5287–5297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schmitz KJ, Helwig J, Bertram S, Sheu SY,
Suttorp AC, Seggewiss J, Willscher E, Walz MK, Worm K and Schmid
KW: Differential expression of microRNA-675, microRNA-139-3p and
microRNA-335 in benign and malignant adrenocortical tumours. J Clin
Pathol. 64:529–535. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schultz NA, Andersen KK, Roslind A,
Willenbrock H, Wøjdemann M and Johansen JS: Prognostic microRNAs in
cancer tissue from patients operated for pancreatic cancer-five
microRNAs in a prognostic index. World J Surg. 36:2699–2707. 2012.
View Article : Google Scholar : PubMed/NCBI
|