Introduction
Hepatocellular carcinoma (HCC) is one of the leading
human malignancies prevalent worldwide, and half of the incident
cases each year occur in China (1,2). In
general, the majority of patients with HCC are diagnosed in the
clinic at a late stage of disease progression, and therefore do not
have the opportunity for a surgical resection. At present,
sorafenib and regorafenib are the only United States of America
Food and Drug Administration-approved molecularly targeted drugs
for patients with HCC (3).
However, the efficacy of HCC treatments remains limited and
unsatisfactory. Therefore, to improve the prognosis and quality of
life of patients with HCC, novel early diagnostic biomarkers and
therapeutic drug targets are urgently required.
SET domain-containing 1B (SETD1B), also known
as KMT2G or Set1B, is an important component of the histone
methyltransferase complex that generates trimethylated histone H3
at Lys4 and has been implicated in multiple biological processes
(4). The SETD1B gene is
located on chromosome 1q12 and encodes a 130-kDa protein with
several functional domains. A previous study demonstrated that
SETD1B associates with a 450-kDa complex that contains all
five noncatalytic components of the SET domain containing 1A
(SET1A) complex: CXXC finger protein 1, AT-rich interaction domain
4A, ASH2 like, histone lysine methyltransferase complex subunit, WD
repeat domain 5 and WD repeat domain 82 (5). The mixed lineage leukemia
(MLL) family of proteins, including MLL1-MLL4, SET1A
and SETD1B, specifically methylates Lys4 of histone H3 and
serves a vital role in the transcriptional regulation of genes
(6). In our previous study, it was
identified that SETD1B was the most frequently mutated gene
in primary hepatic neuroendocrine tumor, and that one of the three
SETD1B mutants, A1054del, promoted cell proliferation, migration
and invasion (7). However, the
underlying role of SETD1B in liver carcinogenesis was not
addressed. Therefore, an investigation into the expression patterns
of SETD1B and its clinical significance in the development and
progression of HCC was warranted.
Materials and methods
Clinical samples
Fresh surgical tumor samples from 76 patients with
HCC were collected from the Hepatobiliary Department of Beijing 302
Hospital (Beijing, China) between October 2013 and March 2018 and
were examined using reverse transcription quantitative polymerase
chain reaction (RT-qPCR) and western blot analysis. For the
immunohistochemical (IHC) analysis, paraffin-embedded HCC samples
were collected between October 2013 and March 2015. The HCC tissues
were fixed in 10% formalin for 24 h at room temperature, embedded
in paraffin and cut into 4-µm sections. Within half an hour of
isolation, HCC tissues and adjacent tissue samples were quickly
placed into liquid nitrogen. Adjacent tissue samples were taken at
a distance >3 cm from the cancer tissues. Written informed
consent was obtained from the enrolled patients with HCC. The
present study was approved by The Ethics Committee of Beijing 302
Hospital.
Cell lines and cell culture
The human liver cancer 97L and HCCLM3 cell lines and
the normal human liver LO2 cell line used in the present study were
obtained from the Experimental Center of Beijing 302 Hospital
(Beijing, China). All these cell lines were maintained and in
Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and incubated
at 37°C with 5% CO2.
RT-qPCR
Total RNA from the frozen tissue samples of 76
patients with HCC was extracted using TRIzol® reagent
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. SETD1B expression levels were quantified by RT-qPCR
methods conducted in an ABI 7500 instrument (Applied Biosystems;
Thermo Fisher Scientific, Inc.) using the Maxima SYBR-Green RT-qPCR
master mix (Thermo Fisher Scientific, Inc.) according to the
protocol of the manufacturer. All experimental samples were
normalized to a human GAPDH control. The sequences of the RT-PCR
primers were as follows: SETD1B forward, 5′-CTGGGTCTACCATCCCTCCA-3′
and reverse, 5′-CTTCCGGAACTTGAGCTGGT-3′; GAPDH forward,
5′-CAGCCTCAAGATCATCAGCA-3′ and reverse, 5′-TGTGGTCATGAGTCCTTCCA-3′.
The amplification procedure consisted of an initial denaturation at
95°C for 5 min, followed by 40 cycles of denaturation at 95°C for
15 sec, and annealing and extension at 60°C for 30 sec. The
2−ΔΔCq method was used to analyze SETD1B expression
levels relative to the GAPDH control (8).
Western blot analysis
The total proteins were extracted from surgical
samples from the patients with HCC using Tissue Protein Extraction
Reagent (Pierce; Thermo Fisher Scientific, Inc.) and quantified
using a Bicinchoninic Acid Protein Assay (Pierce; Thermo Fisher
Scientific, Inc.). A total of 30 µg protein was loaded per lane.
Proteins were separated by 12% SDS-PAGE and were subsequently
transferred onto polyvinylidene fluoride (PVDF) membranes (EMD
Millipore, Billerica, MA, USA) for western blot analysis. The PVDF
membranes were incubated with a primary monoclonal anti-SETD1B
antibody (cat. no. ab113984; Abcam, Cambridge, MA, USA; 1:500) at
2–8°C overnight and were subsequently incubated with a horseradish
peroxidase (HRP)-conjugated secondary antibody (cat. no. RABHRP1;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany; 1:1,000) for 1 h at
room temperature. Targeted SETD1B protein bands were visualized
using an enhanced chemiluminescence kit (Pierce; Thermo Fisher
Scientific, Inc.). The primary antibody for β-actin (cat. no. 3700;
Cell Signaling Technology, Inc., Danvers, MA, USA; 1:1,000) was
used to as a loading control for the western blot analysis.
IHC staining
IHC experiments were conducted as previously
described (9). The deparaffinized
sections were boiled for 2.5 min in citrate buffer, pH 6.0.
Endogenous peroxidase activity was blocked through incubation with
a 3% hydrogen peroxide solution for 20 min at room temperature.
Subsequently, the 4-µm sections were incubated for 24 h at 4°C with
a primary monoclonal anti-SETD1B antibody (Abcam; cat. no.
ab113984; 1:500) and subsequently with an HRP-labeled anti-rabbit
immunoglobulin G secondary antibody (cat. no. AP101P;
Sigma-Aldrich; Merck KGaA; 1:1,000) for 2 h at 37°C. The positive
cells were analyzed in five distinct fields and images were
captured under different magnifications using a light microscope
(Olympus Corporation, Tokyo, Japan; magnification, ×100 and
×200).
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical analyses were performed using the SPSS 20.0 software
(IBM Corp., Armonk, NY, USA). The Wilcoxon test was used to
determine SETD1B expression in HCC paired tissues. A paired t-test
was used to compare SETD1B expression in RNA-Seq studies.
RNA-Seq data of 66 paired HCC samples were retrieved
from The Cancer Genome Atlas (TCGA) Firehose pipeline on the Broad
Institute website (http://gdac.broadinstitute.org/). Kaplan-Meier curves
were conducted to analyze the associations between the SETD1B
expression levels and the overall survival (OS) or disease-free
survival (DFS) rates of the patients with HCC. Univariate and
multivariate cox regression analysis were performed with the Cox
proportional hazards regression model to determine the effects of
prognostic factors on survival. P<0.05 was considered to
indicate a statistically significant difference.
Results
SETD1B is upregulated in HCC samples
and cell lines
To investigate the role of SETD1B in the development
and progression of HCC, RT-qPCR was used to examine the SETD1B
expression levels in 76 pairs of HCC tissues and adjacent nontumor
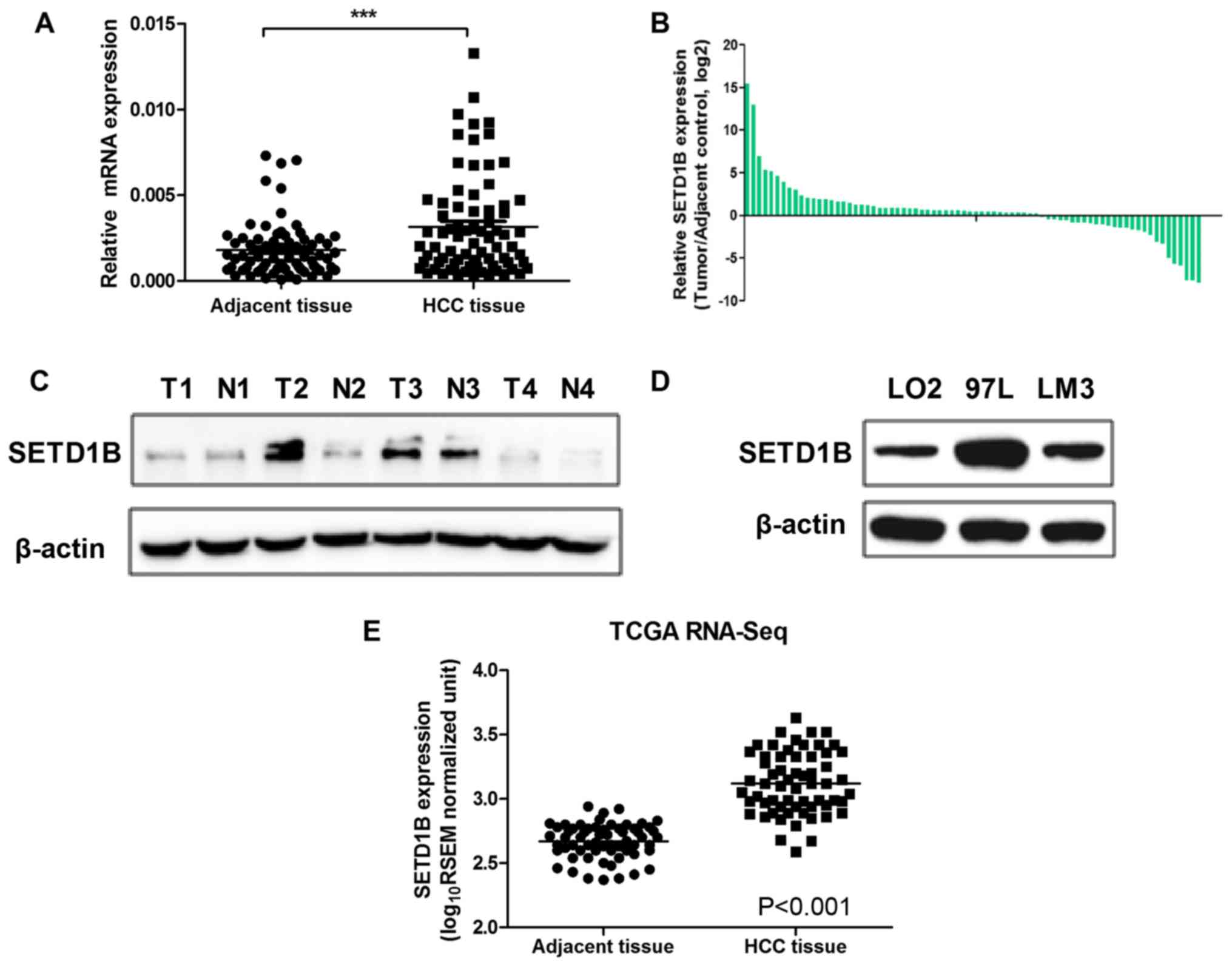
tissue samples. As observed in Fig.
1, SETD1B levels were significantly upregulated in the HCC
tissues compared with those in the adjacent normal tissues
(P<0.001; Fig. 1A-B). To
additionally confirm the overexpression of SETD1B in HCC tissues, 4
pairs of HCC and adjacent normal tissues were selected to evaluate
the protein levels of SETD1B by western blot analysis. As expected,
it was visually observed that the western blot analysis results
were consistent with the RT-qPCR data (Fig. 1C). The expression levels of SETD1B
in 2 human HCC cell lines (97L and HCCLM3) were also determined and
upregulated SETD1B expression levels were observed in the HCC cell
lines compared with that in LO2 cells (Fig. 1D). In addition, the SETD1B
expression levels of 66 paired HCC samples in The Cancer Genome
Atlas (TCGA) RNA-Seq data set were analyzed, and it was identified
that SETD1B levels were also significantly increased in these
samples (Fig. 1E). These results
prompted additional investigation into the potential role of SETD1B
in HCC carcinogenesis.
Upregulation of SETD1B in HCC tissues
determined by IHC
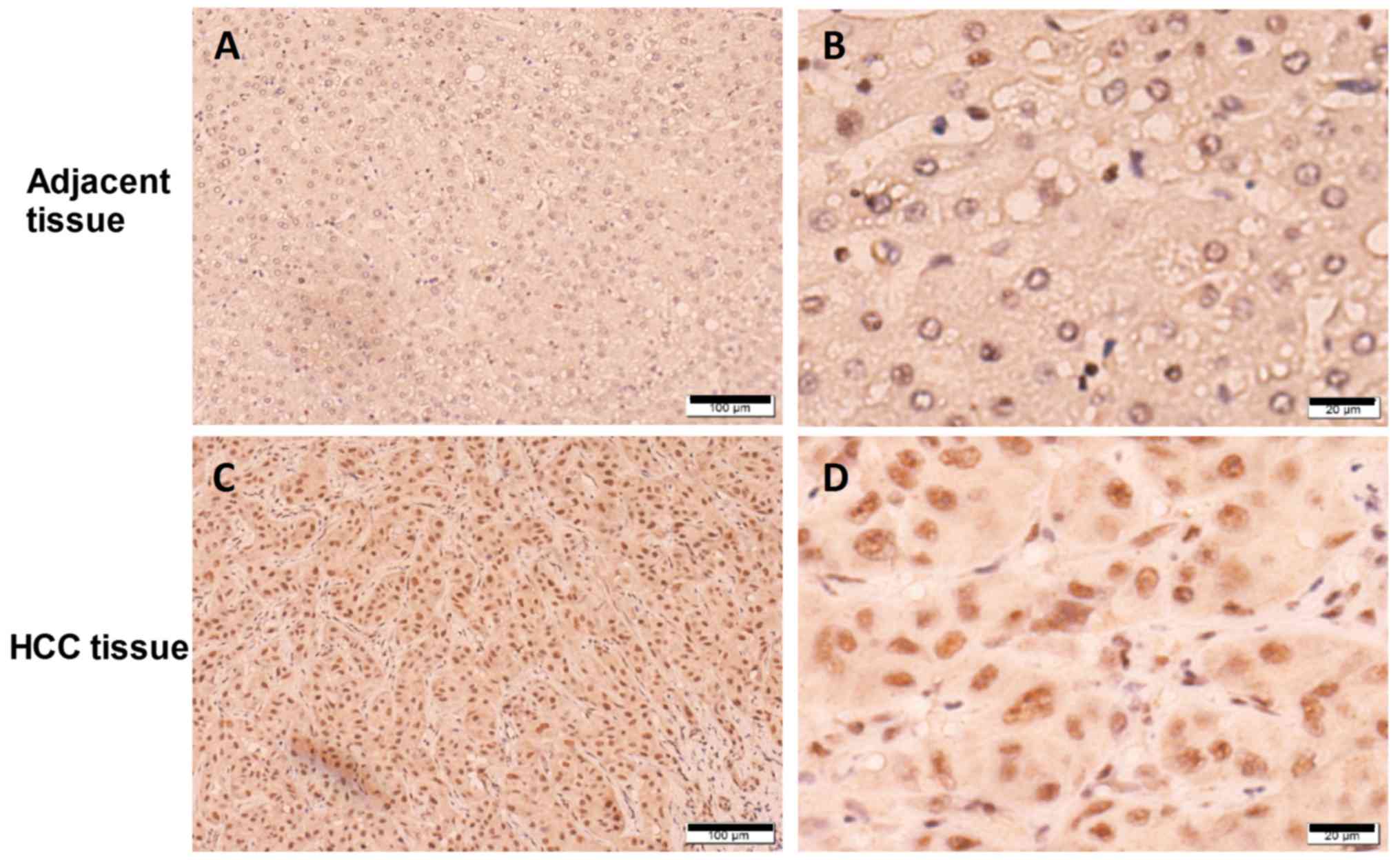
Next, to additionally clarify whether SETD1B was
upregulated at the protein level, IHC was performed to examine
SETD1B expression in sections of paired HCC tissue samples. It was
visually observed that SETD1B expression was primarily located in
the nucleus and was markedly increased in the HCC cells compared to
that in normal adjacent hepatocytes (Fig. 2). Taking together, it was concluded
that SETD1B was upregulated in human HCC tissues at the mRNA and
protein levels.
Association between SETD1B protein
expression and the clinicopathological features of HCC
To additionally explore whether SETD1B expression in
HCC tissues determined the clinical prognosis in patients with HCC,
76 patients with HCC were divided into high SETD1B and low SETD1B
expression groups according to the mean value of the expression
levels of SETD1B in HCC samples. As demonstrated in Table I, it was identified that high
SETD1B expression was closely associated with tumor size
(P<0.05), clinical tumor stage (P<0.01) and whether or not
liver cirrhosis was present (P<0.05). By contrast, no
association was observed between SETD1B expression and other
parameters, including sex, age, the α-fetoprotein level, smoking
status, drinking status, recurrence and portal vein tumor thrombus
(PVTT) (P>0.05).
 | Table I.Association between SETD1B expression
and clinicopathological features in hepatocellular carcinoma. |
Table I.
Association between SETD1B expression
and clinicopathological features in hepatocellular carcinoma.
|
|
| SETH1B
expression |
|
|---|
|
|
|
|
|
|---|
| Parameters | No. of patients | Low | High | P-value |
|---|
| Age, years |
|
<60 | 58 | 31 | 27 | 0.280 |
| ≥60 | 18 | 7 | 11 |
|
| Sex |
| Male | 68 | 33 | 35 | 0.455 |
|
Female | 8 | 5 | 3 |
|
| Tumor size, cm |
|
<5 | 48 | 28 | 20 | 0.028a |
| ≥5 | 28 | 9 | 19 |
|
| AFP |
|
<20 | 24 | 13 | 11 | 0.622 |
| ≥20 | 52 | 25 | 27 |
|
| Histological
grade |
|
Well/moderate | 70 | 34 | 36 | 0.395 |
| Poor | 6 | 4 | 2 |
|
| Clinical stage |
| I–II | 59 | 39 | 20 | 0.007b |
|
III–IV | 17 | 5 | 12 |
|
| Number of tumors |
|
Single | 63 | 30 | 33 | 0.361 |
|
Multiple | 13 | 8 | 5 |
|
| Alcohol
consumption |
| Yes | 32 | 18 | 14 | 0.353 |
| No | 44 | 20 | 24 |
|
| Smoking status |
| Yes | 42 | 20 | 22 | 0.645 |
| No | 34 | 18 | 16 |
|
| Recurrence |
|
Yes | 23 | 12 | 11 | 0.803 |
| No | 53 | 26 | 27 |
|
| Portal vein tumor
thrombus |
|
Yes | 42 | 21 | 21 | 0.896 |
| No | 34 | 17 | 17 |
|
| Microvascular
invasion |
|
Yes | 61 | 32 | 29 | 0.387 |
| No | 15 | 6 | 9 |
|
| Liver
cirrhosis |
|
Absent | 27 | 16 | 11 | 0.039a |
|
Present | 49 | 17 | 32 |
|
SETD1B expression and HCC patient
survival
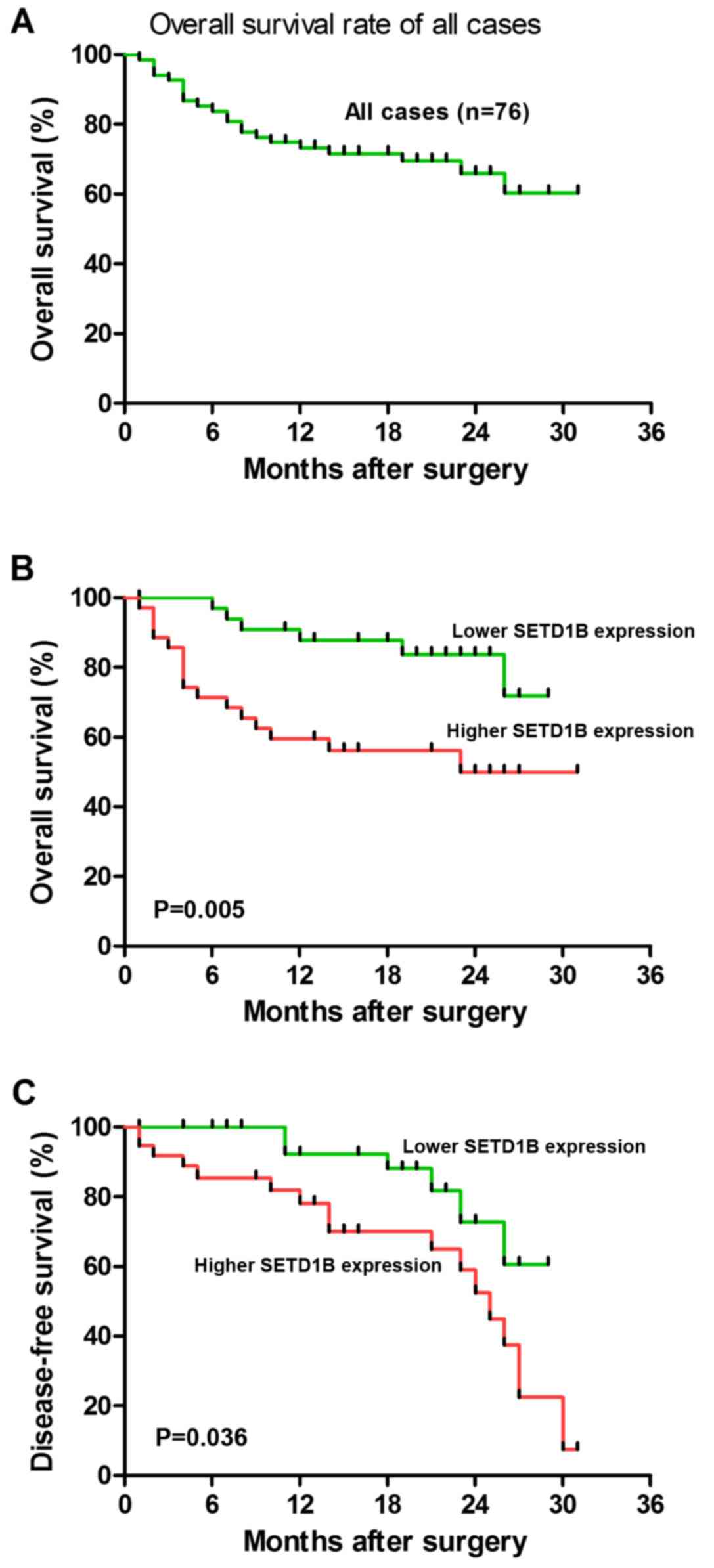
To additionally evaluate whether SETD1B expression
exhibited prognostic potential for the OS of patients with HCC, the
association between SETD1B expression and HCC patient survival
rates was analyzed using Kaplan-Meier analyses. As indicated in
Fig. 3, the 3-year OS rate of the
76 patients with HCC was 60%. In addition, the associations between
SETD1B expression and the survival outcomes of the patients with
HCC were investigated using on Kaplan-Meier analyses. The results
suggested that an increased SETD1B expression level in HCC tissues
was significantly associated with a decrease in OS (P=0.005;
Fig. 3B) and DFS (P=0.036;
Fig. 3C) during the 3-year
follow-up period. In addition, survival benefits were observed in
patients with a small tumor size (P=0.003), with an early clinical
stage (P=0.041), without PVTT (P=0.043) and without liver cirrhosis
(P=0.021). Multivariate Cox regression analysis revealed that
SETD1B expression [relative risk (RR)=4.151; P=0.016], tumor size
(RR=8.639; P=0.001) and clinical stage (RR=6.371; P=0.006) were
independent prognostic markers for OS in patients with HCC
(Table II), indicating that
SETD1B is essential for the development, progression and outcomes
of HCC.
 | Table II.Univariate and multivariate cox
regression analysis for overall survival in 76 patients with
HCC. |
Table II.
Univariate and multivariate cox
regression analysis for overall survival in 76 patients with
HCC.
|
| Univariate
regression analysis | Multivariate
regression analysis |
|---|
|
|
|
|
|---|
| Variable | RR | P-value | RR | P-value |
|---|
| Age |
|
<60 | 1.453 |
| 1 |
|
|
≥60 |
| 0.456 | 1.296 | 0.625 |
| Sex |
|
Male | 0.981 |
| 1 |
|
|
Female |
| 0.956 | 1.032 | 0.968 |
| Tumor size
(cm) | 1 |
|
|
|
| <5
cm | 6.138 |
| 1 |
|
| ≥5
cm |
| 0.003 | 8.639 | 0.001 |
| α-fetoprotein | 1 |
|
|
|
|
<20 | 0.738 |
| 1 |
|
|
≥20 |
| 0.536 | 0.683 | 0.428 |
| Histological
grade | 1 |
|
|
|
|
Well/moderate | 1.956 |
| 1 |
|
|
Poor |
| 0.396 | 4.175 | 0.096 |
| Clinical stage | 1 |
|
|
|
|
I–II | 5.42 |
|
|
|
|
III–IV |
| 0.041 | 6.371 | 0.006 |
| Number of
tumors | 1 |
|
|
|
|
Single | 8.624 |
| 1 |
|
|
Multiple |
| 0.361 | 6.751 | 0.132 |
| Alcohol
consumption | 1 |
|
|
|
|
Yes | 1.435 |
| 1 |
|
| No |
| 0.416 | 1.369 | 0.652 |
| Smoking status | 1 |
|
|
|
|
Yes | 1.466 |
| 1 |
|
| No |
| 0.465 | 1.096 | 0.725 |
| Recurrence | 1 |
|
|
|
|
Yes | 1.853 |
| 1 |
|
| No |
| 0.656 | 1.496 | 0.854 |
| Portal vein tumor
thrombus | 1 |
|
|
|
|
Yes | 1.493 |
|
|
|
| No |
| 0.043 |
|
|
| Microvascular
invasion | 1 |
|
|
|
|
Yes | 6.453 |
| 1 |
|
| No |
| 2.456 | 4.296 | 0.625 |
| Liver
cirrhosis | 1 |
|
|
|
|
Absent | 5.62 |
|
|
|
|
Present |
| 0.021 |
|
|
| SETD1B
expression | 1 |
|
|
|
|
Low | 5.123 |
| 1 |
|
|
High |
Discussion
To the best of our knowledge, the present study
demonstrated for the first time that SETD1B expression is
significantly increased in HCC tissues compared with that in
adjacent normal tissues. Specifically, the increased expression of
SETD1B in HCC was associated with tumor size, a more advanced
clinical stage and the development of liver cirrhosis. In addition,
it was identified that increased SETD1B expression was associated
with decreased OS rates. Furthermore, Cox regression analysis
demonstrated that SETD1B is an independent predictive marker for
the prognosis of HCC. This result was additionally validated with a
larger cohort of HCC samples from the TCGA database. These data
suggested that SETD1B may serve critical roles in HCC development
and progression, and monitoring SETD1B levels may have potential
clinical applications. However, insights into the mechanism of how
overexpression, not mutation, contributes to a poor outcome in HCC
require additional investigation.
SETD1B encodes a histone H3 Lysine 4
(H3K4)-methyltransferase and is a component of the SET1
complex (SET1C)/complex proteins associated with Set1
complex, which participates in a number of biological processes
(10–13). For example, a previous study
demonstrated that tumor cells use the SETD1B-Histone H3 lysine 4
trimethylation (H3K4me3) epigenetic axis to bypass the normal role
of interferon regulatory factor 8 expression in activating
inducible nitric oxide synthase (iNOS) expression in
myeloid-derived suppressor cells under pathological conditions
(14). Setd1b deficiency causes
female sterility in mice and serves as a maternal effect gene by
regulating the oocyte gene expression program (15). The MLL/Setd1b methyltransferase is
required for Spemann's organizer gene activation in Xenopus
(16). A frameshift mutation in
the histone methylation-associated gene SETD1B results in its
regional heterogeneity in gastric and colorectal cancer with high
microsatellite instability (17).
Song et al (18) identified
that SETD1B, as one of the important histone regulator genes, is
frequently altered in esophageal squamous cell carcinoma. The
molecular mechanisms for SETD1B function in carcinogenesis
and cancer progression have not been clearly elucidated. Although a
role of SETD1B as an oncogene in HCC has been suggested in the
present study, the underlying mechanisms of increased SETD1B
expression in HCC progression remain largely elusive. For example,
to verify that the significance of SETD1B in HCC is based on
epigenetic modification, subsequent studies with chromatin
immunoprecipitation PCR in HCC tissues are required to assess
whether the H3K3 modifications are enhanced around the critical
transcription factor genes that have been suggested to
significantly contribute to HCC pathogenesis. In addition, a
limited number of HCC tissue samples were analyzed in the present
study, and SETD1B expression levels require confirmation with
larger cohorts of HCC clinical samples. Whether SETD1B may also be
detected in the plasma or even in circulating exosomes and whether
the circulating SETD1B is also associated with HCC development are
also key issues to consider. Therefore, the role of SETD1B in HCC
progression requires investigation, and will be a focus in
subsequent studies.
SETD1B catalyzes the methylation of H3K4, and
several other enzymes including SETD1A, histone-lysine
N-methyltransferase SETD7, MLL1-4 and histone-lysine
N-methyltransferase SMYD (SMYD) 1–3 catalyze the same reaction
(11). The level of H3K4
methylation affects gene transcription, but this level depends not
only on the activity of these methyltransferases but also on that
of demethylases, or ‘erasers’ (19). In humans, the erasers for H3K4me
are lysine-specific histone demethylase (LSD)1-2, lysine-specific
demethylase 5A-D and ribosomal oxygenase 1. For example, a previous
study described the role of the SMYD3 histone methyltransferase in
tumorigenesis and whether the effects are local or global (19). JARID1B promotes metastasis and the
epithelial-mesenchymal transition via phosphatase and tensin
homolog/protein kinase B signaling in HCC cells (20). While it is true that the role of
SETD1B in HCC has not been described at present, those other
‘writers’ or ‘erasers’ also affect the level of the same epigenetic
marker, H3K4me3, that results from SETD1B action. It has been
suggested that a high level of JARID1B expression was associated
with decreased OS in patients with HCC (20). Considering that SETD1B and JARID1B
induce opposite effects on the level of H3K4me3, it was
hypothesized that the reason for these results is a small sample
size and individual differences in the patients with HCC.
In summary, the results of the present study reveal
that SETD1B expression is markedly upregulated in HCC tissues, and
is associated with a poor prognosis in patients with HCC. The
overexpression of SETD1B was associated with tumor size, clinical
stage and the presence of liver cirrhosis. These data demonstrate
that SETD1B has potential as a predictive marker for prognosis and
as a therapeutic drug target for HCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
National Natural Science Foundation of China (grant no.
81601860).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
DC, CW, GL, RW, ZW and LY performed the experiments.
DC, TL, PZ and JY gathered clinical samples and performed the
clinical analysis. XW, SZ and PY designed the study and wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Informed consent was obtained from the enrolled
patients. The present study was approved by the Ethics Committee of
Beijing 302 Hospital (Beijing, China).
Patient consent for publication
Written informed consent was obtained from the
enrolled patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yang Y, Zhou Y, Hou J, Bai C, Li Z, Fan J,
Ng IOL, Zhou W, Sun H, Dong Q, et al: Hepatic IFIT3 predicts
interferon-α therapeutic response in patients of hepatocellular
carcinoma. Hepatology. 66:152–166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Serag HB: Hepatocellular carcinoma. N
Engl J Med. 365:1118–1127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheng AL, Kang YK, Lin DY, Park JW, Kudo
M, Qin S, Chung HC, Song X, Xu J, Poggi G, et al: Sunitinib versus
sorafenib in advanced hepatocellular cancer: Results of a
randomized phase III trial. J Clin Oncol. 31:4067–4075. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee JH, Tate CM, You JS and Skalnik DG:
Identification and characterization of the human Set1B histone
H3-Lys4 methyltransferase complex. J Biol Chem. 282:13419–13428.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Duncan EM, Chitsazan AD, Seidel CW and
Alvarado AS: Set1 and MLL1/2 target distinct sets of functionally
different genomic loci in vivo. Cell Rep. 17:9302016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Han J, Zhang Y, Cao F, Liu Z, Li S,
Wu J, Hu C, Wang Y, Shuai J, et al: Structural basis for activity
regulation of MLL family methyltransferases. Nature. 530:447–452.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang BL, Ji X, Yu LX, Gao Y, Xiao CH, Liu
J, Zhao DX, Le Y, Diao GH, Sun JY, et al: Somatic mutation
profiling of liver and biliary cancer by targeted next generation
sequencing. Oncol Lett. 16:6003–6012. 2018.PubMed/NCBI
|
|
8
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li J, Wu H, Li W, Yin L, Guo S, Xu X,
Ouyang Y, Zhao Z, Liu S, Tian Y, et al: Downregulated miR-506
expression facilitates pancreatic cancer progression and
chemoresistance via SPHK1/Akt/NF-κB signaling. Oncogene.
35:5501–5514. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Davie JR, Xu W and Delcuve GP: Histone
H3K4 trimethylation: Dynamic interplay with pre-mRNA splicing.
Biochem Cell Biol. 94:1–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang W and Ernst P: Distinct functions of
histone H3, lysine 4 methyltransferases in normal and malignant
hematopoiesis. Curr Opin Hematol. 24:322–328. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hiraide T, Nakashima M, Yamoto K, Fukuda
T, Kato M, Ikeda H, Sugie Y, Aoto K, Kaname T, Nakabayashi K, et
al: De novo variants in SETD1B are associated with intellectual
disability, epilepsy and autism. Hum Genet. 137:95–104. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schmidt K, Zhang Q, Tasdogan A, Petzold A,
Dahl A, Arneth BM, Slany R, Fehling HJ, Kranz A, Stewart AF and
Anastassiadis K: The H3K4 methyltransferase Setd1b is essential for
hematopoietic stem and progenitor cell homeostasis in mice. Elife.
7:e271572018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Redd PS, Ibrahim ML, Klement JD, Sharman
SK, Paschall AV, Yang D, Nayak-Kapoor A and Liu K: SETD1B activates
iNOS expression in myeloid-derived suppressor cells. Cancer Res.
77:2834–2843. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Brici D, Zhang Q, Reinhardt S, Dahl A,
Hartmann H, Schmidt K, Goveas N, Huang J, Gahurova L, Kelsey G, et
al: Setd1b, encoding a histone 3 lysine 4 methyltransferase, is a
maternal effect gene required for the oogenic gene expression
program. Development. 144:2606–2617. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin H, Min Z and Tao Q: The MLL/Setd1b
methyltransferase is required for the Spemann's organizer gene
activation in Xenopus. Mech Dev. 142:1–9. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Choi YJ, Oh HR, Choi MR, Gwak M, An CH,
Chung YJ, Yoo NJ and Lee SH: Frameshift mutation of a histone
methylation-related gene SETD1B and its regional heterogeneity in
gastric and colorectal cancers with high microsatellite
instability. Hum Pathol. 45:1674–1681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Song Y, Li L, Ou Y, Gao Z, Li E, Li X,
Zhang W, Wang J, Xu L, Zhou Y, et al: Identification of genomic
alterations in oesophageal squamous cell cancer. Nature. 509:91–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Medjkane S, Cock-Rada A and Weitzman JB:
Role of the SMYD3 histone methyltransferase in tumorigenesis: Local
or global effects? Cell Cycle. 11:18652012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang B, Qi G, Tang F, Yuan S, Wang Z,
Liang X, Li B, Yu S, Liu J, Huang Q, et al: JARID1B promotes
metastasis and epithelial-mesenchymal transition via PTEN/AKT
signaling in hepatocellular carcinoma cells. Oncotarget.
6:12723–12739. 2015. View Article : Google Scholar : PubMed/NCBI
|