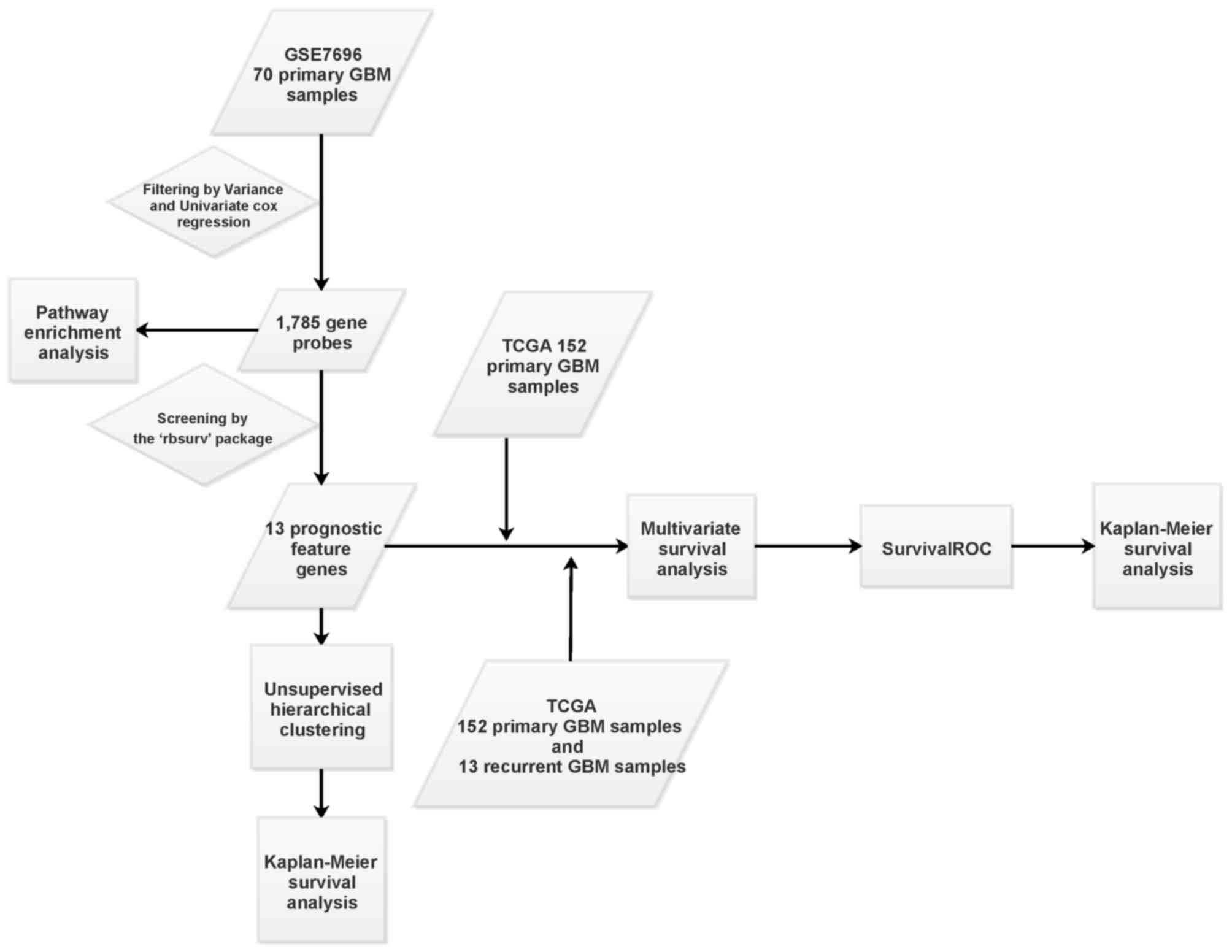
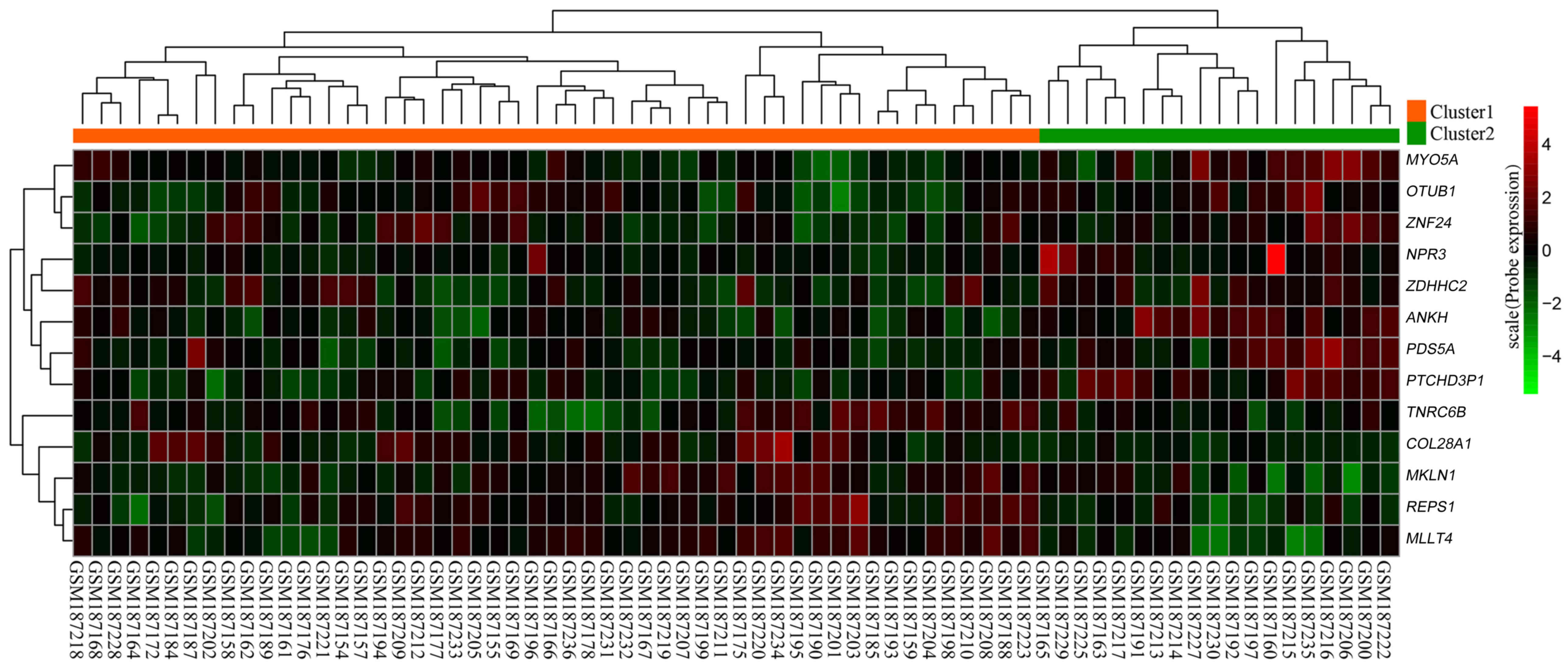
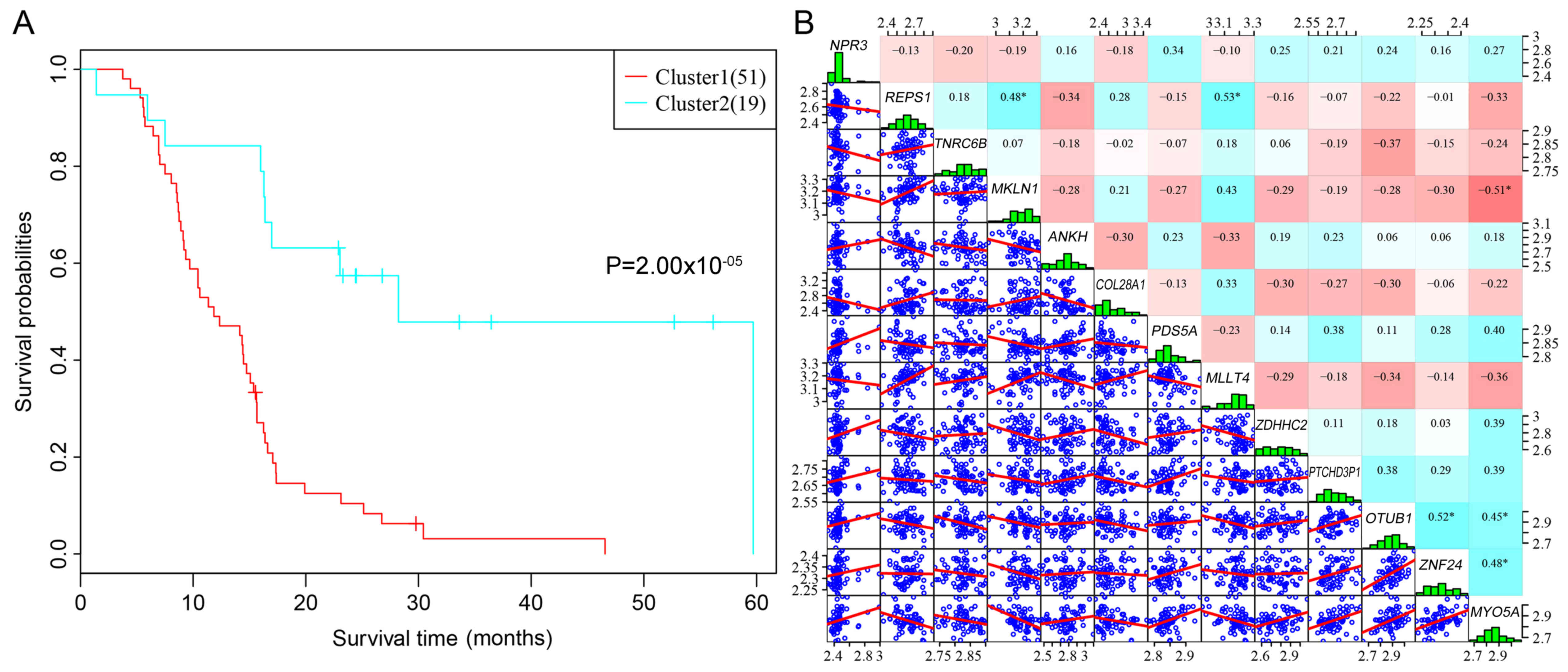
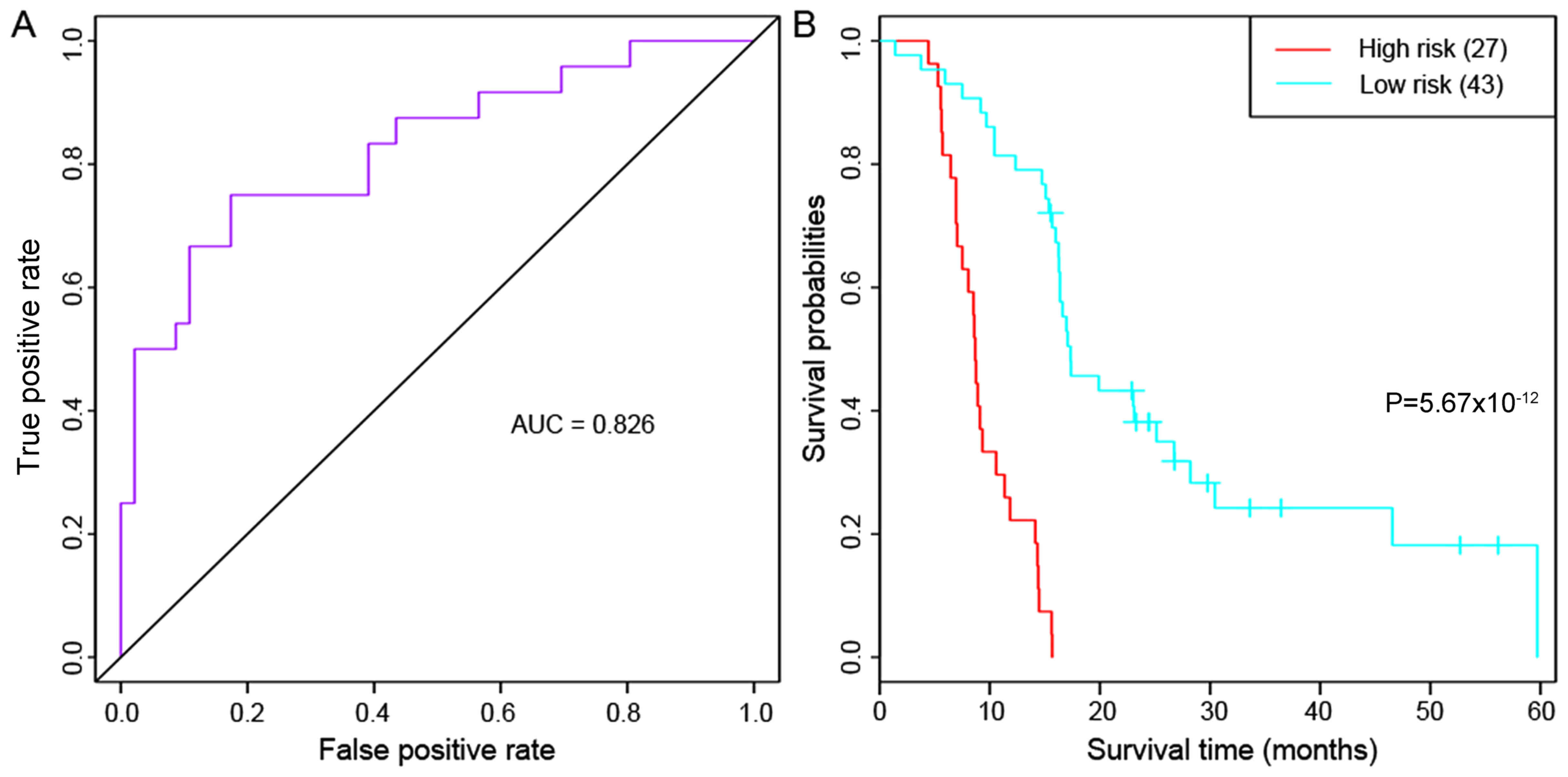
|
1
|
Bleeker FE, Molenaar RJ and Leenstra S:
Recent advances in the molecular understanding of glioblastoma. J
Neurooncol. 108:11–27. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Young RM, Jamshidi A, Davis G and Sherman
JH: Current trends in the surgical management and treatment of
adult glioblastoma. Ann Transl Med. 3:1212015.PubMed/NCBI
|
|
3
|
McGuire S: World Cancer Report 2014.
Geneva, Switzerland: World Health Organization, International
Agency for Research on Cancer, WHO Press, 2015. Adv Nutr.
7:418–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gallego O: Nonsurgical treatment of
recurrent glioblastoma. Curr Oncol. 22:e273–e281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fulci G and Chiocca EA: The status of gene
therapy for brain tumors. Expert Opin Biol Ther. 7:197–208. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alptekin M, Eroglu S, Tutar E, Sencan S,
Geyik MA, Ulasli M, Demiryurek AT and Camci C: Gene expressions of
TRP channels in glioblastoma multiforme and relation with survival.
Tumor Biology. 36:9209–9213. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen J, Luan Y, Yu R, Zhang Z, Zhang J and
Wang W: Transient receptor potential (TRP) channels, promising
potential diagnostic and therapeutic tools for cancer. Biosci
Trends. 8:1–10. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hong Y, Shang C, Xue YX and Liu YH:
Silencing of Bmi-1 gene enhances chemotherapy sensitivity in human
glioblastoma cells. Med Sci Monit. 21:1002–1007. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ye L, Wang C, Yu G, Jiang Y, Sun D, Zhang
Z, Yu X, Li X, Wei W, Liu P, et al: Bmi-1 induces radioresistance
by suppressing senescence in human U87 glioma cells. Oncol Lett.
8:2601–2606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hiddingh L, Tannous BA, Teng J, Tops B,
Jeuken J, Hulleman E, Boots-Sprenger SH, Vandertop WP, Noske DP,
Kaspers GJ, et al: EFEMP1 induces γ-secretase/Notch-mediated
temozolomide resistance in glioblastoma. Oncotarget. 5:363–374.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang J, Chen L, Han L, Shi Z, Zhang J, Pu
P and Kang C: EZH2 is a negative prognostic factor and exhibits
pro-oncogenic activity in glioblastoma. Cancer Lett. 356:929–936.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou X, Ren Y, Zhang J, Zhang C, Zhang K,
Han L, Kong L, Wei J, Chen L, Yang J, et al: HOTAIR is a
therapeutic target in glioblastoma. Oncotarget. 6:8353–8365.
2015.PubMed/NCBI
|
|
13
|
Zhang K, Sun X, Zhou X, Han L, Chen L, Shi
Z, Zhang A, Ye M, Wang Q, Liu C, et al: Long non-coding RNA HOTAIR
promotes glioblastoma cell cycle progression in an EZH2 dependent
manner. Oncotarget. 6:537–546. 2015.PubMed/NCBI
|
|
14
|
Insin P and Prueksaritanond N: Evaluation
of four risk of malignancy indices (RMI) in the preoperative
diagnosis of ovarian malignancy at Rajavithi hospital. Thai J
Obstet Gynaecol. 21:2013.
|
|
15
|
Sim J, Nam DH, Kim Y, Lee IH, Choi JW, Sa
JK and Suh YL: Comparison of 1p and 19q status of glioblastoma by
whole exome sequencing, array-comparative genomic hybridization,
and fluorescence in situ hybridization. Med Oncol. 35:602018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yao Q, Cai G, Yu Q, Shen J, Gu Z, Chen J,
Shi W and Shi J: IDH1 mutation diminishes aggressive phenotype in
glioma stem cells. Int J Oncol. 52:270–278. 2017.PubMed/NCBI
|
|
17
|
Han J and Puri RK: Analysis of the cancer
genome atlas (TCGA) database identifies an inverse relationship
between interleukin-13 receptor α1 and α2 gene expression and poor
prognosis and drug resistance in subjects with glioblastoma
multiforme. J Neurooncol. 136:463–474. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guadagno E, Vitiello M, Francesca P, Calì
G, Caponnetto F, Cesselli D, Camorani S, Borrelli G, Califano M,
Cappabianca P, et al: PATZ1 is a new prognostic marker of
glioblastoma associated with the stem-like phenotype and enriched
in the proneural subtype. Oncotarget. 8:59282–59300. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Murat A, Migliavacca E, Gorlia T, Lambiv
WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven
MC, et al: Stem cell-related ‘self-renewal’ signature and high
epidermal growth factor receptor expression associated with
resistance to concomitant chemoradiotherapy in glioblastoma. J Clin
Oncol. 26:3015–3024. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lambiv WL, Vassallo I, Delorenzi M, Shay
T, Diserens AC, Misra A, Feuerstein B, Murat A, Migliavacca E,
Hamou MF, et al: The Wnt inhibitory factor 1 (WIF1) is targeted in
glioblastoma and has a tumor suppressing function potentially by
induction of senescence. Neuro Oncol. 13:736–747. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stupp R, Dietrich PY, Ostermann Kraljevic
S, Pica A, Maillard I, Maeder P, Meuli R, Janzer R, Pizzolato G,
Miralbell R, et al: Promising survival for patients with newly
diagnosed glioblastoma multiforme treated with concomitant
radiation plus temozolomide followed by adjuvant temozolomide. J
Clin Oncol. 20:1375–1382. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Therneau TM: Survival analysis (R package
survival version 2.39–5). 2015.
|
|
24
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44:D457–D62. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Boil. 8:R1832007. View Article : Google Scholar
|
|
26
|
Cho HJ, Kim S, Kang J and Lee JW: How to
use the rbsurv Package. 2010.
|
|
27
|
Renaud G, Stenzel U, Maricic T, Wiebe V
and Kelso J: deML: Robust demultiplexing of Illumina sequences
using a likelihood-based approach. Bioinformatics. 31:770–772.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang SJ: Unsupervised hierarchical
clustering based on sequential partitioning and merging.
International Symposium on Vlsi Design, Automation and Test.
2016.
|
|
29
|
Porcher R: CORR Insights(®):
Kaplan-meier survival analysis overestimates the risk of revision
arthroplasty: A meta-analysis. Clin Orthop Relat Res.
473:3431–3442. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Smith B, Williams J and Schulze-Kremer S:
The ontology of the gene ontology. AMIA Annu Symp Proc. 609–613.
2003.PubMed/NCBI
|
|
31
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Heagerty PJ, Thomas L and Pepe MS:
Time-dependent ROC curves for censored survival data and a
diagnostic marker. Biometrics. 56:337–344. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bauer R, Ratzinger S, Wales L, Bosserhoff
A, Senner V, Grifka J and Grässel S: Inhibition of collagen XVI
expression reduces glioma cell invasiveness. Cell Physiol Biochem.
27:217–226. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huijbers IJ, Iravani M, Popov S, Robertson
D, Alsarraj S, Jones C and Isacke CM: A role for fibrillar collagen
deposition and the collagen internalization receptor endo180 in
glioma invasion. PLoS One. 5:e98082010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Honma K, Miyata T and Ochiya T: Type I
collagen gene suppresses tumor growth and invasion of malignant
human glioma cells. Cancer Cell Int. 7:122007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Veit G, Kobbe B, Keene DR, Paulsson M,
Koch M and Wagener R: Collagen XXVIII, a novel von Willebrand
factor A domain-containing protein with many imperfections in the
collagenous domain. J Biol Chem. 281:3494–3504. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hagemann C, Weigelin B, Schommer S,
Schulze M, Al-Jomah N, Anacker J, Gerngras S, Kühnel S, Kessler AF,
Polat B, et al: The cohesin-interacting protein, precocious
dissociation of sisters 5A/sister chromatid cohesion protein 112,
is up-regulated in human astrocytic tumors. Int J Mol Med.
27:39–51. 2011.PubMed/NCBI
|
|
38
|
Oyama T, Miyoshi Y, Koyama K, Nakagawa H,
Yamori T, Ito T, Matsuda H, Arakawa H and Nakamura Y: Isolation of
a novel gene on 8p21.3-22 whose expression is reduced significantly
in human colorectal cancers with liver metastasis. Genes
Chromosomes Cancer. 29:9–15. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jen J and Wang YC: Zinc finger proteins in
cancer progression. J Biomed Sci. 23:532016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yan SM, Tang JJ, Huang CY, Xi SY, Huang
MY, Liang JZ, Jiang YX, Li YH, Zhou ZW, Ernberg I, et al: Reduced
expression of ZDHHC2 is associated with lymph node metastasis and
poor prognosis in gastric adenocarcinoma. PLoS One. 8:e563662013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Khalfallah O, Ravassard P, Che SL, Fligny
C, Serre A, Bayard E, Faucon-Biguet N, Mallet J, Meloni R and
Nardelli J: Zinc finger protein 191 (ZNF191/Zfp191) is necessary to
maintain neural cells as cycling progenitor. Stem Cells.
27:1643–1653. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Carrasco-Garcia E, Martinez-Lacaci I,
Mayor-López L, Tristante E, Carballo-Santana M, García-Morales P,
Ventero Martin MP, Fuentes-Baile M, Rodriguez-Lescure Á and Saceda
M: PDGFR and IGF-1R inhibitors induce a G2/M arrest and subsequent
cell death in human glioblastoma cell lines. Cells. 7:E1312018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li JZ, Chen X, Liu Y, Ding L, Qiu L, Hu ZL
and Zhang J: The transcriptional repression of platelet-derived
growth factor receptor-beta by the zinc finger transcription factor
ZNF24. Biochem Biophys Res Commun. 397:318–322. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Beadle C, Assanah MC, Monzo P, Vallee R,
Rosenfeld SS and Canoll P: The role of myosin II in glioma invasion
of the brain. Mol Biol Cell. 19:3357–3368. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ivkovic S, Beadle C, Noticewala S, Massey
SC, Swanson KR, Toro LN, Bresnick AR, Canoll P and Rosenfeld SS:
Direct inhibition of myosin II effectively blocks glioma invasion
in the presence of multiple motogens. Mol Biol Cell. 23:533–542.
2011. View Article : Google Scholar
|
|
46
|
Edimo WE, Ramos AR, Ghosh S, Vanderwinden
JM and Erneux C: The SHIP2 interactor Myo1c is required for cell
migration in 1321 N1 glioblastoma cells. Biochem Biophys Res
Commun. 476:508–514. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu R, Fang XH and Zhong P: Myosin VI
contributes to malignant proliferation of human glioma cells.
Korean J Physiol Pharmacol. 20:139–145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wong SY, Ulrich TA, Deleyrolle LP, MacKay
JL, Lin JM, Martuscello RT, Jundi MA, Reynolds BA and Kumar S:
Constitutive activation of myosin-dependent contractility
sensitizes glioma tumor-initiating cells to mechanical inputs and
reduces tissue invasion. Cancer Res. 75:1113–1122. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Brennan CW, Verhaak RG, McKenna A, Campos
B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ,
Berman SH, et al: The somatic genomic landscape of glioblastoma.
Cell. 155:462–477. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Warrington NM, Sun T, Luo J, Mckinstry RC,
Parkin PC, Ganzhorn S, Spoljaric D, Albers AC, Merkelson A, Stewart
DR, et al: The cyclic AMP pathway is a sex-specific modifier of
glioma risk in type I neurofibromatosis patients. Cancer Res.
75:16–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kiuru M and Busam KJ: The NF1 gene in
tumor syndromes and melanoma. Lab Invest. 97:146–157. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
de Vrij J, Maas SL, Kwappenberg KM,
Schnoor R, Kleijn A, Dekker L, Luider TM, de Witte LD, Litjens M,
van Strien ME, et al: Glioblastoma-derived extracellular vesicles
modify the phenotype of monocytic cells. Int J Cancer.
137:1630–1642. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Leech AO, Cruz RG, Hill AD and Hopkins AM:
Paradigms lost-an emerging role for over-expression of tight
junction adhesion proteins in cancer pathogenesis. Ann Transl Med.
3:1842015.PubMed/NCBI
|
|
54
|
Gu L, Lu LS, Zhou DL and Liu ZC: Long
noncoding RNA BCYRN1 promotes the proliferation of colorectal
cancer cells via Up-regulating NPR3 expression. Cell Physiol
Biochem. 48:2337–2349. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li JK, Chen C, Liu JY, Shi JZ, Liu SP, Liu
B, Wu DS, Fang ZY, Bao Y, Jiang MM, et al: Long noncoding RNA
MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via
inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer.
16:1112017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Coe BP, Henderson LJ, Garnis C, Tsao MS,
Gazdar AF, Minna J, Lam S, Macaulay C and Lam WL: High-resolution
chromosome arm 5p array CGH analysis of small cell lung carcinoma
cell lines. Genes Chromosomes Cancer. 42:308–313. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kloth JN, Oosting J, van Wezel T, Szuhai
K, Knijnenburg J, Gorter A, Kenter GG, Fleuren GJ and Jordanova ES:
Combined array-comparative genomic hybridization and
single-nucleotide polymorphism-loss of heterozygosity analysis
reveals complex genetic alterations in cervical cancer. BMC
Genomics. 8:532007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xu L, Li J, Bao Z, Xu P, Chang H, Wu J,
Bei Y, Xia L, Wu P, Yan K, et al: Silencing of OTUB1 inhibits
migration of human glioma cells in vitro. Neuropathology.
37:217–226. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Dubinski D, Won SY, Gessler F,
Quick-Weller J, Behmanesh B, Bernatz S, Forster MT, Franz K, Plate
KH, Seifert V, et al: Dexamethasone-induced leukocytosis is
associated with poor survival in newly diagnosed glioblastoma. J
Neurooncol. 137:503–510. 2018. View Article : Google Scholar : PubMed/NCBI
|