Introduction
MicroRNAs (miRNAs) are endogenous, evolutionarily
highly conserved, small noncoding RNAs, which are derived from the
genomes of eukaryotic organisms and various viruses and have
multiple functions in the regulation of gene expression in animals
and plants (1,2). miRNA (miR)-210, a prototypical
hypoxamir, is one of the most widely studied miRNAs (3,4).
Mounting evidence has demonstrated that miR-210 is involved in the
sophisticated regulation of various biological processes, and is
associated with the development and progression of different
diseases, including cardiovascular, cerebrovascular and
immunological diseases, in addition to numerous types of cancer.
For example, Mutharasan et al (5) reported that miR-210 was markedly
upregulated through Akt serine/threonine kinase 1- and tumor
protein p53-dependent pathways to protect cardiomyocytes from
hypoxia, leading to apoptosis in vitro. Zeng et al
(6) reported that miR-210 was
significantly reduced in blood samples from stroke patients at
different times compared with healthy controls. miR-210 promotes
cell cycle progression by targeting MAX network transcriptional
repressor and E2F transcription factor 3, disrupts normal DNA
repair and increases genetic instability through a
homology-dependent repair DNA repair pathway (7). miR-210 expression is decreased in
patients with systemic lupus erythematosus and rheumatoid arthritis
compared with healthy controls (8). These above data suggest that miR-210
serves an indispensable role in the pathogenesis of numerous
diseases.
Inflammation is a well-known cause of cancer in
humans. Being one of the most common inflammatory diseases in the
world, hepatitis B virus (HBV) infection is the leading cause of
hepatocellular carcinoma (HCC), particularly in Asian countries
(9–11). The etiological association between
HBV and HCC represents an opportunity for clinicians to prevent HCC
development and alleviate the outcomes of HCC by treating HBV
infection. However, current immunoprophylaxis strategies against
HBV do not completely abolish HBV transmission. The molecular
mechanisms by which HBV infection leads to HCC are comparatively
complex. Therefore, elucidating the mechanism of HBV infection is
of particular importance.
A previous study demonstrated that macrophages serve
a pivotal role in immune hepatitis (12). In addition, it was reported that
miR-210 downregulates lipopolysaccharide (LPS)-stimulated
proinflammatory cytokine production by targeting nuclear factor-κB1
in macrophages (13). Moreover,
hepatitis B e-antigen HBeAg induces miR-155 expression to increase
liver injury by increasing inflammatory cytokine production in
macrophages (10). However,
whether miR-210, which is similar to miR-155, may also affect
HBV-induced hepatitis by altering macrophage function has not been
completely elucidated.
In the present study, it was identified that miR-210
expression levels were significantly reduced in peripheral blood
monocytes (PBMs) and the serum of patients with CHB, although there
was no marked correlation between miR-210 expression and
HBV-associated antigens and DNA in the serum. Moreover, it was
demonstrated that miR-210 expression was not affected by
HBV-associated antigens in different types of macrophages in
vitro. However, it is notable that the serum of patients with
CHB markedly downregulated miR-210 expression in PBMs from healthy
controls.
Patients and methods
Subjects and serological tests
The present study recruited 25 subjects between
April 2016 and March 2017 from Shandong Provincial Hospital
Affiliated to Shandong University (Jinan, China), including 11
healthy controls and 14 patients with chronic hepatitis B (CHB).
These enrolled subjects were all negative for antibodies to
hepatitis A, C, D, E virus and human immunodeficiency virus. None
of them were all autoimmune liver diseases and taking
immunosuppressive drugs or anti-viral therapy more than 3 months.
The standard set by the National Viral Hepatitis Conference of
China in 2015 was used for the diagnosis of CHB (14,15).
The concentrations of alanine aminotransferase (ALT) and aspartate
aminotransferase (AST) in the serum were analyzed with automatic
electrochemical luminescence immunoanalyzer (Roche Cobas E602;
Roche Applied Science, Pleasanton, CA, USA). The hepatitis B
surface antigen (HBsAg), HBeAg, anti-HBe antibody (HBeAb) and
hepatitis B core antibody (HBcAb) content in serum was determined
with a chemiluminescence microparticle immunoassay kit (Abbott
Pharmaceutical Co., Ltd., Lake Bluff, IL, USA) using the Abbott
i2000 ARCHITECT system. The HBV DNA content was detected with a
fluorescent label HBV DNA quantitative kit (PerkinElmer, Inc.,
Waltham, MA, USA) using an ABI-7500 quantitative polymerase chain
reaction (qPCR) analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The clinical characteristics
of these enrolled subjects are summarized in Table I. The research protocol and consent
program were approved by Shandong Provincial Hospital Affiliated to
Shandong University Ethics Committee. Written informed consent was
acquired from each patient for this study.
 | Table I.Clinical characteristics of
subjects. |
Table I.
Clinical characteristics of
subjects.
| Category | Healthy
controls | CHB |
|---|
| No. cases | 11 | 14 |
| Sex, male | 8 (72.73%) | 12 (85.71%) |
| BMIa | 23.72±1.84 | 24.70±1.89 |
| Age,
yearsa | 42±12.87 | 50.5±13.94 |
| ALT,
U/la | 23±9.54 | 308.5±341.16 |
| AST,
U/la | 21±7.14 | 222±254.71 |
| HBV DNA positive,
>103 copies/ml | 0 | 7 (50%) |
| HBsAg positive | 0 | 14 (100%) |
| HBeAg positive | 0 | 8 |
| HBeAb positive | 0 | 7 |
| HBcAb positive | 0 | 4 |
Cells and reagents
Mouse primary peritoneal macrophages (PMs), the
mouse macrophage cell line RAW264.7 (American Type Culture
Collection, Manassas, VA, USA), and human hepatoma cell lines HepG2
and HepG2.2.15 (Chinese Academy of Sciences, Beijing, China) were
acquired and cultured as previously described (16–18).
HBV-associated antigens, including hepatitis B core antigen (HBcAg;
cat. no. ab119441), HBeAg (cat. no. ab91273) and HBsAg (cat. no.
ab167754), were purchased from Abcam (Cambridge, MA, USA) and used
as was previously described (16).
Light microscope analysis
RAW264.7 cells were treated with or without 2 µg/ml
HBcAg, HBeAg, HBsAg or a mixture for 24 h. The cell morphology was
observed with a light microscope (Ti-S; Nikon Corporation, Tokyo,
Japan), and the original magnification was ×100.
Acquisition of mouse primary PMs
Male C57BL/6J mice (n=6), 6–8 weeks old, were
obtained from Shandong University Experimental Animal Center
(Jinan, China). All mice were housed in specific pathogen-free
conditions. The animal room was kept at 20–22°C under a relative
humidity 40–60% and 12 h light/dark cycle. These mice were feed
ad libitum and had access to water. All animal experiments
were undertaken in accordance with the National Institutes of
Health Guide for the Care and Use of Laboratory Animals, with the
approval of the Scientific Investigation Board of the Shandong
Provincial Hospital Affiliated to Shandong University. Mouse
primary PMs were obtained as described previously (16).
miR-210 quantification in cells or
serum
Total RNA in cells and serum was extracted with the
miRNeasy Mini kit, miRNeasy Serum/Plasma kit and miRNeasy
Serum/Plasma Spike-In Control, according to the manufacturer's
protocols, as described previously (16). For the quantification of miR-210,
cDNA was obtained with the miScript II reverse transcription kit.
miR-210 (cat. no. MS00003801) quantification was evaluated with the
miScript SYBR Green PCR kit, according to the manufacturer's
protocol as previously described (17). U6 snRNA (cat. no. MS00033740) and
miR-39 (cat. no. 219610) were used as an internal control. All of
the above kits were purchased from Qiagen, Inc. (Valencia, CA,
USA). qPCR was performed via the following steps: 95°C for 15 min,
followed by 45 cycles of 95°C for 15 sec, 55°C for 30 sec, 70°C for
30 sec and 65°C for 30 sec, using the LightCycler Real-time PCR
System (Roche Diagnostics, Indianapolis, IN, USA), as described
previously (18).
Co-culture and HBV-associated antigen
treatment experiments in vitro
A total of ~4×105/ml macrophages were
seeded and adhered to the lower chamber of a Transwell plate using
Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc.) with 10% fetal bovine serum (Gibco®
Sera; Thermo Fisher Scientific, Inc.). Subsequently,
3×105/ml HepG2/HepG2.2.15 cells were added into the
upper chambers in the same medium. Co-incubation was performed for
12, 24, 36 and 48 h at 37°C in a humidified incubator with 5%
CO2. Mouse primary PMs, mouse macrophage RAW264.7 cells,
and human PBMs were stimulated with individual HBV-associated
antigens (2 µg/ml) or a mixture for 24, 36 or 48 h. These cells
were harvested using TRIzol® (Thermo Fisher Scientific,
Inc.) for the evaluation of miR-210 expression levels by qPCR.
Cross culture of cell and serum
The serum was collected from 15 ml whole blood of
healthy controls and patients with CHB by centrifuging at 1,000 × g
for 5–10 min at 20–25°C. PBMs from healthy controls were obtained
by density gradient centrifugation (780 × g for 20–30 min at
20–25°C) and seeded in a 12-well plate with complete medium of
RPMI-1640 (Gibco®; Thermo Fisher Scientific, Inc.) with
10% FBS overnight for attachment. The following day, the suspended
cells were removed and the adherent cells were incubated with their
own serum or serum from patients with CHB for 36 h. These PBMs were
harvested with TRIzol to extract the total RNA. A reverse
transcription experiment was performed, and the expression of
miR-210 was detected with the corresponding kits, as described
above.
Statistical analysis
Data are presented as the mean ± standard deviation.
All statistical analyses were performed using GraphPad Prism 5
software (GraphPad Software, Inc., La Jolla, CA, USA). Student's
t-tests were used to verify significant differences between two
groups. One-way analysis of variance followed by the least
significant difference post hoc test was used to assess significant
differences between the different treatments. Spearman correlation
analysis was performed between miR-210 expression and serum ALT,
AST, HBsAg, HBeAg, HBeAb, HBcAb or HBV DNA levels. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-210 expression is decreased, and
is negatively correlated with serum ALT and AST
To evaluate the effect of miR-210 on HBV infection,
PBMs and serum were collected from patients with CHB or healthy
controls and miR-210 expression was assessed. As presented in
Figs. 1 and 2, the expression of miR-210 was
significantly decreased in PBMs (Fig.
1A) and serum (Fig. 2A). The
associations between the expression of miR-210 and multiple
clinical parameters were also assessed. miR-210 expression was
negatively correlated with serum ALT and AST; the correlation
coefficients were −0.5868 and −0.7231, and −0.7319 and −0.5692,
respectively (Figs. 1B, C;
2B and C). There was no marked
correlation between miR-210 expression and HBsAg, HBeAg, HBeAb,
HBcAb and HBV-DNA in the serum (Figs.
1D-H; 2D-H). Taken together,
miR-210 expression was reduced in the PBMs and serum of patients
with CHB and was inversely correlated with serum ALT and AST, but
there was no correlation with other HBV infection-associated
clinical indexes.
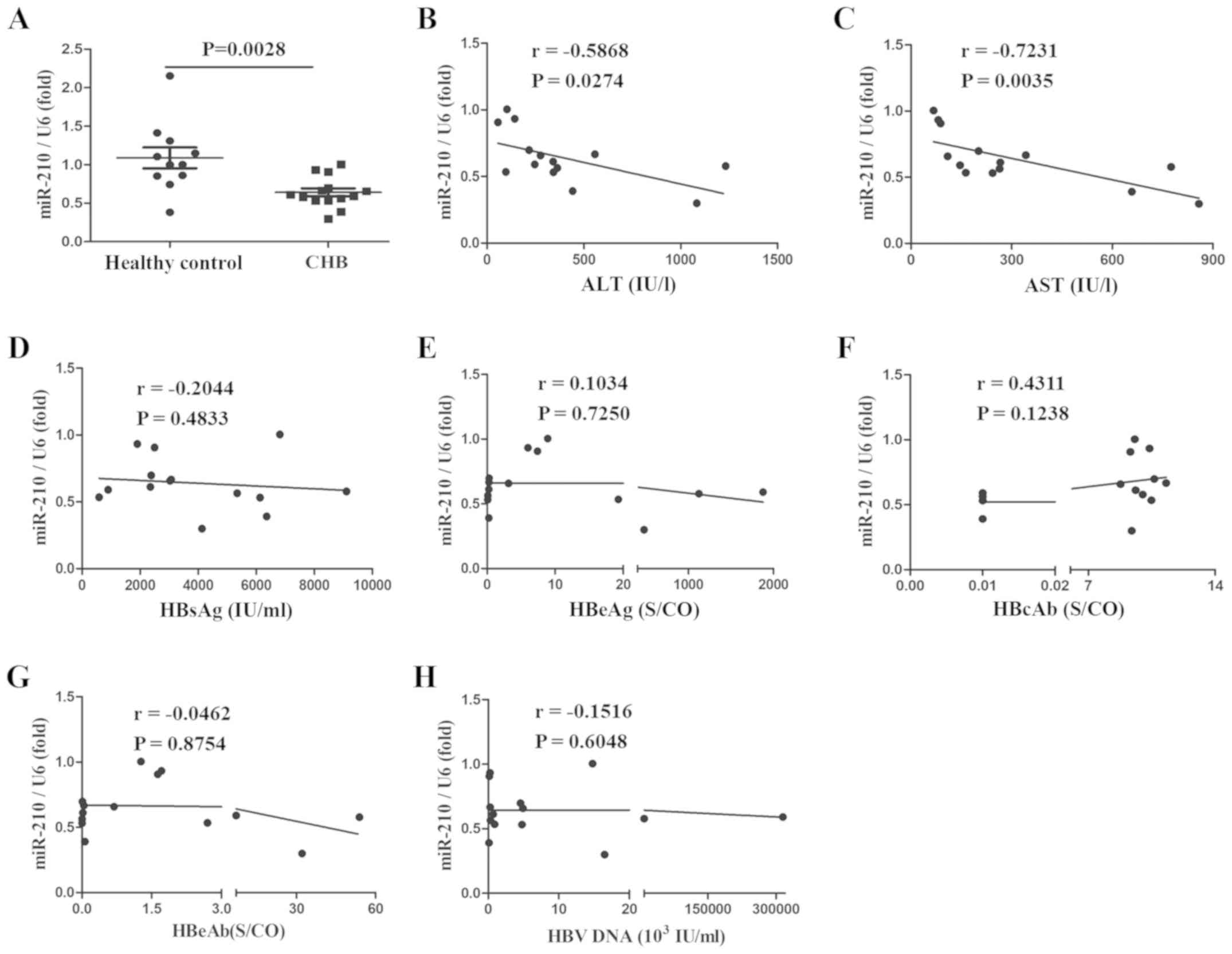 | Figure 1.miR-210 expression is decreased in
the PBMs of patients with CHB, and is negatively correlated with
serum ALT and AST. (A) miR-210 expression was detected by
quantitative polymerase chain reaction in the PBMs of healthy
controls and patients with CHB. Data are presented as the mean ±
standard deviation of triplicate experiments. The correlation
between the expression of miR-210 and the (B) ALT, (C) AST, (D)
HBsAg, (E) HBeAg, (F) HBcAb, (G) HBeAb or (H) HBV-DNA content was
analyzed. r represents the Spearman correlation coefficient. Data
are representative of three independent experiments. PBMs,
peripheral blood monocytes; ALT, alanine aminotransferase; AST,
aspartate aminotransferase; HBsAG, hepatitis B surface antigen;
HBeAg, hepatitis B e-antigen; HBeAb, anti-HBe antibody; HBcAb,
hepatitis B core antibody; CHB, chronic hepatitis B; S/CO,
Sample/Cut Off; miR, microRNA; HBV, hepatitis B virus. |
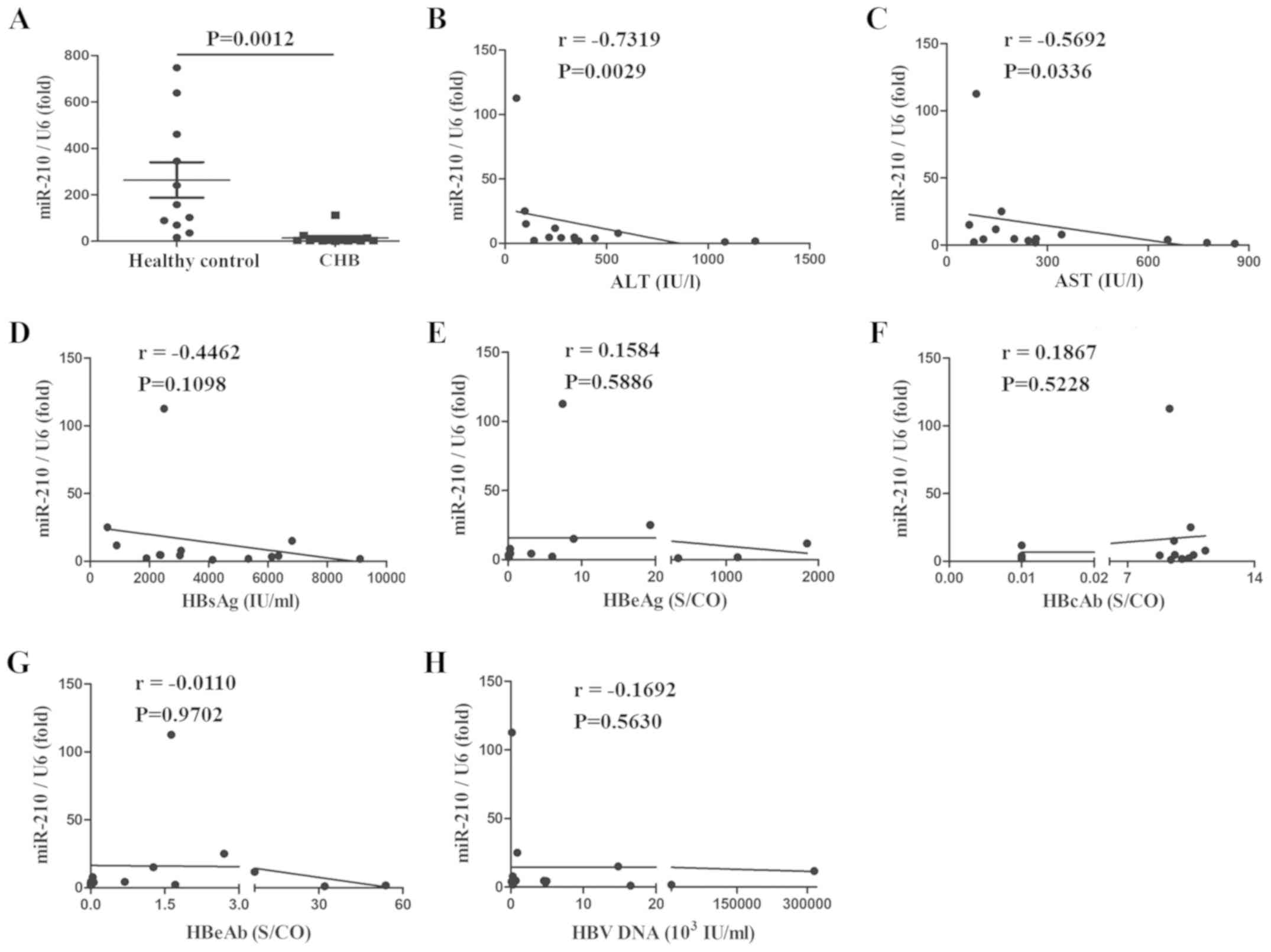 | Figure 2.miR-210 expression is decreased in
the serum of patients with CHB, and is negatively correlated with
serum ALT and AST. (A) miR-210 expression was measured by
quantitative polymerase chain reaction in the serum of healthy
controls and patients with CHB. Data are presented as the mean ±
standard deviation of triplicate experiments. The correlation
between the expression of miR-210 and (B) ALT, (C) AST, (D) HBsAg,
(E) HBeAg, (F) HBcAb, (G) HBeAb or (H) HBV-DNA content was
analyzed. r represents the Spearman correlation coefficient. Data
are representative of three independent experiments. ALT, alanine
aminotransferase; AST, aspartate aminotransferase; HBsAG, hepatitis
B surface antigen; HBeAg, hepatitis B e-antigen; HBeAb, anti-HBe
antibody; HBcAb, hepatitis B core antibody; CHB, chronic hepatitis
B; S/CO, Sample/Cut Off; miR, microRNA; HBV, hepatitis B virus. |
miR-210 expression is not affected by
HBV-associated antigens in different types of macrophages
A previous study demonstrated that HBeAg is able to
promote the production of macrophage inflammatory factors and the
expression of miR-155, by miRNA sequencing and a qPCR approach
(16). miR-210 expression was also
decreased in this miRNA sequencing analysis. However, the effect
and mechanism of miR-210 on macrophage activation has not been
completely elucidated. Therefore, in the present study, RAW264.7
macrophages were stimulated with HBV-associated antigens for 24 h.
As presented in Fig. 3A, as
previously reported, RAW264.7 cells were elongated with multiple
pseudopodia in the HBeAg and mix groups. Subsequently, miR-210
expression was detected in RAW264.7 macrophages with different
HBV-associated antigens at different times. As presented in
Fig. 3B, the expression of miR-210
was not altered by the different antigens or at different time
points in RAW264.7 cells. To eliminate the possibility that the
cell type led to this lack of change, mouse primary PMs and human
PBMs were acquired and stimulated with HBV-associated antigens. It
was observed that HBeAg was able to induce miR-210 expression, but
the increase was not significant in these different types of
macrophages (Fig. 3C and D). Taken
together, although miRNA sequencing (16) indicated that miR-210 expression was
decreased, there was no notable difference between the different
types of macrophages under treatment with HBV-associated antigens,
according to the qPCR analysis.
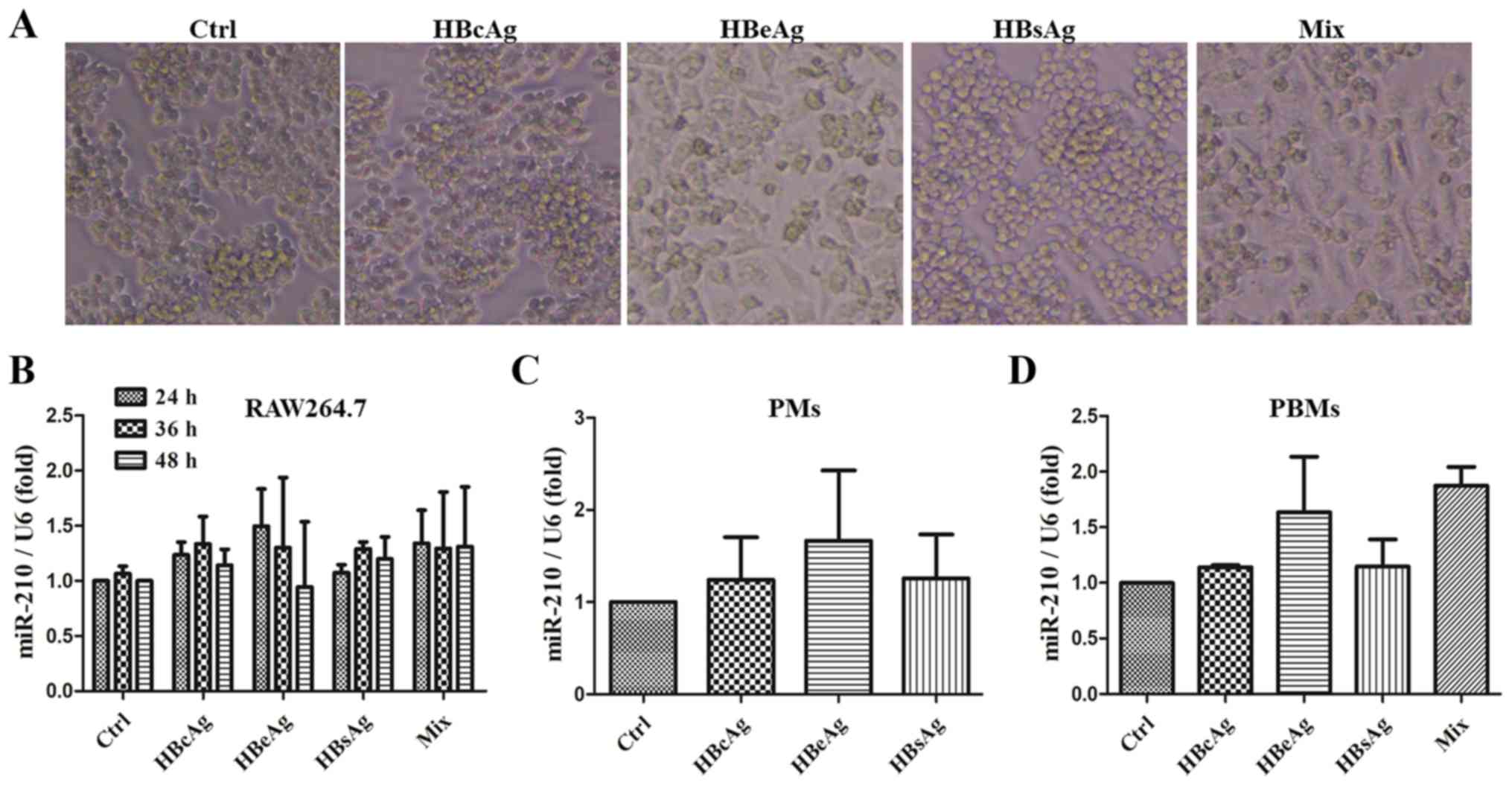 | Figure 3.miR-210 expression is not affected by
HBV-associated antigens in different types of macrophages. RAW264.7
cells were treated with HBcAg, HBeAg, HBsAg or a mixture (2 µg/ml)
for 24, 36 or 48 h. (A) The cell morphology was observed with a
light microscope (original magnification, ×100). (B) The expression
of miR-210 was tested by qPCR. (C) Mouse primary PMs and (D) PBMs
of healthy controls were stimulated with HBcAg, HBeAg, HBsAg or a
mixture (2 µg/ml) for 36 h. The expression of miR-210 was detected
by qPCR. Data are representative of three independent experiments
(mean ± standard deviation). PMs, peritoneal macrophages; PBMs,
peripheral blood monocytes; miR, microRNA; HBcAg, hepatitis B core
antigen; HBeAg, hepatitis B e-antigen; HBsAG, hepatitis B surface
antigen; qPCR, quantitative polymerase chain reaction. |
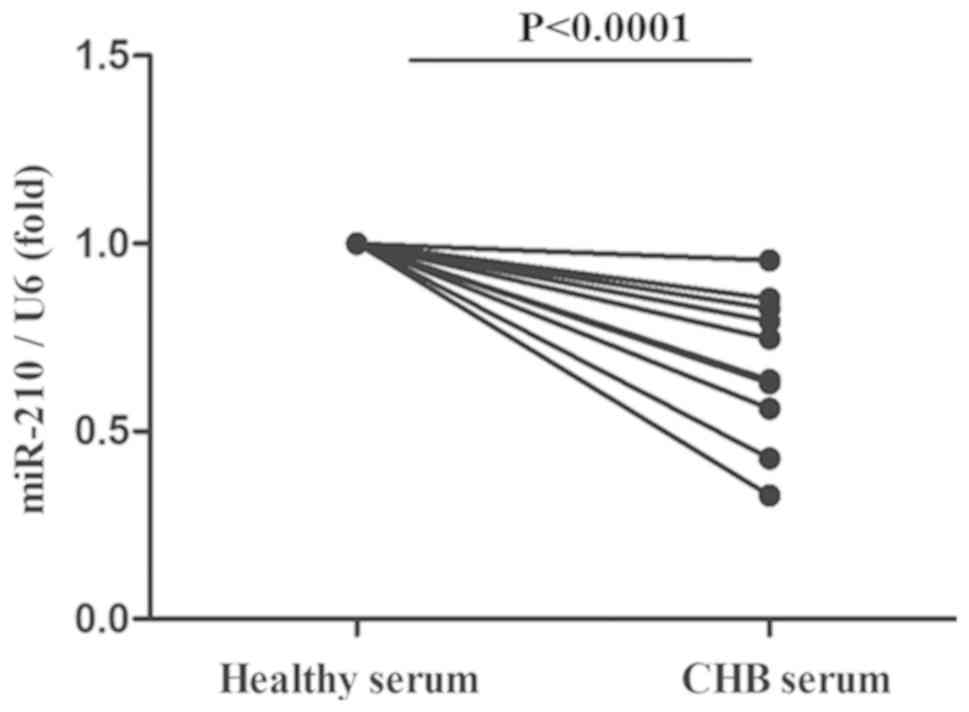
Serum from patients with CHB markedly
downregulated the miR-210 expression of PBMs from healthy
controls
To observe the effect of HBV infection on miR-210
expression in vivo, PBMs were obtained from healthy controls
and treated with the serum of patients with CHB. As illustrated in
Fig. 4, it was identified that the
serum of patients with CHB was able to significantly reduce the
miR-210 expression of PBMs from healthy controls.
Discussion
The precursor of miR-210 forms a stem-loop structure
and is located within the intron region of AK123483 gene, which is
located on chromosome 11p15.5 (19). The gene sequence of miR-210 is
evolutionarily conserved between different species, indicating its
functional importance (20,21).
Convincing evidence suggests that miR-210 is widely expressed in
multiple tissues and cells, including hematopoietic stem cells,
numerous cancer cells, myeloid cells and lymphocytes (22–25).
miR-210 is a pivotal hypoxia-response factor in numerous diseases
(26). Faraonio et al
(27) reported that miR-210 was
aberrantly overexpressed in senescent human diploid fibroblasts,
leading to an increase in DNA damage and the repression of cell
proliferation. Bar et al (28) reported that miR-210 is highly
expressed in cells of the triple-negative breast cancer subtype,
and in the tumor microenvironment, and its overexpression has been
linked to poor prognosis. In addition, miR-210 was induced after
photodynamic therapy in HeLa cells (29). Surprisingly, the increased
expression of miR-210 has an exceptionally long half-life and does
not rapidly return to normal levels after hypoxic exposure for 24 h
(30). In addition, although there
were also reports that miR-210 levels in serum may predict HBV
replication and translation (31)
and the virological response to treatment with interferon-α in
patients with CHB (32), the role
and mechanism of miR-210 in HBV infection was not elucidated.
Notably, it was previously reported that miR-210 in murine
macrophages negatively regulates the LPS-induced production of
proinflammatory cytokines (13).
Therefore, it was hypothesized that HBV-associated antigens may
activate macrophages in a similar manner to LPS and induce the
expression of miR-210, to ultimately regulate their function in HBV
infection.
In the present study, the results demonstrated that
miR-210 expression was significantly diminished in the PBMs and
serum of patients with CHB, but there was not a marked correlation
between miR-210 expression and HBsAg, HBeAg, HBeAb, HBcAb and
HBV-DNA in the serum. A number of studies have shown that the
expression of certain stress proteins (including heat shock
proteins and C-reactive proteins) are increased during HBV
infection (33,34). Moreover, these proteins may affect
macrophage activation (35,36).
Therefore, it was speculated that the expression of macrophage
miR-210 may be affected by these stress proteins or other unknown
factors during HBV infection. The present study used the
traditional adherent separation method, as described previously
(16). This assay is relatively
weak compared with fluorescence-activated cell sorting (FACS), but
it is more readily available. In addition, since the small sample
size was a limitation of the current study, the differences between
male and female subjects were not compared. Therefore, future
studies may expand the sample content and/or use a FACS assay to
detect the effects and mechanisms of miR-210 or other miRs during
HBV infection. The detection of these serum miRs as an important
indicator may be a basis for observing the immune status of
patients with HBV infection.
When liver cells are infected by HBV, they
experience an immune attack, leading to acute and chronic
inflammatory responses in the liver. In CHB, the virus is
frequently maintained at a low level over a long period (37). The number of patients with CHB, and
the gradually increasing trend of liver cirrhosis and HCC, has led
to large expenditure for medical treatment, representing a serious
health burden in worldwide, but particularly in China (38). Therefore, early diagnosis and
treatment may be a good therapeutic strategy to avoid disease
progression, including the prevention of liver fibrosis and HCC.
However, the factors that regulate the development of CHB have not
been clearly identified. It has been indicated that once
individuals are infected with HBV, the HBV DNA converts into
covalently closed circular DNA to promote viral DNA replication. In
addition, various viral proteins are produced, including HBsAg,
HBcAg and HBeAg, resulting in the activation of the immune system
(39). It was previously reported
that HBeAg is able to promote miR-155 expression to enhance liver
damage by elevating inflammatory cytokine production in macrophages
(16). However, less is known
about the effect of HBV-associated antigens on hypoxic miR-210. It
has been reported that PBMs may be directly stimulated in
vitro (16,40–42).
There have also been reports that PBMs may be induced to become M1
or M2 macrophages by exposure to granulocyte-macrophage
colony-stimulating factor or macrophage colony-stimulating factor
(43). Therefore, the PBMs in the
present study were not treated with GM-CSF to avoid the induction
of monocyte activation not by HBV-associated antigens. In the
present study, the results demonstrated that HBV-associated
antigens have no effect on the expression of miR-210 in different
types of macrophages from mice and humans, including cell lines and
primary macrophages. However, it was demonstrated that the serum of
patients with CHB was able to significantly diminish the miR-210
expression of PBMs from healthy controls.
In conclusion, it was observed that miR-210
expression was markedly decreased in the PBMs and serum of patients
with CHB. Subsequently, the association between miR-210 expression
and multiple clinical indexes in HBV infection was assessed, and it
was observed that there was not a marked correlation between them.
In vitro, it was demonstrated that HBV-associated antigens
had no effect on miR-210 expression in macrophages. Notably, the
serum of patients with CHB was able to impair miR-210 expression in
PBMs from healthy controls. Therefore, these findings suggest that
there may be another regulatory mechanism of miR-210 expression in
HBV infection.
Acknowledgements
The authors would like to thank Professor Qiang Zhu
for helping with language editing.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81600469, 81772626,
81871700 and 81401868), the Science and Technology Development
Projects of Shandong Province (grant no. 2017GSF218053), the
Clinical Medical Science and Technology Innovation Program of Jinan
City (grant no. 201704114), the Natural Science Foundation of
Shandong Province (grant no. ZR2018PH003), and the Shandong
Province Medical and Health Science and Technology Development
Project (grant nos. 2017WS194 and 2017WS465).
Availability of data and materials
The authors confirm that all data and materials are
fully available without any restriction. All relevant data are
within the paper.
Authors' contributions
JQ, HB and CQ conceived and designed the
experiments. FL and WW performed the experiments. WR and SS
analyzed the data. LN and MX collected clinical samples, fed the
mice and assisted with the experiments. HB and JQ wrote the
manuscript.
Ethics approval and consent to
participate
The research protocol and consent program were
approved by Shandong Provincial Hospital Affiliated to Shandong
University Ethics Committee. Written informed consent was acquired
from each patient for this study. All animal experiments were
undertaken in accordance with the National Institutes of Health
Guide for the Care and Use of Laboratory Animals, with the approval
of the Scientific Investigation Board of the Shandong Provincial
Hospital Affiliated to Shandong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HBV
|
hepatitis B virus
|
|
HCC
|
hepatocellular carcinoma
|
|
miR-210
|
microRNA-210
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
CHB
|
chronic hepatitis B
|
|
PMs
|
peritoneal macrophages
|
|
PBMs
|
peripheral blood monocytes
|
References
|
1
|
Nguyen TH, Liu X, Su ZZ, Hsu AC, Foster PS
and Yang M: Potential role of MicroRNAs in the regulation of
antiviral responses to influenza infection. Front Immunol.
9:15412018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wei Y, Zhu M and Schober A: Macrophage
MicroRNAs as therapeutic targets for atherosclerosis, metabolic
syndrome, and cancer. Int J Mol Sci. 19:E17562018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chan YC, Banerjee J, Choi SY and Sen CK:
miR-210: The master hypoxamir. Microcirculation. 19:215–223. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chan SY and Loscalzo J: MicroRNA-210: A
unique and pleiotropic hypoxamir. Cell Cycle. 9:1072–1083. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mutharasan RK, Nagpal V, Ichikawa Y and
Ardehali H: microRNA-210 is upregulated in hypoxic cardiomyocytes
through Akt- and p53-dependent pathways and exerts cytoprotective
effects. Am J Physiol Heart Circ Phyiol. 301:H1519–H1530. 2011.
View Article : Google Scholar
|
|
6
|
Zeng L, Liu J, Wang Y, Wang L, Weng S,
Tang Y, Zheng C, Cheng Q, Chen S and Yang GY: MicroRNA-210 as a
novel blood biomarker in acute cerebral ischemia. Front Biosci
(Elite Ed). 3:1265–1272. 2011.PubMed/NCBI
|
|
7
|
Dang K and Myers KAL: The role of
hypoxia-induced miR-210 in cancer progression. Int J Mol Sci.
16:6353–6372. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang Q, Chen SS, Li J, Tao SS, Wang M,
Leng RX, Pan HF and Ye DQ: miR-210 expression in PBMCs from
patients with systemic lupus erythematosus and rheumatoid
arthritis. Ir J Med Sci. 187:243–249. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chan SL, Wong VW, Qin S and Chan HL:
Infection and cancer: The case of hepatitis B. J Clin Oncol.
34:83–90. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wen WH, Huang CW, Chie WC, Yeung CY, Zhao
LL, Lin WT, Wu JF, Ni YH, Hsu HY, Chang MH, et al: Quantitative
maternal hepatitis B surface antigen predicts maternally
transmitted hepatitis B virus infection. Hepatology. 64:1451–1461.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Niller HH, Ay E, Banati F, Demcsák A,
Takacs M and Minarovits J: Wild type HBx and truncated HBx:
Pleiotropic regulators driving sequential genetic and epigenetic
steps of hepatocarcinogenesis and progression of HBV-associated
neoplasms. Rev Med Virol. 26:57–73. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu L, Qi J, Zhao P, Liang X, Ju Y, Liu P,
Liu B, Guo C, Zhang L, Ma C and Gao L: T cell immunoglobulin- and
mucin-domain-containing molecule-4 attenuates concanavalin
A-induced hepatitis by regulating macrophage. J Leukoc Biol.
88:329–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qi J, Qiao Y, Wang P, Li S, Zhao W and Gao
C: MicroRNA-210 negatively regulates LPS-induced production of
proinflammatory cytokines by targeting NF-κB1 in murine
macrophages. FEBS Lett. 586:1201–1207. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chinese Society of Infectious Diseases,
Chinese Medical Association, Chinese Society of Infectious
Diseases, Chinese Medical Association, ; Hou JL and Lai W: The
guideline of prevention and treatment for chronic hepatitis B: A
2015 update. Zhonghua Gan Zang Bing Za Zhi. (In Chinese).
23:888–905. 2015.PubMed/NCBI
|
|
15
|
Hou J, Wang G, Wang F, Cheng J, Ren H,
Zhuang H, Sun J, Li L, Li J, Meng Q, et al: Guideline of prevention
and treatment for chronic hepatitis B (2015 Update). J Clin Transl
Hepatol. 5:297–318. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang W, Bian H, Li F, Li X, Zhang D, Sun
S, Song S, Zhu Q, Ren W, Qin C and Qi J: HBeAg induces the
expression of macrophage miR-155 to accelerate liver injury via
promoting production of inflammatory cytokines. Cell Mol Life Sci.
75:2627–2641. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bian H, Li F, Wang W, Zhao Q, Gao S, Ma J,
Li X, Ren W, Qin C and Qi J: MAPK/p38 regulation of cytoskeleton
rearrangement accelerates induction of macrophage activation by
TLR4, but not TLR3. Int J Mol Med. 40:1495–1503. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qi J, Li T, Bian H, Li F, Ju Y, Gao S, Su
J, Ren W and Qin C: SNAI1 promotes the development of HCC through
the enhancement of proliferation and inhibition of apoptosis. FEBS
Open Bio. 6:326–337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Camps C, Buffa FM, Colella S, Moore J,
Sotiriou C, Sheldon H, Harris AL, Gleadle JM and Ragoussis J:
hsa-miR-210 is induced by hypoxia and is an independent prognostic
factor in breast cancer. Clin Cancer Res. 14:1340–1348. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang X, Ding L, Bennewith KL, Tong RT,
Welford SM, Ang KK, Story M, Le QT and Giaccia AJ:
Hypoxia-inducible mir-210 regulates normoxic gene expression
involved in tumor initiation. Mol Cell. 35:856–867. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ren CX, Leng RX, Fan YG, Pan HF, Wu CH and
Ye DQ: MicroRNA-210 and its theranostic potential. Expert Opin Ther
Targets. 20:1325–1338. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang X, Le QT and Giaccia AJ:
MiR-210-micromanager of the hypoxia pathway. Trends Mol Med.
16:230–237. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiang Y, Li L, Tan X, Liu B, Zhang Y and
Li C: miR-210 mediates vagus nerve stimulation-induced antioxidant
stress and anti-apoptosis reactions following cerebral
ischemia/reperfusion injury in rats. J Neurochem. 134:173–181.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Noman MZ, Buart S, Romero P, Ketari S,
Janji B, Mari B, Mami-Chouaib F and Chouaib S: Hypoxia-inducible
miR-210 regulates the susceptibility of tumor cells to lysis by
cytotoxic T cells. Cancer Res. 72:4629–4641. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen Z, Li Y, Zhang H, Huang P and Luthra
R: Hypoxia-regulated microRNA-210 modulates mitochondrial function
and decreases ISCU and COX10 expression. Oncogene. 29:4362–4368.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li C, Zhao M, Zhang C, Zhang W, Zhao X,
Duan X and Xu W: miR210 modulates respiratory burst in apostichopus
japonicas coelomocytes via targeting toll-like receptor. Dev Comp
Immunol. 65:377–381. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Faraonio R, Salerno P, Passaro F, Sedia C,
Iaccio A, Bellelli R, Nappi TC, Comegna M, Romano S, Salvatore G,
et al: A set of miRNAs participates in the cellular senescence
program in human diploid fibroblasts. Cell Death Differ.
19:713–721. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bar I, Merhi A, Abdel-Sater F, Ben Addi A,
Sollennita S, Canon JL and Delrée P: The microRNA miR-210 is
expressed by cancer cells but also by the tumor microenvironment in
triple-negative breast cancer. J Histochem Cytochem. 65:335–346.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kushibiki T: Photodynamic therapy induces
microRNA-210 and −296 expression in HeLa cells. J Biophotonics.
3:368–372. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fasanaro P, D'Alessandra Y, Di Stefano V,
Melchionna R, Romani S, Pompilio G, Capogrossi MC and Martelli F:
MicroRNA-210 modulates endothelial cell response to hypoxia and
inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol
Chem. 283:15878–15883. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu F, Yang J, Ouyang J, Zheng Y, Chen B,
Li G, Lu Z, Dong P and Zheng J: Serum microRNA-210 levels in
different groups of chronic hepatitis B patients. Clin Chim Acta.
450:203–209. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li J, Zhang X, Chen L, Zhang Z, Zhang J,
Wang W, Wu M, Shi B, Zhang X, Kozlowski M, et al: Circulating
miR-210 and miR-22 combined with ALT predict the virological
response to interferon-alpha therapy of CHB patients. Sci Rep.
7:156582017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wyżewski Z, Gregorczyk KP, Szczepanowska J
and Szulc-Dąbrowska L: Functional role of Hsp60 as a positive
regulator of human viral infection progression. Acta Virol.
62:33–40. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hao S, Wang Y, Gao G and Li Z: Hepatitis B
virus upregulates the expression of C-reactive protein both in vivo
and in vitro. Ann Clin Lab Sci. 47:432–435. 2017.PubMed/NCBI
|
|
35
|
Vega VL, Rodríguez-Silva M, Frey T,
Gehrmann M, Diaz JC, Steinem C, Multhoff G, Arispe N and De Maio A:
Hsp70 translocates into the plasma membrane after stress and is
released into the extracellular environment in a
membrane-associated form that activates macrophages. J Immunol.
180:4299–42307. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pilling D, Galvis-Carvajal E, Karhadkar
TR, Cox N and Gomer RH: Monocyte differentiation and macrophage
priming are regulated differentially by pentraxins and their
ligands. BMC Immunol. 18:302017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Karayiannis P: Hepatitis B virus:
Virology, molecular biology, life cycle and intrahepatic spread.
Hepatol Int. 11:500–508. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
WHO, ; Hepatitis B: http://www.who.int/en/news-room/fact-sheets/detail/hepatitis-bJuly.
2018
|
|
39
|
Stramer SL, Wend U, Candotti D, Foster GA,
Hollinger FB, Dodd RY, Allain JP and Gerlich W: Nucleic acid
testing to detect HBV infection in blood donors. N Engl J Med.
364:236–247. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Justo-Junior AS, Villarejos LM, Lima XTV,
Nadruz W Jr, Sposito AC, Mamoni RL, Abdalla R, Fernandes JL,
Oliveira RTD and Blotta MHSL: Monocytes of patients with unstable
angina express high levels of chemokine and pattern-recognition
receptors. Cytokine. 2018.
|
|
41
|
Gu X, Wei C, Zhu X, Lu F, Sheng B and Zang
X: Effect of interleukin-31 on septic shock through regulating
inflammasomes and interleukin-1β. Exp Ther Med. 16:171–177.
2018.PubMed/NCBI
|
|
42
|
Santoni G, Morelli MB, Amantini C, Santoni
M, Nabissi M, Marinelli O and Santoni A: Immuno-transient receptor
potential ion channels: The role in monocyte- and
macrophage-mediated inflammatory responses. Front Immunol.
9:12732018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Schneider A, Weier M, Herderschee J,
Perreau M, Calandra T, Roger T and Giannoni E: IRF5 is a key
regulator of macrophage response to lipopolysaccharide in newborns.
Front Immunol. 9:15972018. View Article : Google Scholar : PubMed/NCBI
|