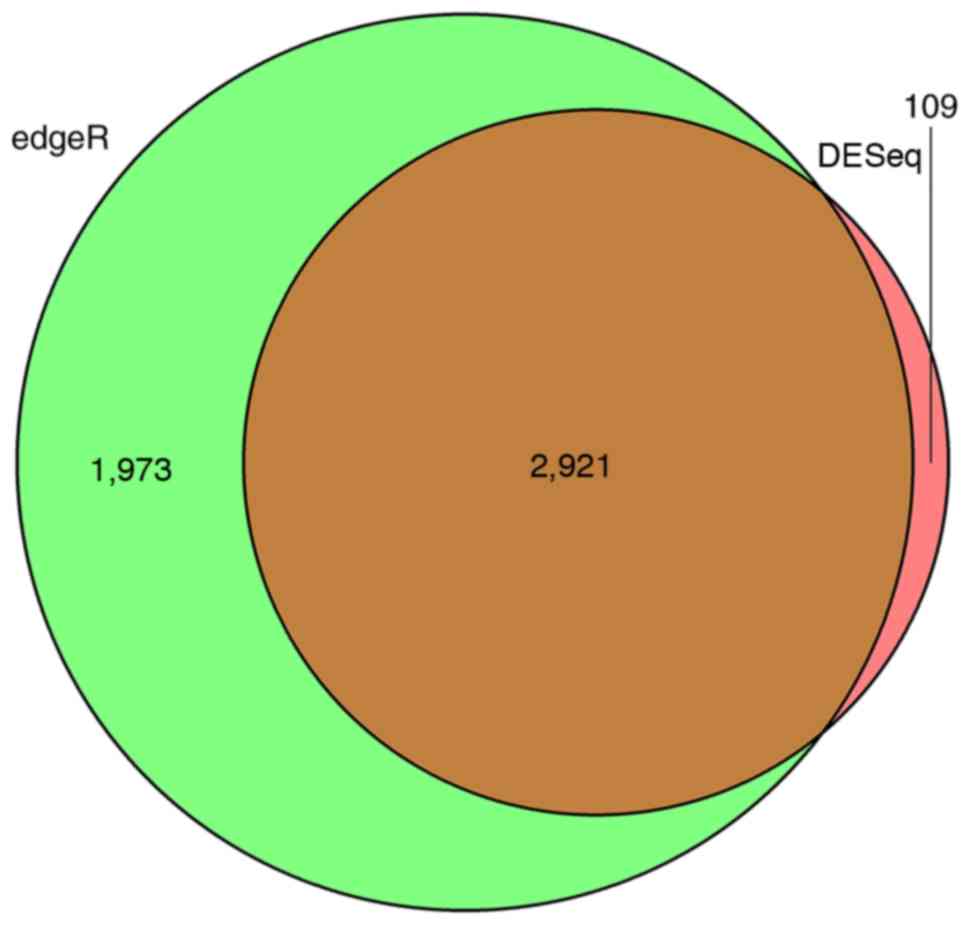
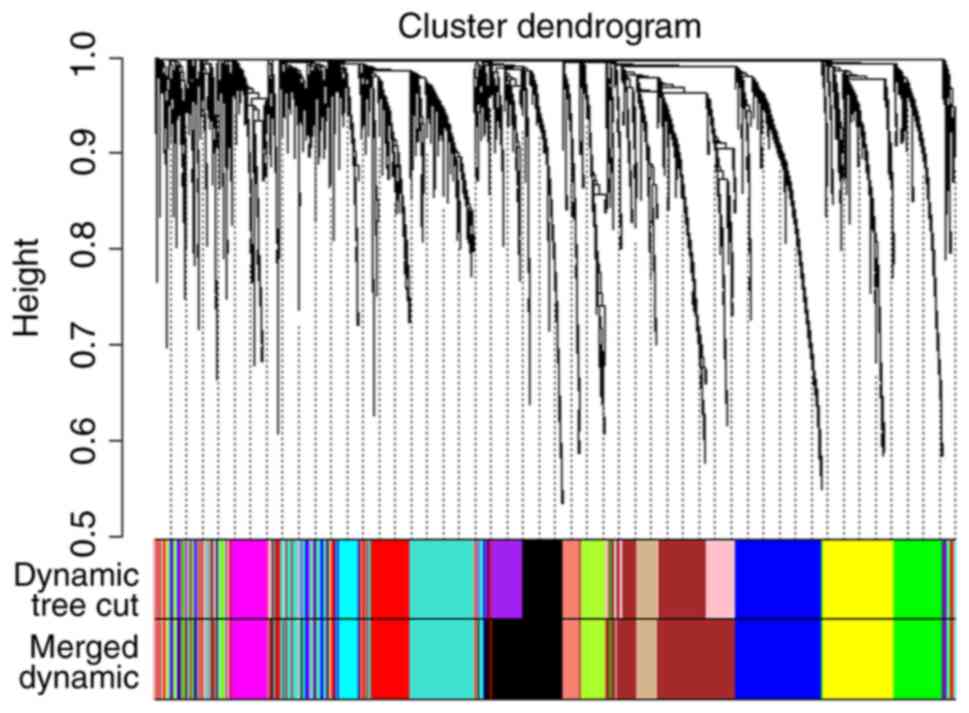
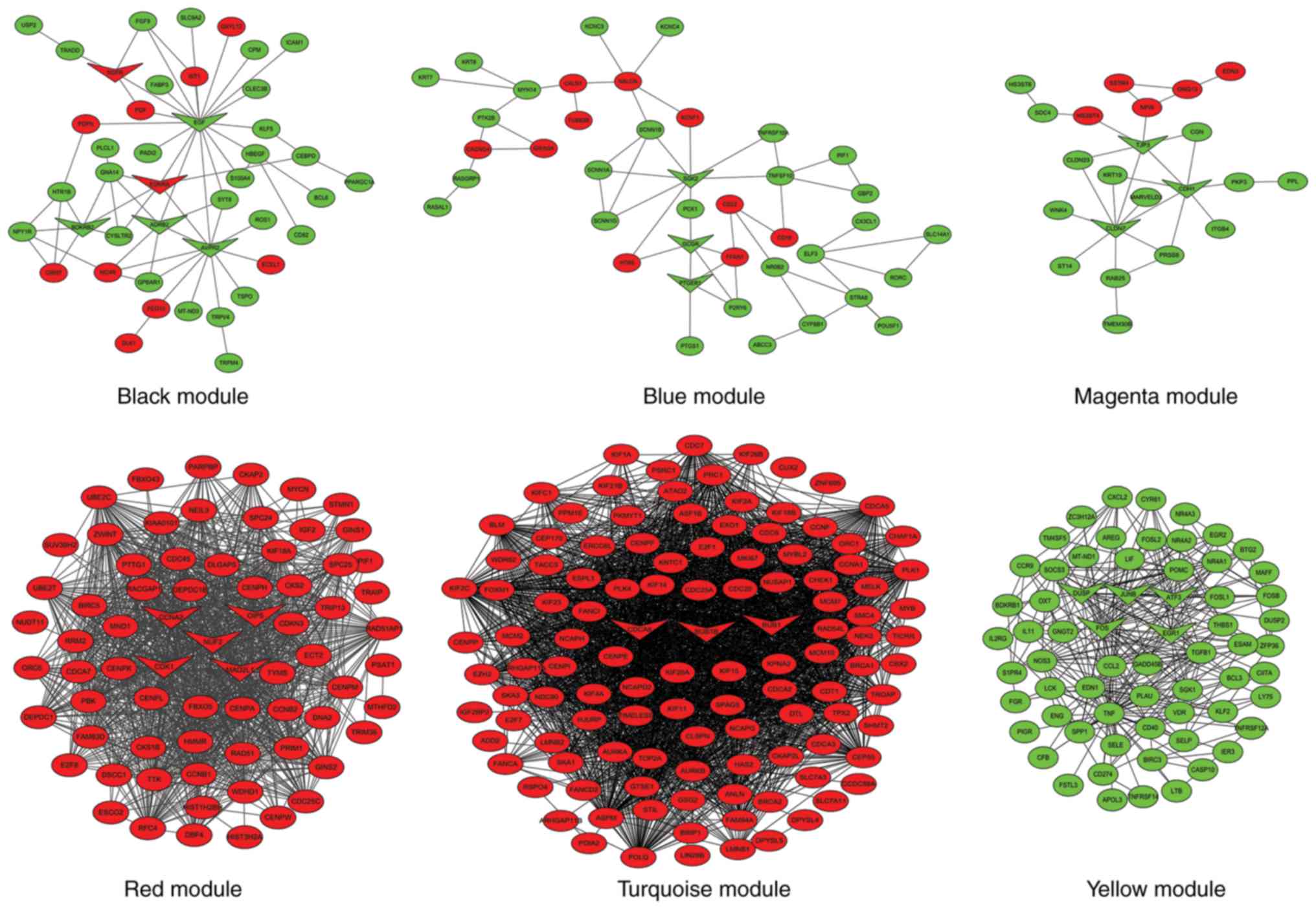
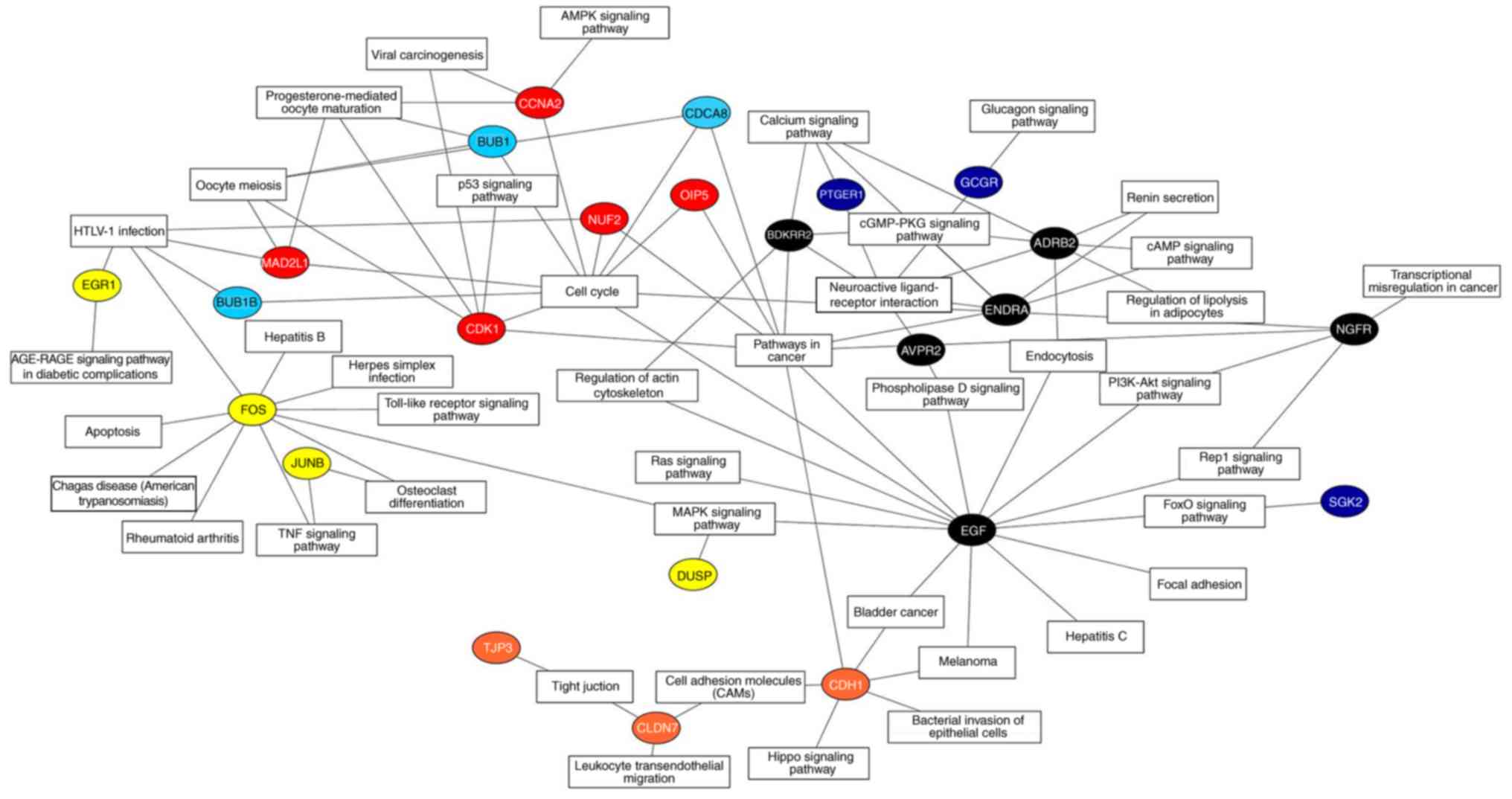
|
1
|
Amirian ES: The role of Hispanic ethnicity
in pediatric Wilms' tumor survival. Pediatr Hematol Oncol.
30:317–327. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Perme MP and Jereb B: Trends in survival
after childhood cancer in Slovenia between 1957 and 2007. Pediatr
Hematol Oncol. 26:240–251. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pastore G, Znaor A, Spreafico F, Graf N,
Pritchard-Jones K and Steliarova-Foucher E: Malignant renal tumours
incidence and survival in European children (1978–1997): Report
from the Automated Childhood Cancer Information System project. Eur
J Cancer. 42:2103–2114. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Geller JI: Current standards of care and
future directions for ‘high-risk’ pediatric renal tumors:
Anaplastic Wilms tumor and Rhabdoid tumor. Urol Oncol. 34:50–56.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
National Cancer Institute: Wilms Tumor and
Other Childhood Kidney Tumors Treatment (PDQ®): Patient
version. PDQ Cancer Information Summaries. National Cancer
Institute (US); Bethesda, MD: 2002
|
|
6
|
Yuan N, Zhang G, Bie F, Ma M, Ma Y, Jiang
X, Wang Y and Hao X: Integrative analysis of lncRNAs and miRNAs
with coding RNAs associated with ceRNA crosstalk network in triple
negative breast cancer. OncoTargets and Ther. 10:5883–5897. 2017.
View Article : Google Scholar
|
|
7
|
Baltimore D: Our genome unveiled. Nature.
409:814–816. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang C, Yuan N, Wu L, Wang X, Dai J, Song
P, Li F, Xu C and Zhao X: An integrated analysis for long noncoding
RNAs and microRNAs with the mediated competing endogenous RNA
network in papillary renal cell carcinoma. Onco Targets and Ther.
10:4037–4050. 2017. View Article : Google Scholar
|
|
9
|
Huang CG, Li FX, Pan S, Xu CB, Dai JQ and
Zhao XH: Identification of genes associated with
castration-resistant prostate cancer by gene expression profile
analysis. Mol Med Rep. 16:6803–6813. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Charlton J, Pavasovic V and
Pritchard-Jones K: Biomarkers to detect Wilms tumors in pediatric
patients: Where are we now? Future Oncol. 11:2221–2234. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
12
|
Lee JS: Exploring cancer genomic data from
the cancer genome atlas project. BMB Rep. 49:607–611. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rao Y, Lee Y, Jarjoura D, Ruppert AS, Liu
CG, Hsu JC and Hagan JP: A comparison of normalization techniques
for microRNA microarray data. Stat Appl Genet Mol Biol.
7:Article222008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li D and Dye TD: Power and stability
properties of resampling-based multiple testing procedures with
applications to gene oncology studies. Comput Math Methods Med
2013. 6102972013.
|
|
16
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang B and Horvath S: A general framework
for weighted gene co-expression network analysis. Stat Appl Genet
Mol Biol. 4:Article172005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Langfelder P and Horvath S: Eigengene
networks for studying the relationships between co-expression
modules. BMC Syst Biol. 1:542007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:W316–W322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:(Database Issue). D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 Suppl 4:S112014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goeman JJ, Oosting J, Cleton-Jansen AM,
Anninga JK and van Houwelingen HC: Testing association of a pathway
with survival using gene expression data. Bioinformatics.
21:1950–1957. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jensen CH, Krogh TN, Støving RK, Holmskov
U and Teisner B: Fetal antigen 1 (FA1), a circulating member of the
epidermal growth factor (EGF) superfamily: ELISA development,
physiology and metabolism in relation to renal function. Clin Chim
Acta. 268:1–20. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lanuti M, Liu G, Goodwin JM, Zhai R, Fuchs
BC, Asomaning K, Su L, Nishioka NS, Tanabe KK and Christiani DC: A
functional epidermal growth factor (EGF) polymorphism, EGF serum
levels, and esophageal adenocarcinoma risk and outcome. Clin Cancer
Res. 14:3216–3222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tanabe KK, Lemoine A, Finkelstein DM,
Kawasaki H, Fujii T, Chung RT, Lauwers GY, Kulu Y, Muzikansky A,
Kuruppu D, et al: Epidermal growth factor gene functional
polymorphism and the risk of hepatocellular carcinoma in patients
with cirrhosis. JAMA. 299:53–60. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Marteil G, Gagne JP, Borsuk E,
Richard-Parpaillon L, Poirier GG and Kubiak JZ: Proteomics reveals
a switch in CDK1-associated proteins upon M-phase exit during the
Xenopus laevis oocyte to embryo transition. Int J Biochem Cell
Biol. 44:53–64. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Russignan A, Spina C, Tamassia N, Cassaro
A, Rigo A, Bagnato A, Rosanò L, Bonalumi A, Gottardi M, Zanatta L,
et al: Endothelin-1 receptor blockade as new possible therapeutic
approach in multiple myeloma. Br J Haematol. 178:781–793. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu J, Cui L, Wang W, Hang XY, Xu AX, Yang
SX, Dou JT, Mu YM, Zhang X and Gao JP: Whole exome sequencing
identifies mutation of ENDRA involved in ACTH-independent
macronodular adrenal hyperplasia. Fam Cancer. 12:657–667. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Canossa M, Twiss JL, Verity AN and Shooter
EM: p75(NGFR) and TrkA receptors collaborate to rapidly activate a
p75(NGFR)-associated protein kinase. EMBO J. 15:3369–3376. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Soland TM, Brusevold IJ, Koppang HS,
Schenck K and Bryne M: Nerve growth factor receptor (p75 NTR) and
pattern of invasion predict poor prognosis in oral squamous cell
carcinoma. Histopathology. 53:62–72. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Williams JM, Chen GC, Zhu L and Rest RF:
Using the yeast two-hybrid system to identify human epithelial cell
proteins that bind gonococcal Opa proteins: Intracellular gonococci
bind pyruvate kinase via their Opa proteins and require host
pyruvate for growth. Mol Microbiol. 27:171–186. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li H, Zhang J, Lee MJ, Yu GR, Han X and
Kim DG: OIP5, a target of miR-15b-5p, regulates hepatocellular
carcinoma growth and metastasis through the AKT/mTORC1 and
β-catenin signaling pathways. Oncotarget. 8:18129–18144.
2017.PubMed/NCBI
|
|
35
|
Koinuma J, Akiyama H, Fujita M, Hosokawa
M, Tsuchiya E, Kondo S, Nakamura Y and Daigo Y: Characterization of
an Opa interacting protein 5 involved in lung and esophageal
carcinogenesis. Cancer Sci. 103:577–586. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li HC, Chen YF, Feng W, Cai H, Mei Y,
Jiang YM, Chen T, Xu K and Feng DX: Loss of the Opa interacting
protein 5 inhibits breast cancer proliferation through
miR-139-5p/NOTCH1 pathway. Gene. 603:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chun HK, Chung KS, Kim HC, Kang JE, Kang
MA, Kim JT, Choi EH, Jung KE, Kim MH, Song EY, et al: OIP5 is a
highly expressed potential therapeutic target for colorectal and
gastric cancers. BMB Rep. 43:349–354. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wigge PA and Kilmartin JV: The Ndc80p
complex from Saccharomyces cerevisiae contains conserved centromere
components and has a function in chromosome segregation. J Cell
Biol. 152:349–360. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kobayashi Y, Takano A, Miyagi Y, Tsuchiya
E, Sonoda H, Shimizu T, Okabe H, Tani T, Fujiyama Y and Daigo Y:
Cell division cycle-associated protein 1 overexpression is
essential for the malignant potential of colorectal cancers. Int J
Oncol. 44:69–77. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dai C, Miao CX, Xu XM, Liu LJ, Gu YF, Zhou
D, Chen LS, Lin G and Lu GX: Transcriptional activation of human
CDCA8 gene regulated by transcription factor NF-Y in embryonic stem
cells and cancer cells. J Biol Chem. 290:22423–22434. 2015.
View Article : Google Scholar : PubMed/NCBI
|