Introduction
Myasthenia gravis (MG) is an acquired neuromuscular
autoimmune disease characterized by fluctuating muscle weakness and
fatigue (1). The primary cause of
MG is acetylcholine receptor antibodies, which destroy
acetylcholine receptors on the posterior synaptic membrane of the
neuromuscular junction with the involvement of cellular immunity
and the complement system, so that it is not able to produce enough
endplate potential, thereby causing the onset of the disease
(2).
In previous years, a number of risk genes of MG have
been identified, including circulating interleukin (IL)-17A, which
was identified to be increased in patients with MG compared with
normal controls, and increases of IL-17A were associated with
general muscle weakness (3). Tumor
necrosis factor-α (TNF-α) is considered to be one of the most
important cytokines in the pathogenesis of autoimmune MG, and the
inhibition of TNF-α may have significant clinical efficacy for MG
(4). Furthermore, chemokine CC
motif receptor (CCR)9 and CCR7 have been demonstrated to be
abnormally expressed at different thymocyte stages of MG, and the
overexpression of CCR9 and CCR7 in CD4-CD8-double negative
thymocytes is associated with abnormal intrathymic T-cell
differentiation in patients with MG (5). These observations suggest an
increasing number of genes are crucial in the pathogenesis of MG.
However, the majority of previous studies investigating MG risk
genes have focused on only one or a few genes in cell lines without
global analysis. Searches for a number of MG risk genes were
performed in our previous studies (6,7),
however, the risk genes identified previously were not sufficiently
comprehensive. The global pathway analysis of MG risk genes in the
present study may assist in further characterizing the pathogenesis
of MG.
Previous studies have demonstrated that microRNAs
(miRNAs/miRs) are increasingly important in the pathogenesis of MG,
and their aberrant expression may contribute towards the specific
mechanism of MG. For example, miR-20b may inhibit nuclear factor of
activated T cells (NFAT) signaling via the repression of NFAT5 and
expression of calmodulin binding transcription activator 1 in
thymoma-associated myasthenia gravis (TAMG) (8). The expression of the forkhead box
(fox) p3 gene was modulated by miR-125a-5p, which is likely to be
involved in the pathogenesis of TAMG (9). The abnormal expression of miR-15a
facilitates pro-inflammatory cytokine production, at least in part
by regulating the expression of C-X-C motif chemokine 10 (CXCL10),
thereby contributing toward the immune response in MG (10). However, the potential mechanisms of
miRNA in MG remain to be fully elucidated.
Certain previous studies have demonstrated that the
pathogenesis of certain immune-associated diseases is associated
with miRNA clusters on chromosomes. miRNAs are distributed across
diverse genomic locations; although certain miRNAs are isolated,
~50% are frequently physically clustered on the human genome to
permit co-regulation, termed miRNA clusters (11). For example, the miR-106a-363
cluster encodes six miRNAs on the X chromosome, which include
miR-18b, miR-106a and miR-363-3p, that were demonstrated to be
involved in T-helper 17 cell differentiation, which further
illustrates the association between the miR-106-363 cluster and
immune-associated diseases (12,13).
The mirn23a gene is located on murine chromosome 8 and codes for
three pre-miRNAs, miR-23a, miR-24-2 and miR-27a (14). The miR-23a-27a-24-2 miRNA cluster
is an inhibitor of B cell development, and bone morphogenetic
protein/mothers against decapentaplegic and Akt/FOXO1 signaling is
critical for mirn23a-mediated immune cell regulation (14). However, there has been no overall
investigation of miRNA clusters in MG. Furthermore, MG-associated
miRNA clusters may have important regulated pathways that can
clarify the mechanism of MG.
The present study identified the risk pathways
regulated by MG risk genes and MG risk miRNAs. Subsequently, each
MG risk miRNA on each chromosome was located and 15 significant
miRNA clusters associated with MG were identified. The risk
pathways of each of the 15 miRNA clusters were identified, which
can further assist in elucidating the potential mechanism of miRNA
clusters in MG at the post-transcriptional regulation level.
Materials and methods
Human MG risk gene data
Human MG risk gene data was acquired using the
following two approaches: i) Gene information was obtained by
searching certain current databases, including DisGeNET (http://www.disgenet.org/web/DisGeNET/menu) (15) and Online Mendelian Inheritance in
Man (http://www.omim.org/) (16); and ii) using the protocols
published in our previous studies (6,7), the
gene was notably differentially expressed in more than five MG
samples using dependable biological laboratorial techniques, and
9,514 items were browsed by manually collecting literature using
the terms [myasthenia gravis (MeSH Terms) and English (Language)]
and the species ‘Homo sapiens’ published prior to March 1st, 2017
on the PubMed database (https://www.ncbi.nlm.nih.gov/pubmed); the eligible
genes were selected. An update was made to the previous catalog of
162 MG risk genes described in our previous study (7) to 245.
Human MG risk miRNA data, miRNA
location data and miRNA target data
Human miRNA information was acquired from miRBase
(http://www.mirbase.org/) (17) and the miRNA location information
was obtained from NCBI-GENE database (https://www.ncbi.nlm.nih.gov/gene/) (18). MG risk miRNAs were additionally
gathered in two ways. To begin with, literature published prior to
March 1st, 2017 was manually searched using the protocols described
in our previous study (7); the
keywords ‘miRNA’ and ‘myasthenia gravis’ or ‘microRNA’ and
‘myasthenia gravis’ or ‘miR’ and ‘myasthenia gravis’ were searched
in PubMed. Additionally, MG risk miRNAs were downloaded from The
Nervous System Disease NcRNAome Atlas database (http://www.bio-bigdata.net/nsdna/) (19). Validated human miRNA target data
was obtained from the miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php)
(20).
Functional enrichment analysis of MG
risk genes
Pathway data were obtained from the Kyoto
Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/) (21) to dissect numerous specific MG risk
pathways. In order to identify the MG risk pathways in which MG
risk genes were enriched, KEGG pathway enrichment analysis was
conducted by applying functional annotation tools in the Database
for Annotation, Visualization, and Integrated Discovery (DAVID)
(https://david.ncifcrf.gov/) (22). The significance level of KEGG
pathway enrichment was calculated using a cut-off of P<0.05.
Gene Ontology (GO) annotation (23) was additionally performed using
DAVID for the MG risk gene catalog. A GO term was considered
significantly enriched if it had a false discovery rate (FDR) value
<0.05.
Calculation of miRNA distances on
chromosomes and definition of miRNA clusters
The location information of all acquired MG risk
miRNAs on chromosomes were obtained using the NCBI-GENE database
(https://www.ncbi.nlm.nih.gov/gene/)
(18) to obtain the detailed
location of each miRNA, in addition to their starting and ending
points. Subsequently, neighboring miRNAs on the same chromosome,
same long arm or short arm, same region, same band and even the
same sub-band were selected, and the distances between these
adjacent miRNAs were calculated. Therefore, a miRNA cluster was
defined as a number of miRNAs with a relative distance of <6 kb
on the same sub-band, same band, same region and same
chromosome.
Functional enrichment analysis of MG
risk miRNA clusters
Enrichment analyses were performed using DAVID with
the target genes of miRNAs on each MG risk miRNA cluster that had
been defined. The significance level of KEGG pathway enrichment was
calculated using a cut-off of P<0.05. However, the GO enrichment
was considered to be significant using a cut-off of FDR
<0.01.
Results
Update of the MG risk gene catalog and
identification of key risk pathways of human MG risk genes
In our previous study (7), a catalog of 162 MG risk genes was
created and 45 MG risk pathways were identified (P<0.05). Using
our previous study, the catalog of MG risk genes was updated and a
new catalog of 245 MG risk genes, which had been confirmed by the
experiments, was created. Of all 245 genes, 131 risk genes were
collected manually from the literature and 114 risk genes were
downloaded from public databases. In view of these MG risk genes,
99 MG risk pathways (P<0.05) were identified using KEGG
enrichment analysis in DAVID. The same P-value (P<0.05) was
selected as that used in the previous study (7). Compared with the previous study
(7), 83 additional MG risk genes
were excavated and 44 additional MG risk pathways were newly
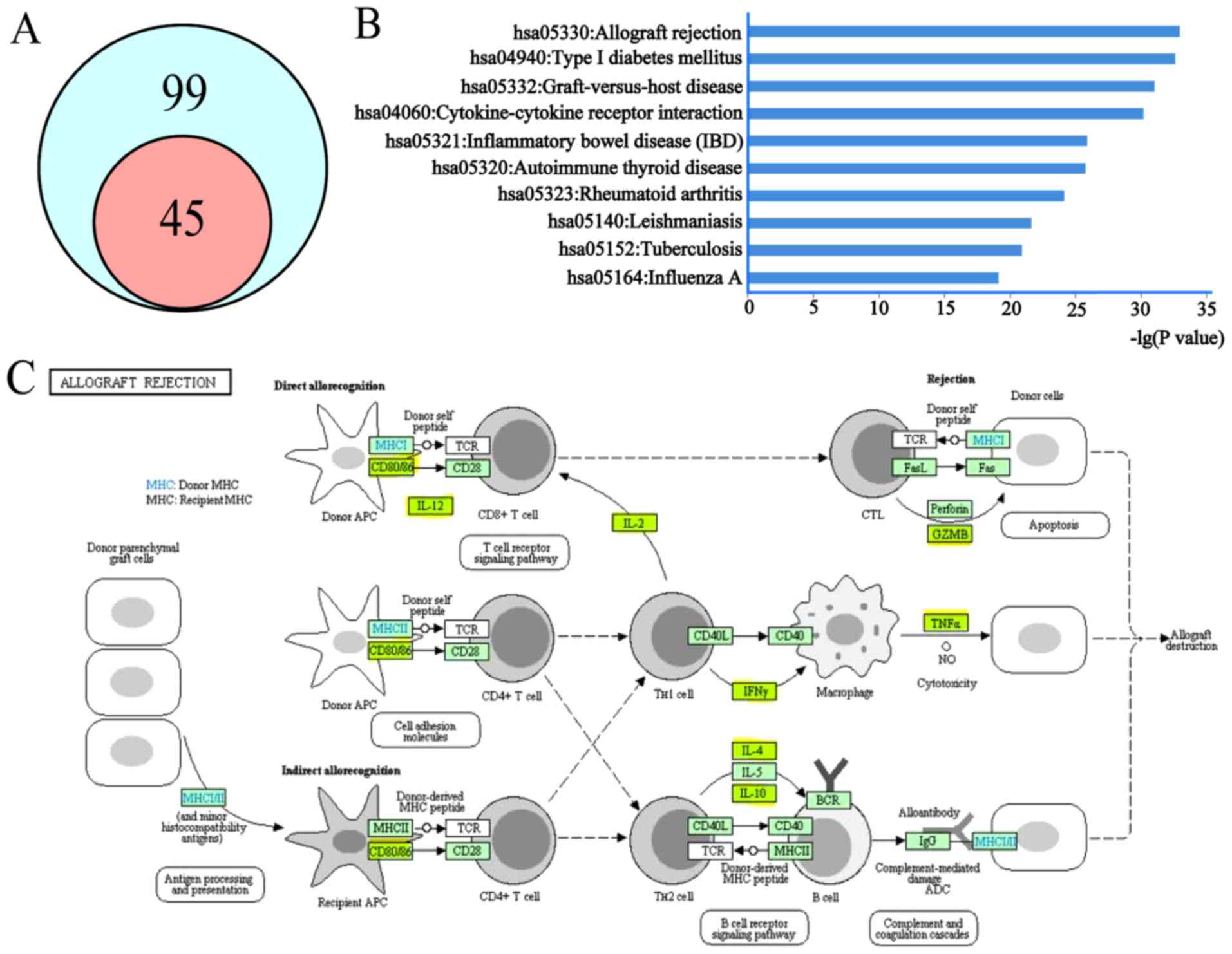
identified. A Wayne diagram was constructed to clarify the
association between pathways that were identified in the present
study and our previous study. As presented in Fig. 1A, it was demonstrated that the
pathways that were enriched in our previous study were all included
in the 99 MG risk pathways identified in the present study. This
was considered an update to our previous findings.
The novel identified top 10 significantly enriched
pathways in the present study are presented in Fig. 1B. It was demonstrated that five of
the top 10 pathways were associated with ‘human disease: Immune
disease’ in the KEGG database, further illustrating the association
between MG and autoimmunity. It was additionally identified that
hsa05330 (allograft rejection) was the most significantly enriched
pathway through KEGG enrichment analysis. A total of eight MG risk
genes were involved in this pathway and important in MG, as
presented in Fig. 1C.
In addition, functional enrichment analysis of these
MG risk genes was performed and 119 GO analysis entries (FDR
<0.05) were obtained, including 92 biological processes (BPs),
11 molecular functions (MFs) and 16 cellular components (CCs). The
top three significant BPs were immune response, inflammatory
response and T cell co-stimulation, further illustrating the
fundamental characteristics of immunity on MG.
Construction of the MG risk miRNA
dataset and its distribution on chromosomes
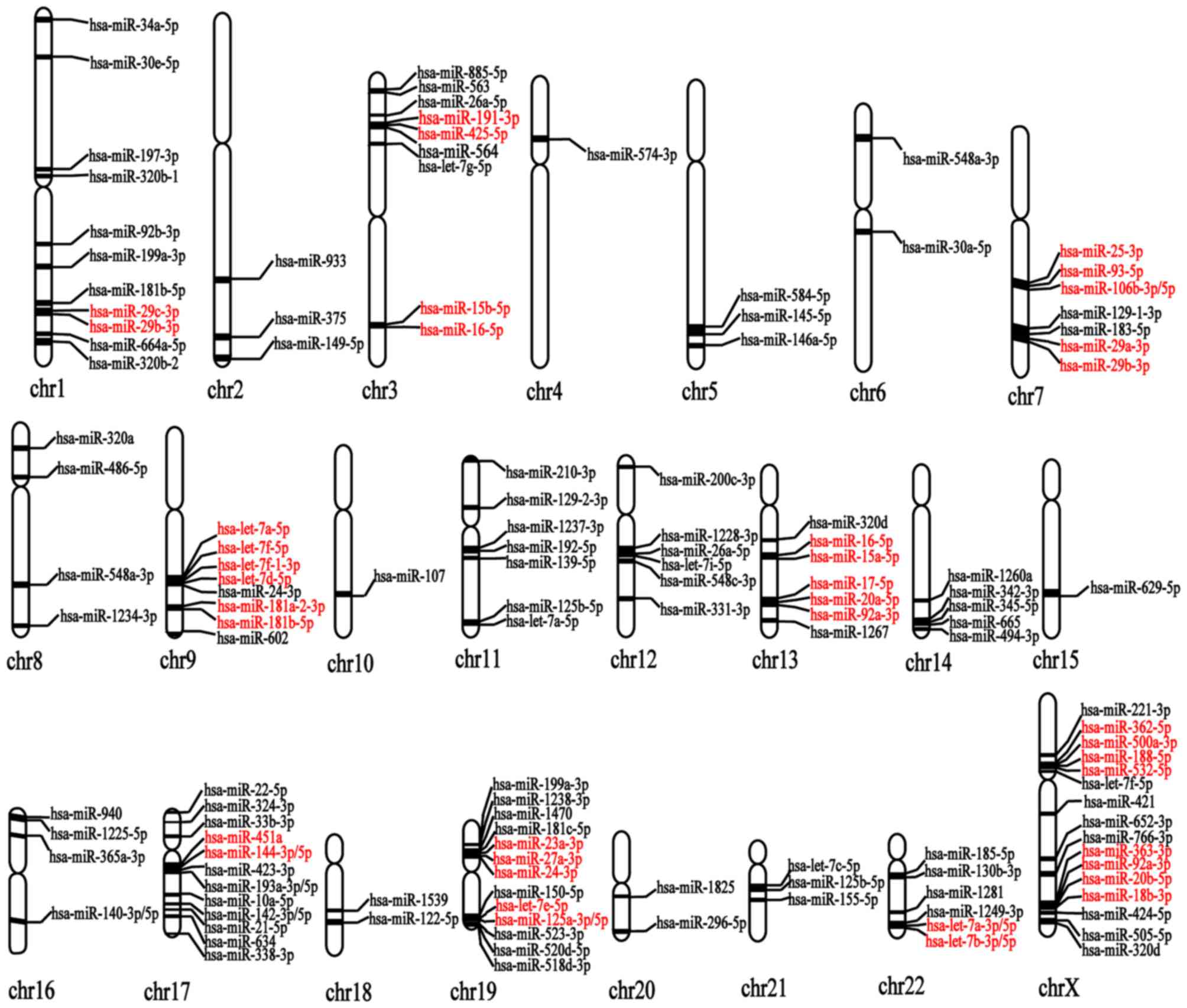
A catalog of 126 miRNAs was constructed and the
positions of each MG risk miRNA on 23 chromosomes were located by
searching information on the mirBase database (17) and NCBI-GENE database (18). The distribution of these 126 MG
risk miRNAs in 23 pairs of chromosomes is presented in Table I. The distribution of MG risk
miRNAs on 23 chromosomes was plotted according to the chromosome
distribution and the position of each miRNA on the chromosomes, as
presented in Fig. 2. It was
demonstrated that the majority of miRNAs are distributed in the X
chromosome, and chromosomes 17, 19, 1, 3, 7, 9 and 22, and the
number of miRNAs on the X chromosome was the largest, with 16
miRNAs on this chromosome. However, no MG risk miRNAs were
identified to be distributed on the Y chromosome, suggesting that
the incidence of MG-related diseases is higher in females compared
with males. This result is consistent with the majority of the
reported proportions of males to females in immunological disease
distribution (24–26).
 | Table I.Distribution of 126 MG risk miRNAs on
23 chromosomes. |
Table I.
Distribution of 126 MG risk miRNAs on
23 chromosomes.
| Chromosome | MG risk miRNAs |
|---|
| 1 | hsa-miR-320b-1,
hsa-miR-197-3p, hsa-miR-30e-5p, hsa-miR-34a-5p, hsa-miR-92b-3p,
hsa-miR-199a-3p, hsa-miR-181b-5p, hsa-miR-29b-3p, hsa-miR-29c-3p,
hsa-miR-664a-5p, hsa-miR-320b-2 |
| 2 | hsa-miR-933,
hsa-miR-375, hsa-miR-149-5p |
| 3 | hsa-let-7g-5p,
hsa-miR-191-3p, hsa-miR-564, hsa-miR-425-5p, hsa-miR-26a-5p,
hsa-miR-563, |
|
| hsa-miR-885-5p,
hsa-miR-15b-5p, hsa-miR-16-5p |
| 4 | hsa-miR-574-3p |
| 5 | hsa-miR-584-5p,
hsa-miR-145-5p, hsa-miR-146a-5p |
| 6 | hsa-miR-30a-5p,
hsa-miR-548a-3p |
| 7 | hsa-miR-25-3p,
hsa-miR-93-5p, hsa-miR-106b-5p, hsa-miR-106b-3p, hsa-miR-129-1-3p,
hsa-miR-183-5p, hsa-miR-29a-3p, hsa-miR-29b-3p |
| 8 | hsa-miR-486-5p,
hsa-miR-320a, hsa-miR-548a-3p, hsa-miR-1234-3p |
| 9 | hsa-let-7a-5p,
hsa-let-7f-5p, hsa-let-7f-1-3p, hsa-let-7d-5p, hsa-miR-24-3p,
hsa-miR-181a-2-3p, |
|
| hsa-miR-181b-5p,
hsa-miR-602 |
| 10 | hsa-miR-107 |
| 11 | hsa-miR-129-2-3p,
hsa-miR-210-3p, hsa-miR-1237-3p, hsa-miR-192-5p, hsa-miR-139-5p,
hsa-miR-125b-5p, hsa-let-7a-5p |
| 12 | hsa-miR-200c-3p,
hsa-miR-1228-3p, hsa-miR-26a-5p, hsa-let-7i-5p, hsa-miR-548c-3p,
hsa-miR-331-3p |
| 13 | hsa-miR-320d,
hsa-miR-16-5p, hsa-miR-15a-5p, hsa-miR-17-5p, hsa-miR-20a-5p,
hsa-miR-92a-3p, |
|
| hsa-miR-1267 |
| 14 | hsa-miR-1260a,
hsa-miR-342-3p, hsa-miR-345-5p, hsa-miR-665, hsa-miR-494-3p |
| 15 | hsa-miR-629-5p |
| 16 | hsa-miR-365a-3p,
hsa-miR-1225-5p, hsa-miR-940, hsa-miR-140-5p, hsa-miR-140-3p |
| 17 | hsa-miR-33b-3p,
hsa-miR-324-3p, hsa-miR-22-5p, hsa-miR-144-3p, hsa-miR-144-5p,
hsa-miR-193a-3p, hsa-miR-193a-5p, hsa-miR-423-3p, hsa-miR-451a,
hsa-miR-10a-5p, hsa-miR-142-3p, hsa-miR-142-5p, |
|
| hsa-miR-21-5p,
hsa-miR-634, hsa-miR-338-3p |
| 18 | hsa-miR-1539,
hsa-miR-122-5p |
| 19 | hsa-miR-24-3p,
hsa-miR-27a-3p, hsa-miR-23a-3p, hsa-miR-181c-5p, hsa-miR-1470,
hsa-miR-1238-3p, hsa-miR-199a-3p, hsa-miR-150-5p, hsa-let-7e-5p,
hsa-miR-125a-3p, hsa-miR-125a-5p, hsa-miR-523-3p, |
|
| hsa-miR-520d-5p,
hsa-miR-518d-3p |
| 20 | hsa-miR-1825,
hsa-miR-296-5p |
| 21 | hsa-let-7c-5p,
hsa-miR-125b-5p, hsa-miR-155-5p |
| 22 | hsa-miR-185-5p,
hsa-miR-130b-3p, hsa-miR-1281, hsa-miR-1249-3p, hsa-let-7a-3p,
hsa-let-7a-5p, |
|
| hsa-let-7b-5p,
hsa-let-7b-3p |
| X | hsa-let-7f-5p,
hsa-miR-188-5p, hsa-miR-362-5p, hsa-miR-500a-3p, hsa-miR-532-5p,
hsa-miR-221-3p, hsa-miR-421, hsa-miR-652-3p, hsa-miR-766-3p,
hsa-miR-18b-3p, hsa-miR-20b-5p, hsa-miR-363-3p, |
|
| hsa-miR-92a-3p,
hsa-miR-424-5p, hsa-miR-320d, hsa-miR-505-5p |
Furthermore, neighboring miRNAs, which are on the
same chromosome, same long arm or short arm, same region, same band
and same sub-band were selected, and the distances between these
adjacent miRNAs were calculated. Subsequently, a miRNA cluster was
defined as a number of adjacent miRNAs with a relative distance of
<6 kb. As a result, 15 miRNA clusters were identified; these are
listed in Table II and these
clusters are marked in red in Fig.
2.
 | Table II.Details of the 15 miRNA clusters
identified with a distance of <6 kb on genomes. |
Table II.
Details of the 15 miRNA clusters
identified with a distance of <6 kb on genomes.
| Chromosome | miRNAs within each
cluster |
|---|
| 1 | hsa-miR-29b-3p,
hsa-miR-29c-3p |
| 3 | hsa-miR-191-3p,
hsa-miR-425-5p |
| 3 | hsa-miR-16-5p,
hsa-miR-15b-5p |
| 7 | hsa-miR-106b-3p,
hsa-miR-106b-5p, |
|
| hsa-miR-93-5p,
hsa-miR-25-3p |
| 7 | hsa-miR-29b-3p,
hsa-miR-29a-3p |
| 9 | hsa-let-7d-5p,
hsa-let-7f-5p, hsa-let-7f-1-3p, |
|
| hsa-let-7a-5p |
| 9 | hsa-miR-181b-5p,
hsa-miR-181a-2-3p |
| 13 | hsa-miR-15a-5p,
hsa-miR-16-5p |
| 13 | hsa-miR-92a-3p,
hsa-miR-20a-5p, |
|
| hsa-miR-17-5p |
| 17 | hsa-miR-144-3p,
hsa-miR-144-5p, |
|
| hsa-miR-451a |
| 19 | hsa-miR-23a-3p,
hsa-miR-27a-3p, |
|
| hsa-miR-24-3p |
| 19 | hsa-miR-125a-3p,
hsa-miR-125a-5p, |
|
| hsa-let-7e-5p |
| 22 | hsa-let-7b-3p,
hsa-let-7b-5p, hsa-let-7a-3p, |
|
| hsa-let-7a-5p |
| X | hsa-miR-532-5p,
hsa-miR-500a-3p, |
|
| hsa-miR-362-5p,
hsa-miR-188-5p |
| X | hsa-miR-92a-3p,
hsa-miR-363-3p, |
|
| hsa-miR-20b-5p,
hsa-miR-18b-3p |
KEGG enrichment analysis of each MG
risk miRNA cluster on 23 chromosomes
Human MG risk miRNA target genes were obtained from
miRTarBase (20). Enrichment
analyses were subsequently performed using the target genes of each
miRNA contained in 15 MG risk miRNA clusters on each chromosome.
For example, there are two clusters on chromosome 3: Cluster
miR-191-3p-425-5p and cluster miR-16-5p-15b-5p. Subsequently, KEGG
enrichment analysis with the target genes of the four MG risk
miRNAs contained in miRNA clusters was performed. A cut-off of
P<0.05 was considered to indicate a statistically significant
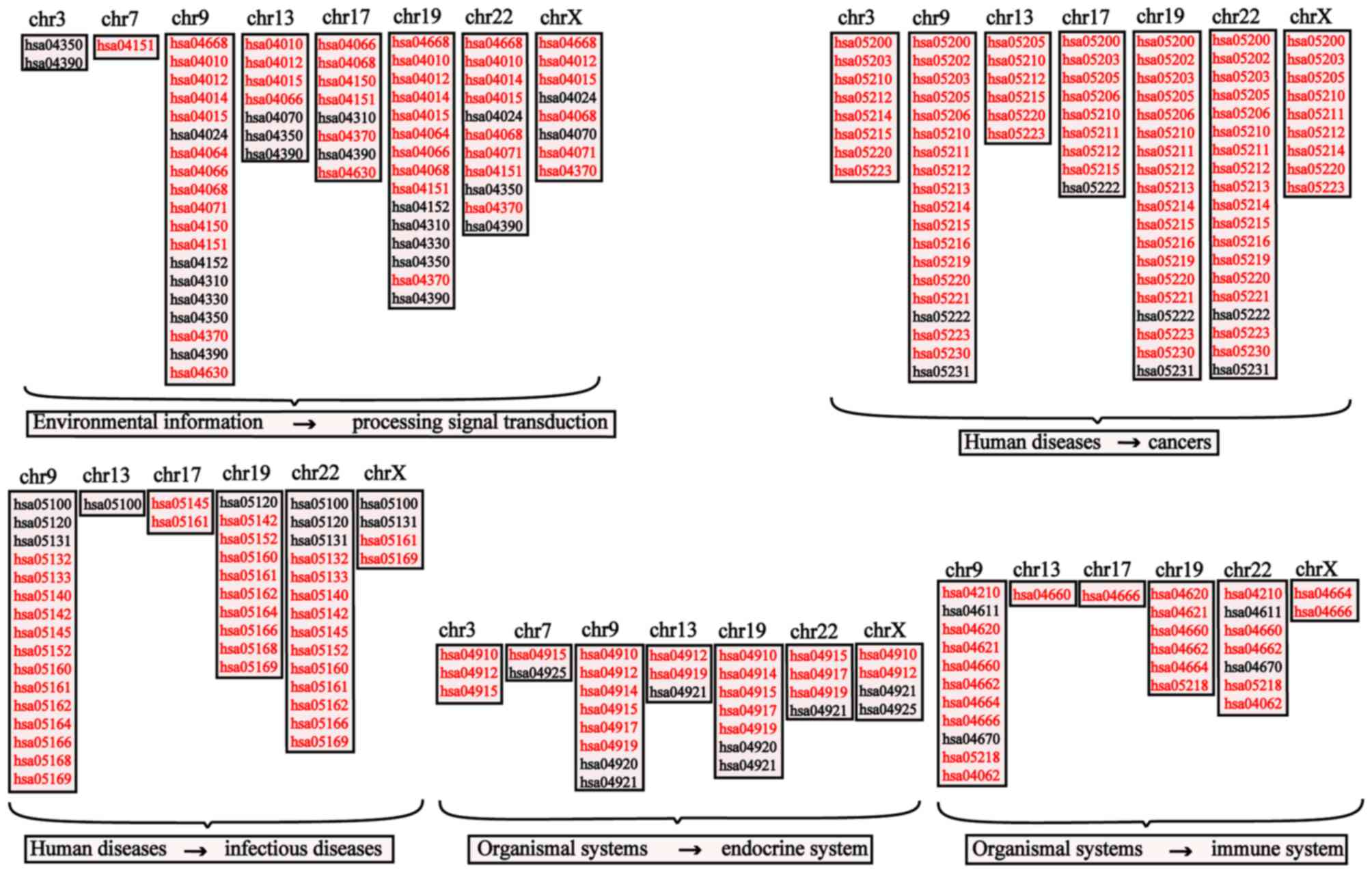
difference. Therefore, numerous significant pathways on chromosome
3 were identified, and a classification of the enriched pathways
was made through KEGG maps. According to the aforementioned steps,
the remaining clusters underwent the same enrichment analysis. The
results of the KEGG enrichment analysis (P<0.05) are presented
in Fig. 3.
In addition, the meaningful pathways (P<0.05)
enriched by MG risk miRNA clusters and the 99 risk pathways
(P<0.05) enriched by MG risk genes in the aforementioned results
were intersected, and the pathways marked in red when enriched in
the two sets of results. The majority of the identified pathways
belonged to the following categories: ‘Environmental information
processing-signal transduction’; ‘Human disease-cancer’; ‘Human
diseases-infectious diseases’; ‘Organismal systems-endocrine
system’; and ‘Organismal systems-immune system’ in the KEGG
database; this suggests that, in addition to the immune system,
cancerous pathways, infectious pathways and endocrine pathways can
also be used to regulate miRNA clusters and thus functionally
characterize MG. The results additionally suggested that the most
significant pathways are enriched on chromosomes 9, 19 and 22,
indicating that the mechanism of MG may be associated with certain
abnormalities of chromosomes 9, 19 and 22.
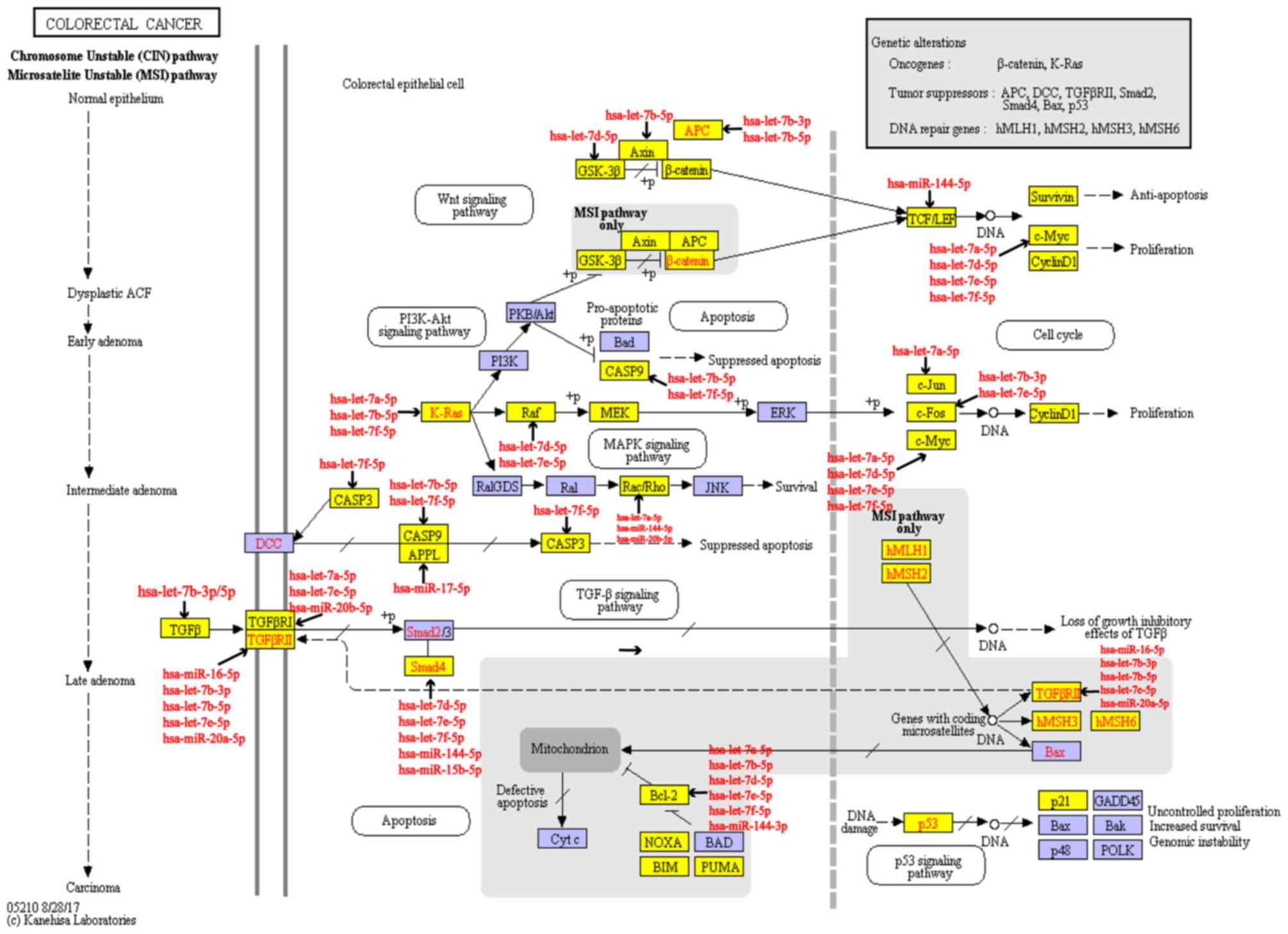
Pathway hsa05210 (colorectal cancer):
A pathway regulated by the majority of MG risk miRNA clusters
A transverse comparison of the pathways of KEGG
enrichment analysis (P<0.05) was subsequently made using the
target genes of each MG risk miRNA cluster. The results
demonstrated that pathway hsa05210 (colorectal cancer) was
regulated by the majority of miRNA clusters (eight miRNA clusters,
including 22 miRNAs). hsa05210 (colorectal cancer) was additionally
a significant pathway that was enriched by the aforementioned MG
risk genes. The miRNA clusters that regulated the hsa05210
(colorectal cancer) pathway are identified in Table III. The hsa05120 (colorectal
cancer) pathway is presented in Fig.
4. The target genes of these eight miRNA clusters are marked in
yellow, and the miRNAs that regulate the target genes are marked in
red, additionally presented in Fig.
4.
 | Table III.miRNA clusters regulating the
hsa05210 (colorectal cancer) pathway. |
Table III.
miRNA clusters regulating the
hsa05210 (colorectal cancer) pathway.
| Chromosome | miRNA clusters |
|---|
| 3 | hsa-miR-16-5p,
hsa-miR-15b-5p |
| 9 | hsa-let-7d-5p,
hsa-let-7f-5p, hsa-let-7f-1-3p, |
|
| hsa-let-7a-5p |
| 13 | hsa-miR-15a-5p,
hsa-miR-16-5p |
| 13 | hsa-miR-92a-3p,
hsa-miR-20a-5p, |
|
| hsa-miR-17-5p |
| 17 | hsa-miR-144-3p,
hsa-miR-144-5p, |
|
| hsa-miR-451a |
| 19 | hsa-miR-125a-3p,
hsa-miR-125a-5p, |
|
| hsa-let-7e-5p |
| 22 | hsa-let-7b-3p,
hsa-let-7b-5p, hsa-let-7a-3p, |
|
| hsa-let-7a-5p |
| X | hsa-miR-92a-3p,
hsa-miR-363-3p, |
|
| hsa-miR-20b-5p,
hsa-miR-18b-3p |
Through analysis, among all the target genes of the
eight miRNA clusters that regulated the hsa05120 pathway,
transforming growth factor β1 (TGFB1), caspase 3 (CASP3), KRAS, MYC
and B-cell lymphoma 2 (BCL2), were MG risk genes. TGFB1 (27,28),
CASP3 (29), KRAS (30), MYC (31) and BCL2 (32) have all been demonstrated to be
associated with miRNA clusters. Overall, hsa05210 (colorectal
cancer) may be a key pathway connecting miRNA clusters with MG.
KEGG enrichment analysis of two MG
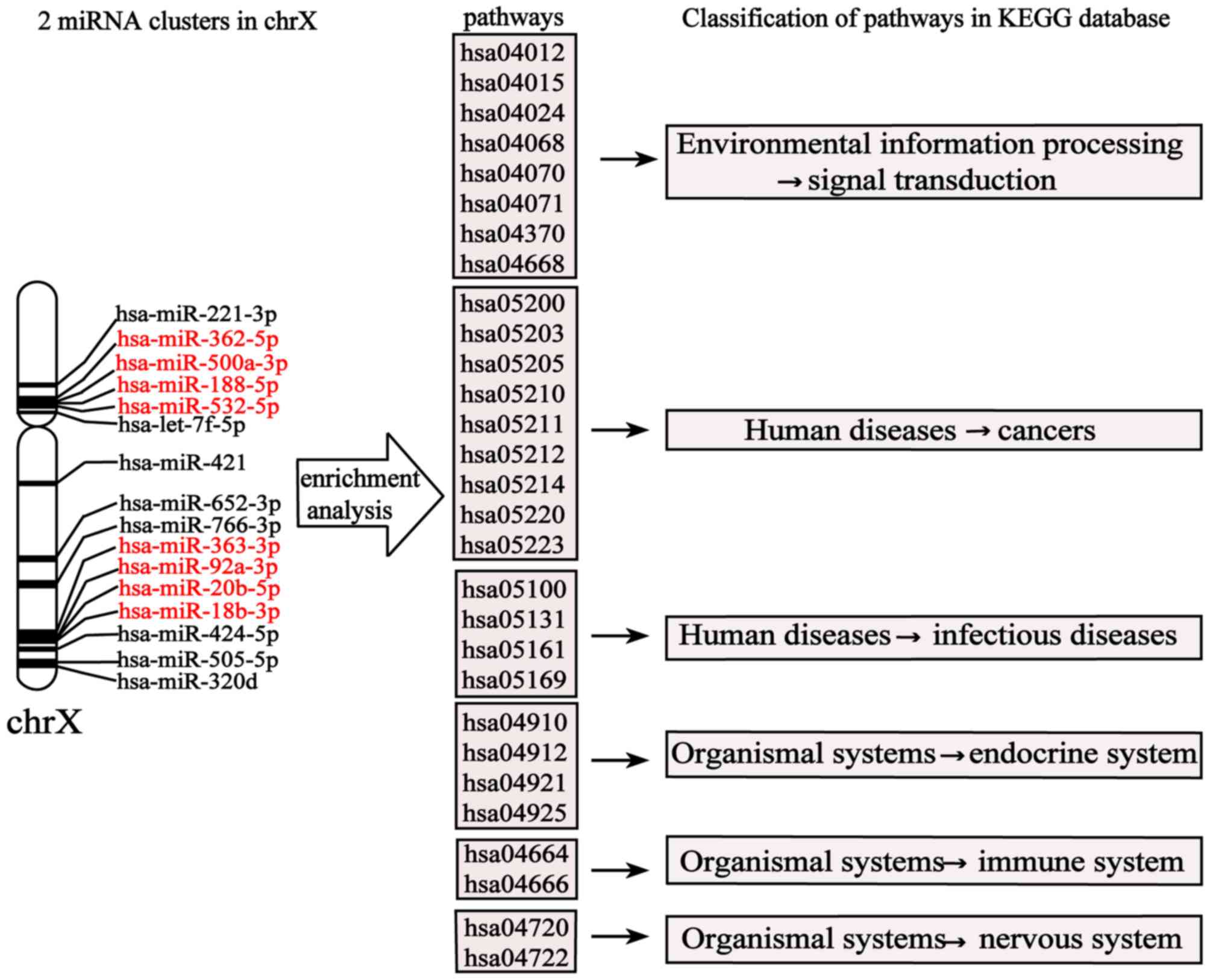
risk miRNA clusters on the X chromosome
As the X chromosome was identified to have the
largest number of MG risk miRNAs and is significant in autoimmune
diseases, the two miRNA clusters on the X chromosome:
Hsa-miR-532-5p, hsa-miR-500a-3p, hsa-miR-362-5p, hsa-miR-188-5p and
hsa-miR-92a-3p, hsa-miR-363-3p, hsa-miR-20b-5p, hsa-miR-18b-3p,
were analyzed. A separate KEGG enrichment analysis (P<0.05) was
performed for the target genes of these two miRNA clusters on the X
chromosome; as a result, 36 pathways were significantly enriched
(P<0.05). Classification of these pathways was performed through
the KEGG database. The results demonstrated that eight pathways
were associated with ‘Environmental information processing-signal
transduction’, nine were associated with ‘Human diseases-cancers’,
four were associated with ‘Human diseases-infectious diseases’,
four were associated with ‘Organismal systems-endocrine system’,
two were associated with ‘Organismal systems-immune system’, and
two were associated with ‘Organismal systems nervous system’, as
presented in Fig. 5. These results
were consistent with the results described for the KEGG enrichment
analysis of target genes of miRNA clusters on 23 chromosomes.
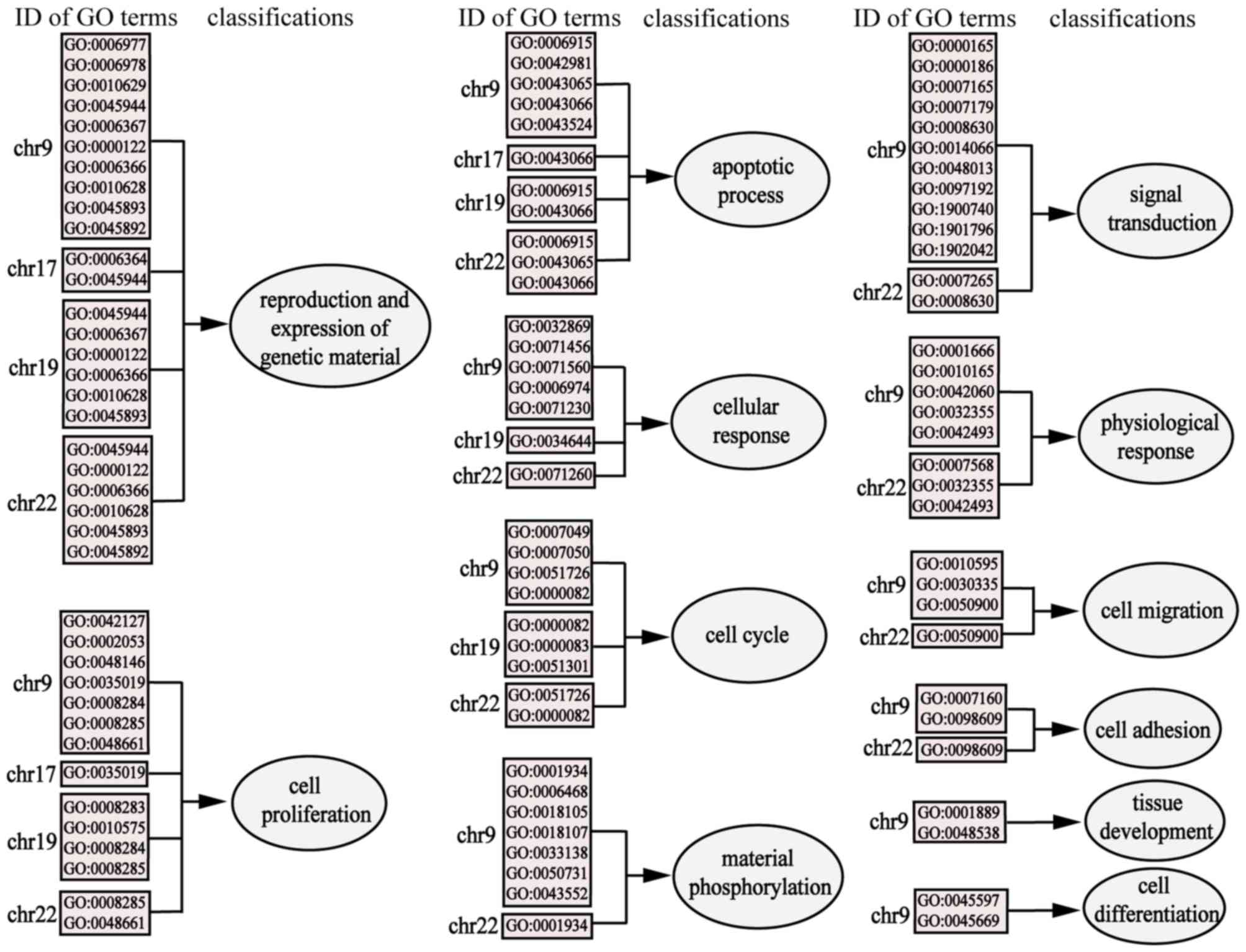
GO enrichment analysis of each MG risk
miRNA cluster on 23 chromosomes
In addition to KEGG analysis, functional enrichment
analysis using the target genes of 15 MG risk miRNA clusters on
each chromosome was performed, and a total of 74 GO_BP terms were
obtained (FDR <0.01). However, with the exception of chromosomes
9, 17, 19 and 22, no enriched pathways were identified on the other
chromosomes. Subsequently, these entries were classified and
divided into the following categories: ‘Apoptotic process’, ‘cell
adhesion’, ‘cell cycle’, ‘cell differentiation’, ‘cell migration’,
‘cell proliferation’, ‘cellular response’, ‘material
phosphorylation’, ‘physiological response’, ‘reproduction and
expression of genetic material’, ‘signal transduction’ and ‘tissue
development’. These results are summarized in Fig. 6. The results of GO_BP enrichment
were additionally concentrated on chromosomes 9, 19 and 22.
Combined with the results of KEGG enrichment analysis, these
results indicated that the pathogenesis of MG may be closely
associated with chromosomes 9, 19 and 22.
Discussion
MG is a neuromuscular autoimmune disease; however,
the specific mechanism of MG remains to be fully elucidated. The
analysis of MG risk miRNA clusters and their regulation of relevant
pathways may assist in elucidating their potential involvement in
the pathogenesis of MG. However, at present, there are few studies
on miRNA clusters and pathways of MG. In the present study, global
analysis of MG-associated miRNA clusters and their potential
mechanism was performed based on the enrichment analysis of MG risk
genes and miRNAs.
MG risk genes were collected and the MG risk
pathways enriched by these genes were identified. Among the top 10
pathways enriched by the MG risk genes, it was identified that the
majority of these pathways were associated with immunity when
searching the KEGG database. The MG risk miRNAs were subsequently
obtained and each of the 126 MG risk miRNAs from each chromosome
were located; 15 miRNA clusters were identified to be significantly
associated with MG. In addition, enrichment analysis was performed
using the target genes of the 15 MG risk miRNA clusters with a
distance of <6 kb on each chromosome. Furthermore, a number of
risk pathways of each of the 15 miRNA clusters were identified and
the most significant pathways were demonstrated to be enriched on
chromosomes 9, 19 and 22. This suggests that the mechanism of MG
may be associated with certain abnormalities of chromosomes 9, 19
and 22.
The top 10 pathways identified using the data of MG
risk genes provides an overview of the pathogenesis of MG. Among
the top 10 MG risk pathways, five were associated with immunity and
three were revealed to be associated with ‘Human diseases:
Infectious diseases’, suggesting that microorganism infection may
be involved in the pathogenesis of MG, providing novel insight into
MG. For example, an active Epstein-Barr virus (EBV) infection in
the thymus of patients with MG has been reported previously,
suggesting that EBV may contribute towards the onset or maintenance
of the autoimmune response by targeting toll-like receptor (TLR)7
and TLR9 in the intrathymic pathogenesis of MG (33). Through pathway analysis, 99
significant pathways were identified that characterized MG in
different aspects. The most significant pathway identified was
hsa05330 (allograft rejection). MG has been reported as a rare
complication of chronic graft-vs.-host disease (GVHD) following
allogeneic hematopoietic stem cell transplantation in several case
reports (34–36), which is consistent with the results
of the present study in which hsa05330 (allograft rejection) had
the most significant association with MG.
miRNA clusters are a novel concept and indicates
that miRNAs are clustered on the human genome to permit
co-regulation. miRNA clusters have the ability to regulate more
biological pathways than normal miRNAs, as one cluster often
contains several miRNAs that have the same biological function. The
present study located each MG risk miRNA on each chromosome and
identified 15 significant miRNA clusters, with distances <6 kb,
associated with MG. Among the 15 miRNA clusters, at least five
miRNA clusters were identified that have been reported to be
associated with immunity, including the miR-25-93-106b cluster
(37), which comprises
hsa-miR-106b-3p, hsa-miR-106b-5p, hsa-miR-93-5p and hsa-miR-25-3p
on chromosome 7; the let-7 family cluster (38), comprising hsa-let-7d-5p,
hsa-let-7f-5p, hsa-let-7f-1-3p and hsa-let-7a-5p on chromosome 9;
the miR-17-92 cluster (39–42),
comprising hsa-miR-92a-3p, hsa-miR-20a-5p and hsa-miR-17-5p on
chromosome 13; the miR-23a-27a-24-2 cluster (14,43–45),
comprising hsa-miR-23a-3p, hsa-miR-27a-3p and hsa-miR-24-3p on
chromosome 19; and the miR-106a-363 cluster (12,13),
comprising hsa-miR-92a-3p, hsa-miR-363-3p, hsa-miR-20b-5p and
hsa-miR-18b-3p on the X chromosome.
The present study identified numerous examples
associated with miRNA clusters that may regulate the immune system.
The miR-25-93-106b cluster has been demonstrated to regulate tumor
metastasis and immune evasion via the modulation of CXCL12 and
programmed death ligand 1 (37).
The miR-17-92 cluster is critical in early B cell development and T
cell differentiation (46,47), and its functions involve autoimmune
disorders, including GVHD and leukemia (39), experimental autoimmune
encephalomyelitis (41), chronic
infections (48) and cancer
(49). The mirn23a gene, which
encodes miR-23a, −24-2 and −27a, transforms into the mirn23a
cluster and is also central to T cell differentiation (44); however, antagonizes B cell
development (45) unlike the
miR-17-92 cluster. The mirn23a cluster was previously reported to
be involved in the pathogenesis of immune-associated diseases,
including hematopoiesis (14),
CKD-induced muscle atrophy (50),
leukemia (51) and a number of
types of carcinoma (52). These
results further support the potential association between miRNA
clusters and immunity, and even the mechanism of MG. Additionally,
10 miRNA clusters that have not been reported previously were
identified through the global analysis of MG risk miRNA clusters in
the present study. These novel identified clusters may provide
novel insight into future experimental studies of miRNA clusters in
MG and immune-associated diseases.
In conclusion, the present study created a catalog
of MG risk genes and miRNAs, acquired MG risk pathways, located
each MG risk miRNA on each chromosome, obtained 15 significant
miRNA clusters associated with MG, and identified the risk pathways
of each of the 15 miRNA clusters. This involved the integration of
a number of single miRNA studies into a global study of miRNA
clusters in MG. As the number of studies on high-throughput data
continues to increase, the results of the present study provide
supporting evidence and offer novel insight for further
investigations on miRNA clusters in the pathogenesis of MG.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81571166, 81771361,
81701190 and 81820108014), the Applied Technique Research and
Development Project of Harbin (grant no. 2016RAXYJ067), the Health
and Family Planning Commission of Heilongjiang Province (grant nos.
2016-052 and 2016-072) and the Fundamental Research Funds for the
Provincial Universities (grant nos. 2017LCZX57, 2017LCZX65 and
2017LCZX48).
Availability of data and materials
All data generated and/or analyzed during the
present study are included in this published article.
Authors' contributions
LW, SN, JW, CB and HZ designed the study. TW, YW and
SL collected the data. CB, HZ, YC, XL, XK, XS and ZL were involved
in the interpretation of data. CB and JW analyzed and visualized
the data. CB drafted the manuscript. SN and LW revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Conti-Fine BM, Milani M and Kaminski HJ:
Myasthenia gravis: Past, present and future. J Clin Invest.
116:2843–2854. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cavalcante P, Barberis M, Cannone M, Baggi
F, Antozzi C, Maggi L, Cornelio F, Barbi M, Dido P, Berrih-Aknin S,
et al: Detection of poliovirus-infected macrophages in thymus of
patients with myasthenia gravis. Neurology. 74:1118–1126. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xie Y, Li HF, Jiang B, Li Y, Kaminski HJ
and Kusner LL: Elevated plasma interleukin-17A in a subgroup of
Myasthenia Gravis patients. Cytokine. 78:44–46. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee JS, Joo IS and Seok JI: Widely varying
TNF-alpha levels in patients with myasthenia gravis. Neurol Sci.
30:259–262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Q, Liu P, Xuan X, Zhang J, Zhang Y, Zhu
Z, Gao F, Zhang Q and Du Y: CCR9 AND CCR7 are overexpressed in CD4
CD8 thymocytes of myasthenia gravis patients. Muscle Nerve.
55:84–90. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang L, Wang J, Sun X, Cao Y, Ning S,
Zhang H, Chen L, Li R, Tian Q, Wang L, et al: Identifying a
polymorphic ‘switch’ that influences miRNAs' regulation of a
myasthenia gravis risk pathway. PLoS One. 9:e1048272014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cao Y, Lu X, Wang J, Zhang H, Liu Z, Xu S,
Wang T, Ning S, Xiao B and Wang L: Construction of an
miRNA-regulated drug-pathway network reveals drug repurposing
candidates for myasthenia gravis. Int J Mol Med. 39:268–278. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xin Y, Cai H, Lu T, Zhang Y, Yang Y and
Cui Y: miR-20b Inhibits T cell proliferation and activation via
NFAT signaling pathway in thymoma-associated myasthenia gravis.
Biomed Res Int 2016. 95957182016.
|
|
9
|
Li J, Qiu D, Chen Z, Du W, Liu J and Mo X:
Altered expression of miR-125a-5p in thymoma-associated myasthenia
gravis and its down-regulation of foxp3 expression in Jurkat cells.
Immunol Lett. 172:47–55. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu XF, Wang RQ, Hu B, Luo MC, Zeng QM,
Zhou H, Huang K, Dong XH, Luo YB, Luo ZH and Yang H: MiR-15a
contributes abnormal immune response in myasthenia gravis by
targeting CXCL10. Clin Immunol. 164:106–113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Leonardo TR, Schultheisz HL, Loring JF and
Laurent LC: The functions of microRNAs in pluripotency and
reprogramming. Nat Cell Biol. 14:1114–1121. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kästle M, Bartel S, Geillinger-Kästle K,
Irmler M, Beckers J, Ryffel B, Eickelberg O and Krauss-Etschmann S:
microRNA cluster 106a~363 is involved in T helper 17 cell
differentiation. Immunology. 152:402–413. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kuppers DA, Schmitt TM, Hwang HC, Samraj
L, Clurman BE and Fero ML: The miR-106a~363Xpcl1 miRNA
cluster induces murine T cell lymphoma despite transcriptional
activation of the p27Kip1 cell cycle inhibitor.
Oncotarget. 8:50680–50691. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kurkewich JL, Hansen J, Klopfenstein N,
Zhang H, Wood C, Boucher A, Hickman J, Muench DE, Grimes HL and
Dahl R: The miR-23a~27a~24-2 microRNA cluster buffers transcription
and signaling pathways during hematopoiesis. PLoS Genet.
13:e10068872017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pinero J, Queralt-Rosinach N, Bravo A,
Deu-Pons J, Bauer-Mehren A, Baron M, Sanz F and Furlong LI:
DisGeNET: A discovery platform for the dynamical exploration of
human diseases and their genes. Database (Oxford) 2015. bav0282015.
View Article : Google Scholar
|
|
16
|
Amberger JS, Bocchini CA, Schiettecatte F,
Scott AF and Hamosh A: OMIM.org: Online mendelian inheritance in
man (OMIM®), an online catalog of human genes and
genetic disorders. Nucleic Acids Res. 43:(Database Issue).
D789–D798. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:(Database Issue). D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brown GR, Hem V, Katz KS, Ovetsky M,
Wallin C, Ermolaeva O, Tolstoy I, Tatusova T, Pruitt KD, Maglott DR
and Murphy TD: Gene: A gene-centered information resource at NCBI.
Nucleic Acids Res. 43:(Database Issue). D36–D42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J, Cao Y, Zhang H, Wang T, Tian Q, Lu
X, Lu X, Kong X, Liu Z, Wang N, et al: NSDNA: A manually curated
database of experimentally supported ncRNAs associated with nervous
system diseases. Nucleic Acids Res. 45:D902–D907. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic Acids Res.
42:(Database Issue). D78–D85. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Smith-Bouvier DL, Divekar AA, Sasidhar M,
Du S, Tiwari-Woodruff SK, King JK, Arnold AP, Singh RR and Voskuhl
RR: A role for sex chromosome complement in the female bias in
autoimmune disease. J Exp Med. 205:1099–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chitnis T and Khoury SJ: Sex influences in
autoimmune disease. Clin Immunol. 149:1692013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu ML, Bakhru P, Conley B, Nelson JS,
Free M, Martin A, Starmer J, Wilson EM and Su MA: Sex bias in CNS
autoimmune disease mediated by androgen control of autoimmune
regulator. Nat Commun. 7:113502016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cao M, Seike M, Soeno C, Mizutani H,
Kitamura K, Minegishi Y, Noro R, Yoshimura A, Cai L and Gemma A:
MiR-23a regulates TGF-β-induced epithelial-mesenchymal transition
by targeting E-cadherin in lung cancer cells. Int J Oncol.
41:869–875. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kitamura K, Seike M, Okano T, Matsuda K,
Miyanaga A, Mizutani H, Noro R, Minegishi Y, Kubota K and Gemma A:
MiR-134/487b/655 cluster regulates TGF-β-induced
epithelial-mesenchymal transition and drug resistance to gefitinib
by targeting MAGI2 in lung adenocarcinoma cells. Mol Cancer Ther.
13:444–453. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Suffert G, Malterer G, Hausser J,
Viiliainen J, Fender A, Contrant M, Ivacevic T, Benes V, Gros F,
Voinnet O, et al: Kaposi's sarcoma herpesvirus microRNAs target
caspase 3 and regulate apoptosis. PLoS Pathog. 7:e10024052011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Naidu S, Shi L, Magee P, Middleton JD,
Lagana A, Sahoo S, Leong HS, Galvin M, Frese K, Dive C, et al:
PDGFR-modulated miR-23b cluster and miR-125a-5p suppress lung
tumorigenesis by targeting multiple components of KRAS and NF-kB
pathways. Sci Rep. 7:154412017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li SG, Shi QW, Yuan LY, Qin LP, Wang Y,
Miao YQ, Chen Z, Ling CQ and Qin WX: C-Myc-dependent repression of
two oncogenic miRNA clusters contributes to triptolide-induced cell
death in hepatocellular carcinoma cells. J Exp Clin Cancer Res.
37:512018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Llobet-Navas D, Rodriguez-Barrueco R,
Castro V, Ugalde AP, Sumazin P, Jacob-Sendler D, Demircan B,
Castillo-Martin M, Putcha P, Marshall N, et al: The miR-424
(322)/503 cluster orchestrates remodeling of the epithelium in the
involuting mammary gland. Genes Dev. 28:765–782. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cavalcante P, Galbardi B, Franzi S,
Marcuzzo S, Barzago C, Bonanno S, Camera G, Maggi L, Kapetis D,
Andreetta F, et al: Increased expression of Toll-like receptors 7
and 9 in myasthenia gravis thymus characterized by active
Epstein-Barr virus infection. Immunobiology. 221:516–527. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Grauer O, Wolff D, Bertz H, Greinix H,
Kuhl JS, Lawitschka A, Lee SJ, Pavletic SZ, Holler E and Kleiter I:
Neurological manifestations of chronic graft-versus-host disease
after allogeneic haematopoietic stem cell transplantation: Report
from the Consensus Conference on Clinical Practice in chronic
graft-versus-host disease. Brain. 133:2852–2865. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Unal S, Sag E, Kuskonmaz B, Kesici S,
Bayrakci B, Ayvaz DC, Tezcan I, Yalnizoglu D and Uckan D:
Successful treatment of severe myasthenia gravis developed after
allogeneic hematopoietic stem cell transplantation with plasma
exchange and rituximab. Pediatr Blood Cancer. 61:928–930. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Heidarzadeh Z, Mousavi SA, Ostovan VR and
Nafissi S: Muscle-specific kinase antibody associated myasthenia
gravis after bone marrow transplantation. Neuromuscul Disord.
24:148–150. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cioffi M, Trabulo SM, Vallespinos M, Raj
D, Kheir TB, Lin ML, Begum J, Baker AM, Amgheib A, Saif J, et al:
The miR-25-93-106b cluster regulates tumor metastasis and immune
evasion via modulation of CXCL12 and PD-L1. Oncotarget.
8:21609–21625. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Punga T, Bartoccioni E, Lewandowska M,
Damato V, Evoli A and Punga AR: Disease specific enrichment of
circulating let-7 family microRNA in MuSK+ myasthenia gravis. J
Neuroimmunol. 292:21–26. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu Y, Heinrichs J, Bastian D, Fu J, Nguyen
H, Schutt S, Liu Y, Jin J, Liu C, Li QJ, et al: MicroRNA-17-92
controls T-cell responses in graft-versus-host disease and leukemia
relapse in mice. Blood. 126:1314–1323. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu D, Bi X, Qu L, Han L, Yin C, Deng J,
Dong Z, Mi QS and Zhou L: miRNA miR-17-92 cluster is differentially
regulated in the imiqumod-treated skin but is not required for
imiqumod-induced psoriasis-like dermatitis in mice. Exp Dermatol.
26:82–84. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
de Kouchkovsky D, Esensten JH, Rosenthal
WL, Morar MM, Bluestone JA and Jeker LT: microRNA-17-92 regulates
IL-10 production by regulatory T cells and control of experimental
autoimmune encephalomyelitis. J Immunol. 191:1594–1605. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Geng XR, Qiu SQ, Yang LT, Liu ZQ, Yang G,
Liu JQ, Zeng L, Li XX, Mo LH, Liu ZG and Yang PC: Allergen-specific
immune response suppresses interleukin 10 expression in B cells via
increasing micro-RNA-17-92 cluster. Cell Biochem Funct. 34:449–454.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chandran PA, Keller A, Weinmann L, Seida
AA, Braun M, Andreev K, Fischer B, Horn E, Schwinn S, Junker M, et
al: The TGF-β-inducible miR-23a cluster attenuates IFN-γ levels and
antigen-specific cytotoxicity in human CD8+ T cells. J
Leukoc Biol. 96:633–645. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cho S, Wu CJ, Yasuda T, Cruz LO, Khan AA,
Lin LL, Nguyen DT, Miller M, Lee HM, Kuo ML, et al: miR-23
approximately 27 approximately 24 clusters control effector T cell
differentiation and function. J Exp Med. 213:235–249. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kurkewich JL, Bikorimana E, Nguyen T,
Klopfenstein N, Zhang H, Hallas WM, Stayback G, McDowell MA and
Dahl R: The mirn23a microRNA cluster antagonizes B cell
development. J Leukoc Biol. 100:665–677. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ventura A, Young AG, Winslow MM, Lintault
L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone
JR, et al: Targeted deletion reveals essential and overlapping
functions of the miR-17 through 92 family of miRNA clusters. Cell.
132:875–886. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jiang S, Li C, Olive V, Lykken E, Feng F,
Sevilla J, Wan Y, He L and Li QJ: Molecular dissection of the
miR-17-92 cluster's critical dual roles in promoting Th1 responses
and preventing inducible Treg differentiation. Blood.
118:5487–5497. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Khan AA, Penny LA, Yuzefpolskiy Y, Sarkar
S and Kalia V: MicroRNA-17~92 regulates effector and memory CD8
T-cell fates by modulating proliferation in response to infections.
Blood. 121:4473–4483. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Olive V, Li Q and He L: mir-17-92: A
polycistronic oncomir with pleiotropic functions. Immunol Rev.
253:158–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang B, Zhang C, Zhang A, Cai H, Price SR
and Wang XH: MicroRNA-23a and MicroRNA-27a mimic exercise by
ameliorating CKD-induced muscle atrophy. J Am Soc Nephrol.
28:2631–2640. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Su R, Dong L, Zou D, Zhao H, Ren Y, Li F,
Yi P, Li L, Zhu Y, Ma Y, et al: microRNA-23a, −27a and −24
synergistically regulate JAK1/Stat3 cascade and serve as novel
therapeutic targets in human acute erythroid leukemia. Oncogene.
35:6001–6014. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li X, Liu X, Xu W, Zhou P, Gao P, Jiang S,
Lobie PE and Zhu T: c-MYC-regulated miR-23a/24-2/27a cluster
promotes mammary carcinoma cell invasion and hepatic metastasis by
targeting Sprouty2. J Biol Chem. 288:18121–18133. 2013. View Article : Google Scholar : PubMed/NCBI
|