Introduction
Chronic or recurring stress is associated with
overproduction of reactive oxygen species (ROS) and reactive
nitrogen species (RNS), highly unstable molecules with unpaired
electrons, leading to oxidative stress and development of
depression through various biological change (1–3).
Oxidative stress causes progressive oxidative damage to lipids,
proteins and DNA in neurons and impairs synaptic function. The
neurons are especially vulnerable to ROS because neuronal activity
increases oxygen utilization for energy production. The neuronal
activity is closely related to brain metabolism and enables the
proper synthesis of neurotransmitters, primarily glutamate and
γ-aminobutyric acid (GABA) (4–6).
Because entry of neuroactive compounds into the brain is highly
restricted by the blood-brain barrier (BBB), these compounds must
be synthesized from glucose (5).
The brain has high energy requirements; excitatory
neurotransmission accounts for most of the energy requirements at
the cortical level. The high demand for energy is mainly achieved
by the production of ATP during metabolism of glucose or oxidative
phosphorylation in the mitochondria (6–8). The
impaired of glucose metabolism results in decreased pyruvate
production, which in turn can lead to mitochondrial DNA (mtDNA)
mutations, and disorders in the mitochondrial respiratory chain
function, mitochondrial defense systems and hence impaired energy
metabolism, and can lead to impaired neuroplasticity and apoptosis
(1). Thus, the brain under chronic
stress show deficits in glucose metabolism, so alternative energy
sources may help to prevent the cell death. Studies showed that
when the brain cannot catabolize glucose efficiently, it may become
reliant upon amino acid oxidation for energy production (9,10).
In the long period of time, effective mechanisms
take place, which entail the transcriptional activation of genes
and gene networks that function to control glucose homeostasis. In
this context, glucose may regulate metabolic genes by modulating
the activity of nuclear factors toward their target genes (11). Thus the molecular basis for
physiological and pathological regulation of glucose metabolism, as
potential adaptive responses in mitochondrial injury, can be
determined at the level of transcription. Due to the restrictive
permeability of the BBB, the brain relies heavily upon glucose
transporters for the delivery of key nutrients (12). Several glucose transporters have
been identified in the brain, of which GLUT3, encoded by
Slc2a3 is of major importance; it ensures that neurons
receive a constant supply of glucose even when interstitial glucose
concentrations are low (13).
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is an enzyme that
plays an important role in energy metabolism; in the glycolysis
process it catalyzes the reversible phosphorylation of
glyceraldehyde-3-phosphate (G3P) to 1,3-bisphosphoglycerate (BPG)
using NAD+ as a cofactor (14). Lactate dehydrogenase (LDH) is the
enzyme catalyzing the final step of the anaerobic metabolic
pathway, glycolysis. LDH is a tetrameric enzyme made up of two
different subunits A and B, encoded by the LDHA and LDHB genes,
respectively. LDHA catalyzes the conversion of pyruvate to lactate
with concomitant inter-conversion of NADH to NAD+, while
LDHB catalyzes the conversion of lactate to pyruvate. Mitochondrial
transcription factor A (TFAM) is a major mtDNA-binding protein that
packages it into nucleoprotein complexes, called nucleoids
(15). TFAM has been shown to
regulate transcription through specific binding to the promoter
region and the copy number of mtDNA (16).
The in vivo studies have shown that the acute
stress changes amino acid metabolism (17,18).
Under chronic stress conditions, the major aim of protein
catabolism is to provide the glucogenic amino acids that serve as
substrates for endogenous glucose production. The glucogenic amino
acids, especially alanine and glutamine, can be converted into
glucose in an enzymatic process of gluconeogenesis. The
gluconeogenesis pathway is normally present in the liver, kidney,
and intestine. However, the emerging reports show an evidence that
gluconeogenic activity can also occur in brain, although the
studies on cerebral gluconeogenesis are limited (19). In the hypermetabolic/stress state,
gluconeogenesis increases dramatically and in proportion to the
degree of the insult to increase the supply of glucose (the major
fuel of reparation). Glucose is the only fuel that can be utilized
by hypoxic tissues (anaerobic glycolysis) (20). This pathway found is becoming more
recognized as an important alternative glucose source for neurons,
specifically in the pathological conditions (19,21).
Amino acids also play an important role as the transmitters of
synaptic excitation and the modulators of synaptic function in the
central nervous system and they can serve a special role as the
precursors for neurotransmitter synthesis (neuroactive amino acids)
(22).
The disruption of pathways of glucose delivery and
metabolism in cells exposed to chronic stress may lead to
debilitation of brain disease through various biological changes;
however, molecular mechanism and cerebral gluconeogenesis in
depression are not established. In this study, we evaluated the
changes in expression of selected genes involved in glycolytic
pathway and the levels of glucogenic and neuroactive amino acids in
the brain of rats exposed to chronic variable stress (CVS). In an
attempt to mimic the excessive human day-to-day stress, several
animal models have been developed. CVS, a well-validated animal
model, has been used widely for studying clinical depression. The
method, which multiple stressors relies on the ability of a
sequence of relatively mild stressors, may lead to behavioral
changes. According to in vivo studies, CVS model of
depression, based on a 40 day treatment procedure with variables
stressors involved, has high validity, since a large number of
recent publications have confirmed that CVS induces behavioral and
neurochemical changes in animals that are similar to the symptoms
and presumed neurochemical changes accompanying depression in
humans (4,23,24).
This protocol, unlike other protocols of chronic stress which use
only one stressor, uses variable stressors, which prevents
adaptation to stress. Studies have shown that 40-day variable
stress causes alterations in feeding behavior and disturbs
neurotransmission in the rodent brain (23). Moreover, it was proved that animals
submitted to CVS may present some effects, such as increased
adrenals after just 2 weeks of stress, while behavioral and
neurochemical changes here may need more time to develop (23).
Materials and methods
Animals
The experiment was performed using male Wistar rats
which were approximately 6 weeks old and weighed 200–250 g at the
time of arrival. The rodents were purchased from a licensed breeder
(Brwinów, Poland) and were housed in standard rectangular
polypropylene cages with free access to standard diet and water.
The standard laboratory conditions included maintaining a constant
temperature (20±2°C) and a 12 h day/12 h night cycle as well as
constant environment (humidity, noise). The study design was
approved by the Local Ethics Committee on Animal Experimentation of
the Medical University of Lublin (no. 12/2015). The procedures were
also performed in accordance with obligatory Polish and European
standards related to the experimental studies on animals (Act from
January 15, 2015 on the Protection of Animals Used for Scientific
or Educational Purposes; Directive 2010/63/eu of the European
Parliament and of the council of 22 September 2010 on the
protection of animals used for scientific purposes).
CVS model of depression
A CVS protocol was based on the description of
Gamaro et al having regard to a few modifications (4,23). A
variable-stressor paradigm was used in the study because the
different stressors used diminish adaptation to stress (4). To avoid predictability, rats were
exposed to these stressors not at the same time each day. A
specification has been presented in detail in part of our published
study (2). In brief, two groups of
rats, each consisted of 10 randomly selected animals, were used in
the study. Rodents in the control (CTL) group were kept undisturbed
in their colony cages (5 rats in each cage of dimensions 65×25 cm,
18 cm-high), while those of the stressed group (CVS) were subjected
to different external stressors for 40 days. The procedure implied
using the following stress agents: food deprivation lasting 24 h,
water deprivation lasting 24 h, restraint lasting 1–3 h at room
temperature or 1.5–2 h at 4°C, forced swimming for 10 or 15 min,
flashing light for 120–210 min, and isolation (2–3 days). The names
of stressors including time of application are presented in
Table I. In the end of the
experiment, the rats were sacrificed by decapitation at 24 h
following the last stressor action. The brain sample from each
animal was obtained, washed with 20 ml of saline and stored at
−75°C until use for further analyses.
 | Table I.Stress factors applied during the
chronic variable stress procedure. Data adapted from Herbet et
al (2). |
Table I.
Stress factors applied during the
chronic variable stress procedure. Data adapted from Herbet et
al (2).
| Day | Stress factor | Time of
duration |
|---|
| 1 | Water
deprivation | 24 h |
| 2 | Food
deprivation | 24 h |
| 3 | Isolation | 24 h |
| 4 | Isolation | 24 h |
| 5 | Isolation | 24 h |
| 6 | Flashing light | 3 h |
| 7 | Food
deprivation | 24 h |
| 8 | Forced
swimming | 10 min |
| 9 | Restraint | 1 h |
| 10 | Water
deprivation | 24 h |
| 11 | No stressor | – |
| 12 | No stressor | – |
| 13 | Restraint and
cold | 2 h |
| 14 | Flashing light | 2.5 h |
| 15 | Food
deprivation | 24 h |
| 16 | Forced
swimming | 15 min |
| 17 | Isolation | 24 h |
| 18 | Isolation | 24 h |
| 19 | Isolation | 24 h |
| 20 | Water
deprivation | 24 h |
| 21 | Food
deprivation | 24 h |
| 22 | Flashing light | 3 h |
| 23 | Restraint | 2 h |
| 24 | Isolation | 24 h |
| 25 | Isolation | 24 h |
| 26 | Restraint and
cold | 1.5 h |
| 27 | Forced
swimming | 10 min |
| 28 | Flashing light | 3.5 h |
| 29 | No stressor | – |
| 30 | Food
deprivation | 24 h |
| 31 | Restraint | 3 h |
| 32 | Flashing light | 2 h |
| 33 | Water
deprivation | 24 h |
| 34 | Restraint and
cold | 2 h |
| 35 | Forced
swimming | 15 min |
| 36 | Isolation | 24 h |
| 37 | Isolation | 24 h |
| 38 | No stressor | – |
| 39 | Flashing light | 3 h |
| 40 | Forced
swimming | 10 min |
Forced swimming test
Forced swim test (FST) was performed according to
Porsolt et al (25), in
order to confirm the ability of stressors an indicative of
depressive behavior. Rats were individually placed in a glass
cylinder (40 cm tall, 25 cm in diameter) containing water at
22–23°C to a height of 30 cm. Water in the tank was changed after
each rat swimming test section. According to the procedure, for the
first exposure, rats were placed in the water for 15 min, so in our
experiment, the last stressor of CVS counted as pre-test session.
Twenty-four h later, rodents were placed in the water again for a 5
min test session, and the immobility time was recorded.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
The samples of isolated hippocampus were rinsed with
20 µl of saline and stored at −75°C before the process of isolation
of RNA. According to the manufacturer's instructions, the nucleic
acid was separated from 30 mg of tissue using Syngen Tissue RNA
Mini kit (Syngen Biotech, Poland) and reverse transcription was
performed using NG dART RT-PCR kit (EURx, Poland). The assessment
of the chosen genes expression (Slc2a3, GAPDH, Ldha, Ldhb
and Tfam) was performed using a quantitative real-time PCR
(qPCR) method. The relative expression of genes was measured with
the ΔΔCt method, using Hprt (Mn00446968_m1) as an endogenous
control. The reaction was carried out in octuplicate by q-PCR using
the SmartChip Real-Time PCR System (WaferGen Biosystems, Inc.,
Fremont, CA, USA), and TaqMan Fast Universal PCR Master Mix (2X)
(Applied BioSystems; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) according to manufacturer's instructions. Sample quality
screening based on amplification, Tm, and Ct values was performed
to remove any outlier data points before ΔΔCt calculation and to
determine fold change in mRNA levels. The data was presented as RQ
value (RQ=2−ΔΔCt).
Amino acids analysis
Reagents used to prepare ninhydrin solution:
Ninhydrin, hydrindantine dihydrate, ethylene glycol monomethyl
ether, 4 M acetate buffer pH 5.6. Reagents used for preparation of
lithium-based buffers are: citric acid monohydrate, lithium citrate
tetrahydrate, lithium chloride, thiodiglycol, sodium azide and
lithium hydroxide monohydrate. Physiological amino acid standard,
Asn+Gln standard and all reagents mentioned above (used for amino
acid analysis) were obtained from Ingos (Prague, Czech Republic).
Sulphosalicylic acid (SSA) were purchased from POCH S.A. (Gliwice,
Poland).
To assess the concentrations of the glucogenic and
neuroactive amino acids, approximately 100 mg of prefrontal cortex
of rat brain from each animal from each group were deproteinized
and homogenized in 6% SSA in lithium citrate buffer (pH 2.8) in
1:10 ratio. The homogenised samples were centrifuged 20 min at
12,000 rpm. The concentration of free amino acids in the obtained
supernatants was determined using an automatic analyzer Ingos AAA
500. Amino acids were separated by the ion-exchange chromatography
using analytic column Ostion LG FA (Ingos) with five lithium-based
buffers (pH 2.6, 3.1, 3.35, 4.05 and 4.65). The original software
Clarity version 6.1.0.130 (DataApex Ltd., Prague, Czech Republic)
was used to estimate the concentrations of free amino acids
(26).
Statistical analysis
Data was analyzed by STATISTICA v. 10 application
(StaftSoft, Cracow, Poland). The results are presented as mean ±
SEM and expressed as percentage of CTL group. Comparisons between
groups were done using an unpaired Student's t-test and P<0.05
was considered to indicate a statistically significant
difference.
Results
Body weight
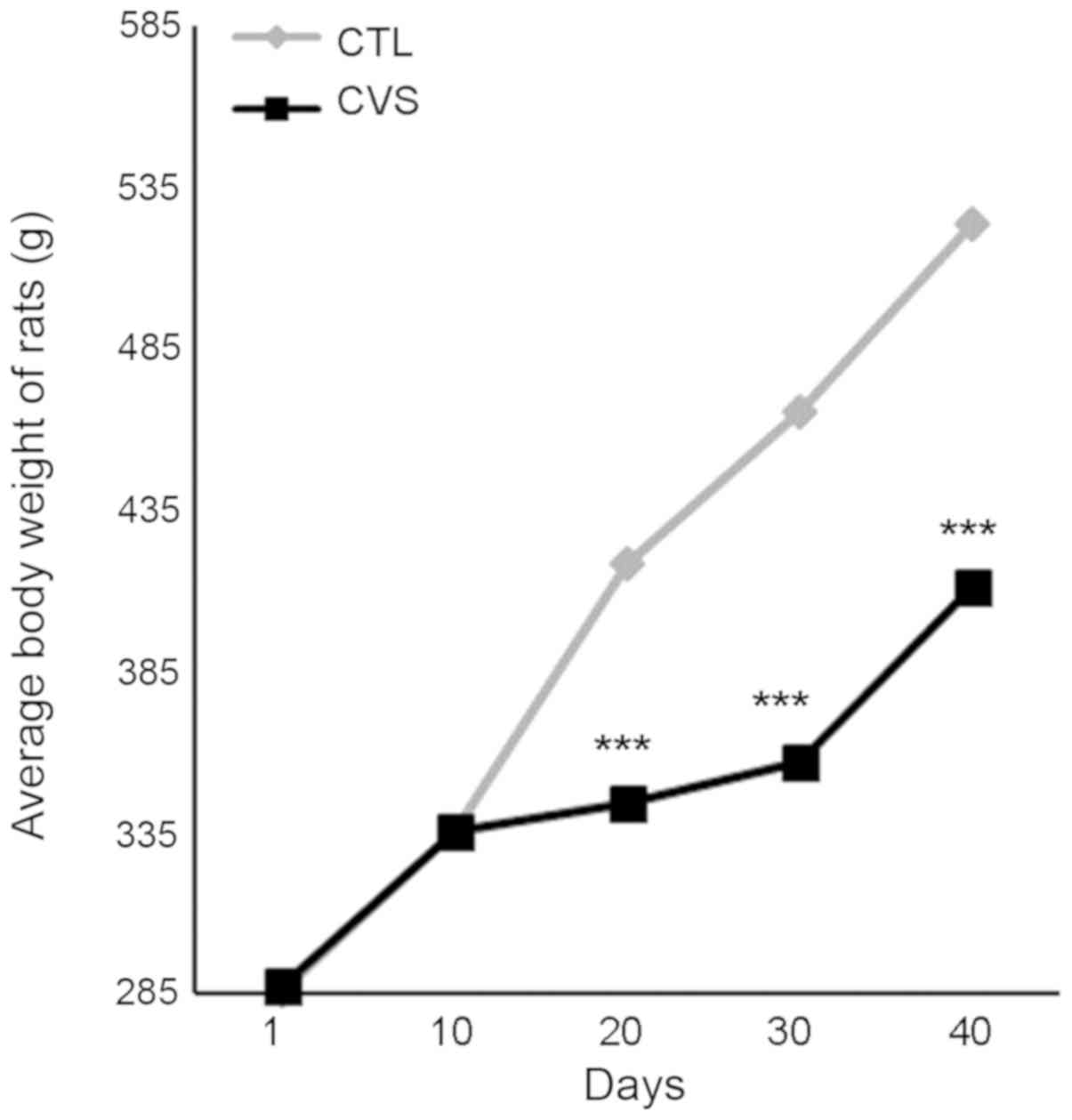
Body weight was measured at 1, 10, 20, 30 days of
the procedure and at the end of the experiment. Our study indicated
that stressed rats have presented a lower body weight than
unstressed [day 1: CTL, 285.7±2.504 g; CVS, 287.0±5.175 g; P=0.823;
t=0.226 (18); F=4.272; day
10: CTL, 335.00±2.687 g; CVS, 335.0±6.708 g; P>0.999; t=0.00
(18); F=6.231; day 20: CTL,
418.00±4.89 g; CVS, 344.00±4.76 g; P<0.001; t=10.832 (18); F=1.059; day 30: CTL, 465.00±7.03 g;
CVS, 357.00±6.50 g; P<0.001; t=11.273 (18); F=1,168; day 40: CTL, 524.5±10.71 g;
CVS, 411.0±7.219 g; P<0.01; t=8.788 (18); F=2.201]. The effect of CVS on body
weight is shown in Fig. 1.
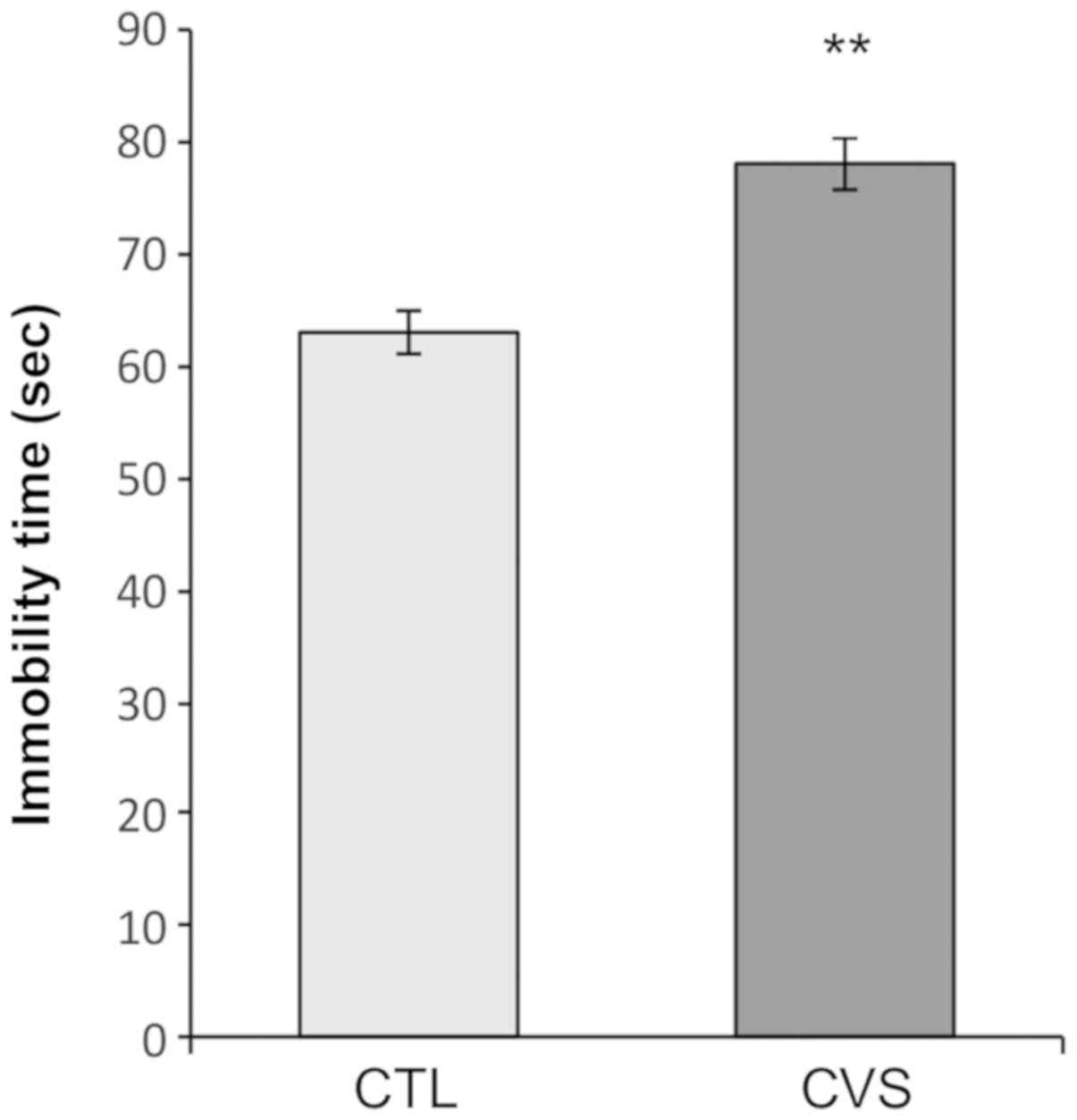
Forced swimming test
After 40 days of stress procedure, forced swimming
test was performed. As shown in Fig.
2, immobility time was increased in animals exposed to CVS
conditions, when compared to the CTL group, indicating that these
animals have presented depressive behavior [CTL, 63±1.88 sec; CVS,
78±2.376 sec; P<0.01; t=4.951 (18); F=1.597].
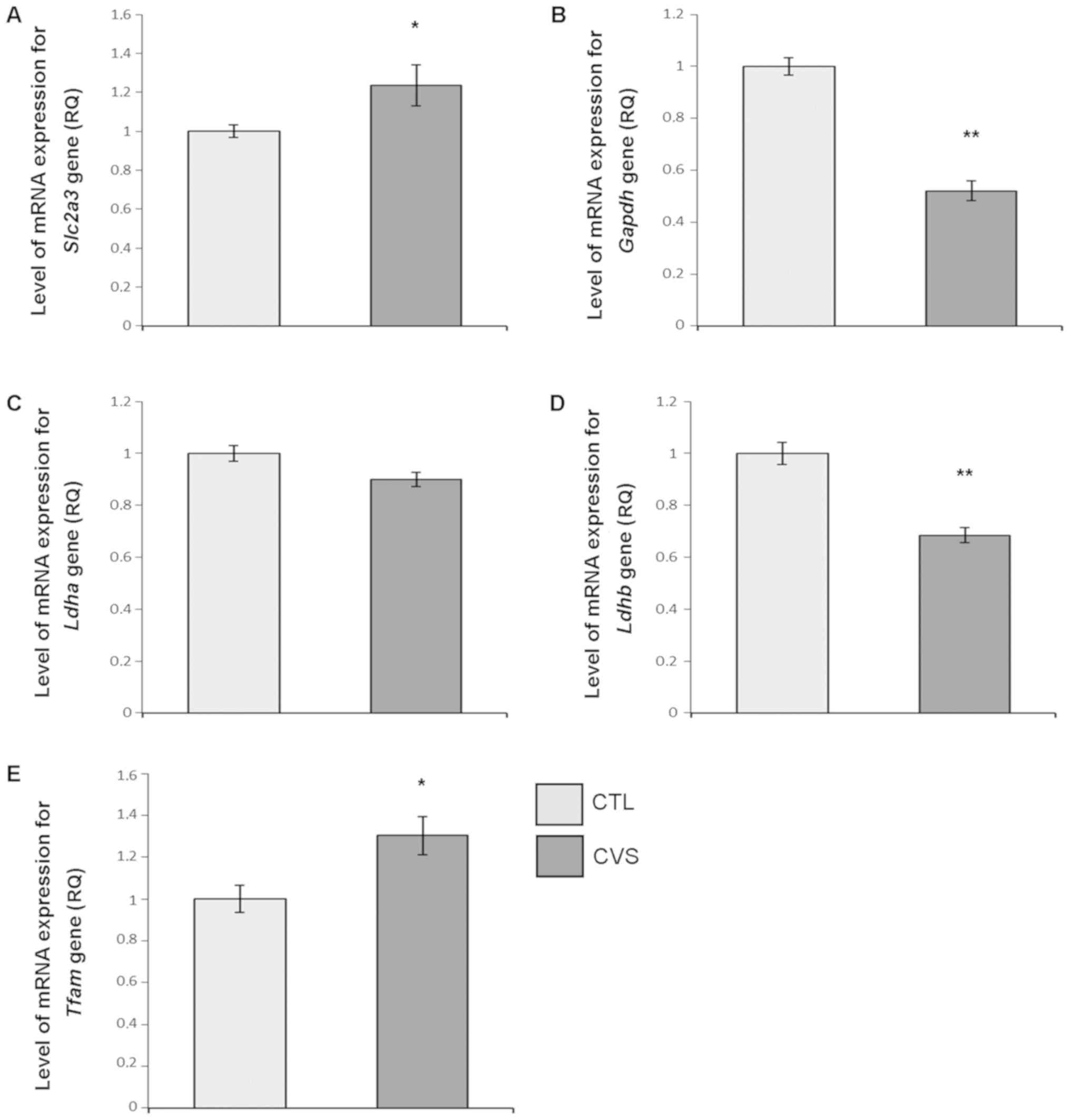
The level of mRNA expression
The expression levels of selected genes involved in
brain glucose metabolism were examined (Table II). Sample variation was accounted
for by comparison to expression levels of Hprt, which is a
housekeeping gene responsible for metabolism of nucleotide.
Expression of mRNA was measured in reference to the CTL group,
where the expression level is estimated as RQ=1. The mRNA levels of
Slc2a3 [CTL, 1.0±0.057; CVS, 1.236±0.104; P<0.05;
t=2.1 (57); F=2.522] and
Tfam [CTL, 1.0±0.064; CVS, 1.303±0.94; P<0.05;
t=2.583 (60); F=2.285]
were significantly increased, while an decrease in the expression
of Ldhb [CTL, 1.0±0.043; CVS, 0.685±0.028; P<0.01;
t=5.533 (60); F=1.904] and
GAPDH [CTL, 1.0±0.045; CVS, 0.52±0.038; P<0.01;
t=7.531 (59); F=2.017] was
noticed in the hippocampus of rats submitted to CVS in comparison
to the CTL. There were no changes in Ldha mRNA level as
compared to CTL [CTL, 1.0±0.048; CVS, 0.9±0.027; P=0.870
t=1.739 (61); F=4.083].
The results of qPCR experiments are shown in Fig. 3.
 | Table II.Description of the primers used in
this study for reverse transcription-quantitative polymerase chain
reaction. |
Table II.
Description of the primers used in
this study for reverse transcription-quantitative polymerase chain
reaction.
| Gene | Primers
(5′-3′) | Accession
number |
|---|
| Slc2a3 |
| NM_017102.2 |
| Forward
primer |
ATGGGGACAGCGAAGGTGA |
|
| Reverse
primer |
CCCAAGGATGGCAATCAGGT |
|
| GAPDH |
| NM_017008.4 |
| Forward
primer |
AGTGCCAGCCTCGTCTCATA |
|
| Reverse
primer |
GATGGTGATGGGTTTCCCGT |
|
| Ldha |
| NM_017025.1 |
| Forward
primer |
ATCTGGATTCGGCTCGGTTC |
|
| Reverse
primer |
AACACAACTGGACCAACTGGA |
|
| Ldhb |
| NM_012595.2 |
| Forward
primer |
TTGTCTGGACAAGATGGCAAC |
|
| Reverse
primer |
TGCCGTACATTCCCTTCACC |
|
| Tfam |
| NM_031326.1 |
| Forward
primer |
ATCTCATCCGTCGCAGTGTG |
|
| Reverse
primer |
CTTCACAAACCCGCACGAAA |
|
The level of amino acids
The automatic amino acid analyzer system was
calibrated with an intact standard of amino acids. Amino acids in
standard were at 2.5 µmol/ml in 0.1 N HCl, except L-cystine at 1.25
µmol/ml. The concentrations of amino acids were determined on the
basis of a three-point curve. The analysis showed a statistically
significant decrease in the concentration of assayed amino acids in
the prefrontal cortex of rats subjected to chronic stress. Detailed
data are presented in Table
III.
 | Table III.The concentration of G and N amino
acids in the prefrontal cortex of rats submitted to chronic
variable stress. Data is displayed as mean ± SEM. |
Table III.
The concentration of G and N amino
acids in the prefrontal cortex of rats submitted to chronic
variable stress. Data is displayed as mean ± SEM.
|
| Concentration
(µmol/ml) x ± SEM |
|
|
|---|
|
|
|
|
|
|---|
| Amino acid | CTL | CVS | t | F |
|---|
| Alanine (ALA) | 0.58±0.07 |
0.21±0.01a | 5.64 (25) | 18.19 |
| G N |
| Arginine (ARG) | 0.08±0.01 |
0.03±0.002a | 5.19 (25) | 17.80 |
| G
N |
| Aspargine
(ASN) | 1.69±0.21 |
0.53±0.03a | 5.86 (25) | 24.71 |
| G |
| Cysteine (CYS) | 0.02±0.002 |
0.01±0.0007a | 3.88 (25) | 9.82 |
| G |
| γ-amino-butylic
acid (GABA) | 2.42±0.27 |
1.31±0.03a | 4.50 (25) | 44.39 |
| N |
| Glutamate
(GLU) | 9.98±0.98 |
6.07±0.31a | 4.55 (30) | 5.75 |
| G
N |
| Glutamine
(GLN) | 4.31±0.54 |
2.04±0.13a | 4.50 (25) | 13.27 |
| G
N |
| Glycine (GLY) | 0.75±0.09 |
0.2±0.01a | 5.95 (25) | 23.03 |
| G
N |
| Histidine
(HIS) | 0.06±0.006 |
0.02±0.001a | 7.49 (25) | 29.63 |
| G |
| Methionine
(MET) | 0.04±0.005 |
0.01±0.001a | 5.75 (25) | 13.67 |
| G |
| Proline (PRO) | 0.13±0.009 |
0.11±0.002a | 2.68 (24) | 9.87 |
| G |
| Serine (SER) | 0.77±0.09 |
0.26±0.02a | 5.48 (25) | 9.87 |
| G
N |
| Taurine (TAU) | 5.86±0.63 |
2.64±0.11a | 5.59 (25) | 25.63 |
| N |
| Tyrosine (TYR) | 0.07±0.003 |
0.02±0.001a | 12.89 (25) | 3.08 |
| N |
| Valine (VAL) | 0.07±0.007 |
0.02±0.001a | 6.54 (25) | 12.01 |
| G |
Discussion
After subjecting the animals to 40 days of CVS, we
acknowledged a depressive-like state. Our findings demonstrated
that the weight gain of the rats was significantly slower during
CVS as compared to non-stressed rats. Literature data support the
fact that stressed rats have a severe loss of body weight as well
as behavioral alterations (23,27).
Emotional changes, such as exposure to stress situations can
influence feeding behavior, and several studies have demonstrated
that chronic exposure to stressors may alter the body weight of
rats (4,28). Likewise, the observed increase in
the immobility time in stressed animals in the forced swimming
test, currently the most popular animal model in antidepressant
drug screening, which is also used in modelling of depression,
suggests that these animals presented depressive behavior.
In the present study, we evaluated the changes in
expression of selected genes involved in glycolytic pathway and the
levels of glucogenic and neuroactive amino acids in the brain of
rats exposed to CVS. We used hippocampus for molecular tests and
prefrontal cortex to determine the levels of glucogenic and
neuroactive amino acids because depressed patients present
alterations in these cerebral structures and relevant research most
often relates to these regions of the brain. Evidence has emerged
in the past decade that depression is associated with small
hippocampal volumes and that this structure of the brain is
particularly associated with stress-related depression (29). Thus, for the molecular tests have
been used the hippocampus-a brain structure particularly associated
with depression. A disturbed metabolism in the prefrontal cortex,
especially dorsolateral and dorsoventral brain regions, is a
frequently replicated finding in depression (30). Thus, this brain structure has been
used to determine the level of amino acids.
Given its high metabolic demands and negligible
intrinsic energy stores, the brain depends upon a continuous influx
of substrates from the blood. Glucose is the main fuel for most
cells, and its transport is tightly regulated. The current study
revealed that in the hippocampus of rats subjected to CVS, the mRNA
level of Slc2a3 was significantly increased as compared to
CTL. These results are consistent with the hypothesis that
overexpression of GLUT3 results from an increased demand of the
cell for glucose (31). Detka
et al (32) showed that
chronic stress enhances the concentration of glucose transporters
in the brain, especially in the hippocampus. These transporters are
always saturated with glucose, even in states of mild
hypoglycaemia, which ensures delivery of the right amount of
substrate to neurons. The pathological changes such as hypoxia,
chronic hypoglycemia and starvation have been shown to induce GLUT3
protein in the immature rat brain and an increase of GLUT3 mRNA in
the mouse brain has also been demonstrated (33). Thus, the expression of glucose
transporters can be raised in the brain as a response to an
enhanced demand for glucose resulting in the support of energy
metabolism. The research has shown that stress events have a
significant impact on ROS generation in brain and it can cause an
increase in energy demand (2,34).
Repeated and unpredictable stress situations increase ROS
generation, which can damage a variety of cell macromolecules,
including those that constitute the electron transport system,
therefore disrupting mitochondrial function (2,34,35).
It is possible that in chronic stress, we can observe a disturbance
in energy production due to mitochondrial damage by excess ROS,
which may activate signaling pathways involved in cellular
adaptation to various types of stress. One of these pathways is the
stimulation of glucose uptake (36). The increase in GLUT3 expression
associated with increased cerebral glucose utilization provides
further confirmation of the central role of GLUT3. There are also
assumptions that glucose uptake plays a role in regulating the
balance between ROS production and scavenging, suggesting that
glucose uptake must be tightly controlled to maintain cellular
energy homeostasis and redox status (36).
Studies have indicated that mRNA levels of GAPDH are
highly regulated in a variety of cell types in certain metabolic
conditions (37–39). In our study, a significant decrease
in mRNA levels of GAPDH was observed in the hippocampus of
rats submitted to chronic stress in comparison to the CTL group.
However, considering the fact that during stress there is an
increased need for glucose, an increase in glycolysis would be
expected. NAD+ is required to enable the sixth step of
glycolysis and it is usually regenerated through oxidative
phosphorylation by the electron transport chain. When the oxygen
supply is restricted or mitochondrial function is reduced by ROS,
NAD+ is regenerated from NADH by LDHA in order to
maintain glycolysis, generating lactate as a by-product. It should
also be emphasized that when cells are exposed to ROS, they need
excessive amounts of the antioxidant cofactor NADPH, which is
reduced from NADP+. In this way, the ROS excess can
cause inactivation of GAPDH. This inactivation temporally routes
the metabolic flux from glycolysis to the pentose phosphate
pathway, allowing the cell to generate more NADPH (40). Therefore, it can be inferred that,
under oxidative conditions, GAPDH can act as a switch to redirect
glucose metabolism to more appropriate defensive pathways.
Unfortunately, permanent inhibition of glycolysis and antioxidant
defense mechanisms are ultimately overcome by the ever-increasing
amount of ROS generated in the brain, which may be eliminating the
possibility of cell survival. That would indicate GAPDH apoptotic
function, confirmed by considerable evidence (14,37,38,41).
GAPDH accumulates in mitochondria during apoptosis and induces
proapoptotic, increased permeability of mitochondrial membranes
(42). Recent research suggests
that GAPDH possesses highly diverse, non-glycolytic functions, such
as nuclear, perinuclear, cytosolic, and membrane-related (14). It affects the release of
Ca2+ from the mitochondria to the cytosol, thereby
inducing the release of neurotransmitters (14,43).
The decreased mRNA level of GAPDH in our study may indicate
Ca2+ retention in mitochondria and consequently reduced
neurotransmitter release that leads to apoptosis. GAPDH can also
undergo many different oxidative modifications, which is of great
interest when neurodegenerative disorders are considered. Several
parallel investigations indicated a relationship between GAPDH and
Alzheimer's and Parkinson's diseases (42–45).
It has been proven that decreased GAPDH glycolytic activity, in
addition to oxidative and post-translational modifications, are
distinct markers of cellular stress in pathology of these diseases
that significantly impact intracellular homeostasis (14,37,38,41).
Taking into account GAPDH functions and locations within the cell
and the above results, it can be inferred that similar mechanisms
may play a role in depression.
The ability of the brain to produce and use lactate
can be regulated by changes in the LDHA and LDHB gene activity,
which allows the brain to optimize the use of energy and metabolism
(46). The current study revealed
that in the hippocampus of rats subjected to CVS, the mRNA levels
of Ldha (responsible for conversion pyruvate to lactate)
remained unchanged but Ldhb (revers reaction) levels were
significantly decreased. Lactate is a substrate for the
mitochondrial TCA cycle via pyruvate, and its oxidation can produce
a significant amount of ATP (47).
Under stress conditions, glucose biotransformation via G6PDH is
defective and if the brain is under prolonged stress, neurons are
more likely to depend on lactate which constitutes an alternative
source of energy. The physiological significance of a decreased
mRNA level of Ldhb in chronic stress could be an
optimization use of lactate for energy production instead
converting into pyruvate. These results provide data for the
relative contribution of lactate probably driving its usage as
energy fuel in conditions of chronic stress. The above results may
also be related to the NAD+/NADH with excess of ROS.
Because LDHB uses NAD+, also needed for NADH regeneration, the
effect observed in our study may be due to the defense mechanism,
enabling overproduction of NADH in response to oxidative stress.
There is a hypothesis that LDH plays a role in the control of the
intracellular redox status (46).
According to this hypothesis, an increase of mitochondrial ROS
causes mitochondrial dysfunction in the brain and leads to a
metabolic shift from aerobic respiration to glycolytic metabolism,
resulting in robustly increased brain lactate levels and in
expression changes of the LDH genes. On the other hand, it has been
shown that serotonin systems are involved in the control of brain
glucose delivery, transport, and uptake (48). Glucose metabolism in the brain can
also be disturbed by downregulation of these systems, which may
result from a decrease in the mRNA level of GAPDH and occur
as a consequence of the weakening of glycolysis.
TFAM is a key regulator of the maintenance and
distribution of mtDNA. It is highly susceptible to the damage
generated by ROS, because of its close proximity to ROS generation
through the respiratory chain and its paucity of protective
histones. Furthermore, there is little capacity for DNA repair in
the mitochondria (49). Our study
revealed a significant increase in the mRNA level of Tfam in
the hippocampus of the rats subjected to CVS as compared to
control. Studies show that TFAM binds preferentially to oxidatively
damaged DNA containing 8-oxoguanine (8-oxo-Gua), which is one of
the most abundant DNA alterations induced by exposure to ROS
(50). In our previous studies, we
have shown a significant increase in AP-site accumulation in
isolated DNA from the hippocampus of the rats subjected to CVS,
which indicates an increase in oxidative damage of DNA (2). At the same time, we noticed
upregulation of Ogg1 (the gene encodes the enzyme
responsible for the excision of 8-oxoguanine) as increased
efficiency of DNA repair. Therefore, it can be assumed that our
study is consistent with earlier results because it may indicate
the contribution of mtTFA in oxidative damage through its binding
to OGG1. Through the regulation of mtTFA, the maintenance of mtDNA
copy number and expression are considered to be essential for
preservation of mitochondrial function (51,52).
It was hypothesized that mtDNA and TFAM stabilize each other
(53). Because ROS excess
decreases the amount of mtDNA, the increased TFAM may enhance the
steady state levels of mtDNA in response to oxidative stress.
Considering the above mentioned results, it can be assumed that the
overexpression of TFAM can prevent the decline in mtDNA, which is
important for maintaining cellular functions under chronic stress.
In addition, Tfam overexpression may explain the need for
increased mitochondrial activity. The results we obtained in this
experiment-upregulation of Slc2a3, but downregulation of
GAPDH and Ldhb, indicate a decrease in glycolysis
with an increased glucose requirement. Therefore, a disruption in
glucose metabolism may be a direct determinant of synaptic
dysfunction, while mitochondrial changes and consequential cell
death appear to be related with atrophy of the hippocampus and
prefrontal cortex observed in depressed patients (4,48,54,55).
In our study, it was observed a statistically
significant reduction in the level of all examined amino acids in
the brains of rats exposed to chronic stress. During stress
exposure, the mitochondrial ATP production is disturbed. Under
these circumstances, the glycolysis becomes the primary source of
energy, increasing the generation of lactate. However, neither
mitochondrial oxidative phosphorylation nor anaerobic glycolysis
alone can produce ATP at a sufficient rate to maintain brain
function. As described above, glucose is diverted towards the
pentose phosphate pathway, resulting in a high antioxidant status
at the expense of energy production. At contingency of high energy
demand, the glucogenic amino acids can be a source of energy
production in gluconeogenesis (19). Astrocytes are competent in
gluconeogenesis, serving as a potential energy pathway for neurons,
activated at times of stress (56). The decreased levels of amino acids
may confirm the mechanism by which in brain the gluconeogenesis
occurs, in order to meet the energy needs of this organ. At the
same time, the above changes may provide a capacity to stimulate
this pathway under chronic stress conditions.
It is worth noting that one of the stressors used in
the procedure of chronic stress in rodents was the deprivation of
water and food. The shortage of food and water can affect the
metabolic pathways in the brain. In mammals, plasma concentration
of metabolic products related to energy balance varies with a
well-established dynamic, depending on whether the organism is well
fed or starved (57). The brain
engages in gluconeogenesis to convert glucogenic amino acids into
glucose for metabolism. The body's glycogen stores are consumed in
about 24 h (58). Unless we're
talking about the case of long-term starvation, brain energy
requirements are covered in part by the metabolism of ketone bodies
(58). Studies showed that rodents
are physiologically equipped to tolerate acute food and water
deprivation for periods of as long as 24 h without overt signs of
physiologic distress or behavioral abnormalities (59). Therefore, we assume that short-term
deprivation of water and food (24 h) did not significantly affect
the energy metabolism of the rats brain due to adaptive properties.
We can therefore consider the deprivation of water and food as
appropriate stress factors, especially that patients suffering from
stress-related depression show concurrent disordered eating
behaviors; they consume excessive amounts of food or they use
starvation as a means of coping with the seemingly intolerable
feelings of depression. However, it is not clear whether this data
can be directly related to people; it is unknown whether short-term
starvation of a few days' duration leads to reduced brain glucose
metabolism in human. Rodents have endogenous nycthemeral rhythms
that make them particularly adaptable to once-daily occurrences,
such as food or water access. During water deprivation in rats, the
increases in plasma osmolality is only a modest approximately 2%
after 24 h; the corresponding decreases in plasma volume is
approximately 4% and may be less than in humans (59). Studies showed that the human brain
adapts to the changes in energy supply as 3 days following
initiation of starvation, at which time ketone bodies account for
approximately one-fourth of the cerebral energy requirements
(58).
A number of assumption regarding the molecular
mechanism of neurodegeneration involves the neuroactive amino acids
(60). Aspartate and glutamate are
the major excitatory neurotransmitters in the brain, whereas,
glycine, taurine and γ-aminobutyric acid are inhibitory. Taurine
has also a protective role in the brain. In addition, glutamine is
a precursor molecule for glutathione, which protects against ROS
toxicity (19). Amino acids such
as tryptophan, tyrosine, histidine, and arginine are used by the
brain for the synthesis of various neurotransmitters and
neuromodulators (22). Abnormal
metabolism of neuroactive amino acids has been demonstrated in the
plasma of patients with major depression and also in the brain
samples of rats in depression model (61–63).
Ni et al (61) showed that
tyrosine, glutamine and proline were significantly downregulated,
while glycine was upregulated in the cerebella of depressed rats.
The decrease of neuroactive amino acids levels observed in our
study can result from a close relationship between the brain
activity, neurotransmission, energy requirements, and glucose
utilization. During activation, glutamate uptake into astrocytes
leads to the increased glucose use and lactate production,
therefore, it can be subsequently used by neurons to meet their
energy needs (56). Glutamate and
aspartate, the major excitatory neurotransmitters in the brain, do
not cross the ΒΒΒ and are synthesized from glucose. Therefore, the
observed reduction in their level provides further support for the
hypothesis of redirecting glucose metabolism to defense pathways in
conditions of chronic stress. The changes in concentration of
neuroactive amino acids could be also a consequence of an imbalance
between excitation and inhibition, as well as alterations of ion
channels during stress. NMDA receptors are highly permeable to
calcium ions which enter cells via NMDA receptor channels, leading
to mitochondrial damage. Recent studies provide the impaired or
reduced glutamatergic neurotransmission in neurological diseases
involving hypofunction of NMDA (64). The above data are consistent with
the reduction of GAPDH mRNA levels noted in our study, which
can provide Ca2+ retention in mitochondria and
consequently reduced neurotransmitter release.
In conclusion, the changes in gene expression and
amino acids levels confirm that the brain energy metabolism
involves complex cellular and molecular mechanisms, which are
necessary to meet the increased energy requirements of the brain
during stress-related depression. The alterations indicate the
support of the energy metabolism by stimulation of gluconeogenesis
pathway and redirection of glucose metabolism to appropriate
defensive pathways under chronic stress conditions. The
overexpression of Tfam may confirm the oxidative damage of
DNA and, at the same time, it may indicate an increased ability to
maintain mitochondrial function as potential adaptive response. We
assume that these changes are coupled with various signaling
mechanisms, which need to be controlled by the cell to avoid
oxidative stress. Our findings provide a novel insight into the
biochemical and molecular events that lead to the development of
depression under chronic stress conditions and they may identify
novel targets for therapeutic intervention.
Acknowledgments
Not applicable.
Funding
The present study was supported by Funds for
Statutory Activity of Medical University of Lublin, Poland (grant
no. DS 38/2015-2016). This research did not receive any specific
grant from funding agencies in the public, commercial, or
not-for-profit sectors.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MH designed and directed the experiment, wrote the
manuscript, analyzed the data, performed the experiments,
corresponding author; DN-C performed the analysis of amino acids,
edited the manuscript; AK and MO performed the molecular study; MI
analyzed and collected the data; MG-G and IP-C performed the
statistical analysis; KP performed the experiments; BŚ supervised
the molecular study, EP supervised the in vivo experiments,
JD supervised the analysis of amino acids; BŚ, EP and JD critically
revised the final version of the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All procedures were conducted in accordance with the
European Communities Council Directive of 22 September 2010
(2010/63/EU) and Polish legislation acts concerning animal
experimentations. The experimental procedures and protocols were
approved by the First Local Ethics Committee at the Medical
University of Lublin.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bansal Y and Kuhad A: Mitochondrial
dysfunction in depression. Curr Neuropharmacol. 14:610–618. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Herbet M, Korga A, Gawrońska-Grzywacz M,
Izdebska M, Piątkowska-Chmiel I, Poleszak E, Wróbel A, Matysiak W,
Jodłowska-Jędrych B and Dudka J: Chronic variable stress is
responsible for lipid and DNA oxidative disorders and activation of
oxidative stress response genes in the brain of rats. Oxid Med Cell
Longev 2017. 73130902017.
|
|
3
|
Khan S and Khan RA: Chronic stress leads
to anxiety and depression. Ann Psychiatry Ment Health.
5:10912017.
|
|
4
|
Tagliari B, Noschang CG, Ferreira AG,
Ferrari OA, Feksa LR, Wannmacher CM, Dalmaz C and Wyse AT: Chronic
variable stress impairs energy metabolism in prefrontal cortex and
hippocampus of rats: Prevention by chronic antioxidant treatment.
Metab Brain Dis. 25:169–176. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mergenthaler P, Lindauer U, Dienel GA and
Meisel A: Sugar for the brain: The role of glucose in physiological
and pathological brain function. Trends Neurosci. 36:587–597. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Detka J, Kurek A, Kucharczyk M, Głombik K,
Basta-Kaim A, Kubera M, Lasoń W and Budziszewska B: Brain glucose
metabolism in an animal model of depression. Neuroscience.
295:198–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ishida A, Noda Y and Ueda T: Synaptic
vesicle-bound pyruvate kinase can support vesicular glutamate
uptake. Neurochem Res. 34:807–818. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rezin GT, Cardoso MR, Gonçalves CL, Scaini
G, Fraga DB, Riegel RE, Comim CM, Quevedo J and Streck EL:
Inhibition of mitochondrial respiratory chain in brain of rats
subjected to an experimental model of depression. Neurochem Int.
53:395–400. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bélanger M, Allaman I and Magistretti PJ:
Brain energy metabolism: Focus on astrocyte-neuron metabolic
cooperation. Cell Metab. 14:724–738. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Griffin JW and Bradshaw PC: Amino acid
catabolism in Alzheimer's disease brain: Friend or foe? Oxid Med
Cell Longev. 2017.54727922017.PubMed/NCBI
|
|
11
|
Chiefari E, Foti DP, Sgarra R, Pegoraro S,
Arcidiacono B, Brunetti FS, Greco M and Manfioletti G:
Transcriptional regulation of glucose metabolism: The emerging role
of the HMGA1 chromatin factor. Front Endocrinol (Lausanne).
9:3572018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shah K, Desilva S and Abbruscato T: The
role of glucose transporters in brain disease: Diabetes and
alzheimer's disease. Int J Mol Sci. 13:12629–12655. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Camandola S and Mattson MP: Brain
metabolism in health, aging, and neurodegeneration. EMBO J.
36:1474–1492. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Butterfield DA, Hardas SS and Lange ML:
Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) and Alzheimer's disease: Many pathways to
neurodegeneration. J Alzheimers Dis. 20:369–393. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kasashima K and Endo H: Interaction of
human mitochondrial transcription factor A in mitochondria: Its
involvement in the dynamics of mitochondrial DNA nucleoids. Genes
Cells. 20:1017–1027. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Campbell CT, Kolesar JE and Kaufman BA:
Mitochondrial transcription factor A regulates mitochondrial
transcription initiation, DNA packaging, and genome copy number.
Biochim Biophys Acta 1819. 921–929. 2012.
|
|
17
|
Murakami T, Yamane H, Tomonaga T and
Furuse M: Forced swimming and imipramine modify plasma and brain
amino acid concentrations in mice. Eur J Pharmacol. 602:73–77.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nagasawa M, Ogino Y, Kurata K, Otsuka T,
Yoshida J, Tomonaga S and Furuse M: Hypothesis with abnormal amino
acid metabolism in depression and stress vulnerability in Wistar
Kyoto rats. Amino Acids. 43:2101–2111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yip J, Geng X, Shen J and Ding Y: Cerebral
gluconeogenesis and diseases. Front Pharmacol. 7:5212017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Heimburger DC and Ard JD: Handbook of
Clinical Nutrition. 4th. Elsevier Inc.; 2006, https://doi.org/10.1016/C2009-0-45871-9
|
|
21
|
Ghosh A, Cheung YY, Mansfield BC and Chou
JY: Brain contains a functional glucose-6-phosphatase complex
capable of endogenous glucose production. J Biol Chem.
280:11114–11119. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Maher T: The role of amino acid precursors
on neurotransmission. Eur J Pharmacol. 668:e82011. View Article : Google Scholar
|
|
23
|
Gamaro GD, Manoli LP, Torres IL, Silveira
R and Dalmaz C: Effects of chronic variate stress on feeding
behavior and on monoamine levels in different rat brain structures.
Neurochem Int. 42:107–114. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao Z, Wang W, Guo H and Zhou D:
Antidepressant-like effect of liquiritin from Glycyrrhiza uralensis
in chronic variable stress induced depression model rats. Behav
Brain Res. 194:108–113. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Porsolt RD, Bertin A and Jalfre M:
Behavioral despair in mice: A primary screening test for
antidepressants. Arch Int Pharmacodyn Ther. 229:327–336.
1977.PubMed/NCBI
|
|
26
|
Iłżecka J, Stelmasiak Z, Solski J,
Wawrzycki S and Szpetnar M: Plasma amino acids concentration in
amyotrophic lateral sclerosis patients. Amino Acids. 25:69–73.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bekris S, Antoniou K, Daskas S and
Papadopoulou-Daifoti Z: Behavioural and neurochemical effects
induced by chronic mild stress applied to two different rat
strains. Behav Brain Res. 161:45–59. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Packard AE, Ghosal S, Herman JP, Woods SC
and Ulrich-Lai YM: Chronic variable stress improves glucose
tolerance in rats with sucrose-induced prediabetes.
Psychoneuroendocrinology. 47:178–188. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
MacQueen G and Frodl T: The hippocampus in
major depression: Evidence for the convergence of the bench and
bedside in psychiatric research? Mol Psychiatry. 16:252–264. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pandya M, Altinay M, Malone DA Jr and
Anand A: Where in the brain is depression? Curr Psychiatry Rep.
14:634–642. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dienel GA: Fueling and imaging brain
activation. ASN Neuro. 4(pii): e000932012.PubMed/NCBI
|
|
32
|
Detka J, Kurek A, Basta-Kaim A, Kubera M,
Lasoń W and Budziszewska B: Elevated brain glucose and glycogen
concentrations in an animal model of depression.
Neuroendocrinology. 100:178–190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Khayat ZA, McCal AL and Klip A: Unique
mechanism of GLUT3 glucose transporter regulation by prolonged
energy demand: increased protein half-life. Biochem J. 333:713–718.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fontella FU, Siqueira IR, Vasconcellos AP,
Tabajara AS, Netto CA and Dalmaz C: Repeated restraint stress
induces oxidative damage in rat hippocampus. Neurochem Res.
30:105–111. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lucca G, Comim CM, Valvassori SS, Réus GZ,
Vuolo F, Petronilho F, Gavioli EC, Dal-Pizzol F and Quevedo J:
Increased oxidative stress in submitochondrial particles into the
brain of rats submitted to the chronic mild stress paradigm. J
Psychiatr Res. 43:864–869. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liemburg-Apers DC, Willems PH, Koopman WJ
and Grefte S: Interactions between mitochondrial reactive oxygen
species and cellular glucose metabolism. Arch Toxicol.
89:1209–1226. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ishitani R, Kimura M, Sunaga K, Katsube N,
Tanaka M and Chuang DM: An antisense oligodeoxynucleotide to
glyceraldehyde-3-phosphate dehydrogenase blocks age-induced
apoptosis of mature cerebrocortical neurons in culture. J Pharmacol
Exp Ther. 278:447–454. 1996.PubMed/NCBI
|
|
38
|
Ishitani R, Sunaga K, Hirano A, Saunders
P, Katsube N and Chuang DM: Evidence that
glyceraldehyde-3-phosphate dehydrogenase is involved in age-induced
apoptosis in mature cerebellar neurons in culture. J Neurochem.
66:928–935. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vilà MR, Nicolás A, Morote J, de Torres I
and Meseguer A: Increased glyceraldehyde-3-phosphate dehydrogenase
expression in renal cell carcinoma identified by RNA-based,
arbitrarily primed polymerase chain reaction. Cancer. 89:152–164.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Grant CM: Metabolic reconfiguration is a
regulated response to oxidative stress. J Biol. 7:12008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chuang DM, Hough C and Senatorov VV:
Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and
neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 45:269–290.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tarze A, Deniaud A, Le Bras M, Maillier E,
Molle D, Larochette N, Zamzami N, Jan G, Kroemer G and Brenner C:
GAPDH, a novel regulator of the pro-apoptotic mitochondrial
membrane permeabilization. Oncogene. 26:2606–2620. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mazzola JL and Sirover MA: Alteration of
intracellular structure and function of glyceraldehyde-3-phosphate
dehydrogenase: A common phenotype of neurodegenerative disorders?
Neurotoxicology. 23:603–609. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sirover MA: Role of the glycolytic
protein, glyceraldehyde-3-phosphate dehydrogenase, in normal cell
function and in cell pathology. J Cell Biochem. 66:133–140. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tatton WG, Chalmers-Redman RM, Elstner M,
Leesch W, Jagodzinski FB, Stupak DP, Sugrue MM and Tatton NA:
Glyceraldehyde-3-phosphate dehydrogenase in neurodegeneration and
apoptosis signaling. J Neural Transm Suppl. 77–100. 2000.PubMed/NCBI
|
|
46
|
Ross JM, Öberg J, Brené S, Coppotelli G,
Terzioglu M, Pernold K, Goiny M, Sitnikov R, Kehr J, Trifunovic A,
et al: High brain lactate is a hallmark of aging and caused by a
shift in the lactate dehydrogenase A/B ratio. Proc Natl Acad Sci
USA. 107:20087–20092. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schurr A: Lactate: The ultimate cerebral
oxidative energy substrate? J Cereb Blood Flow Metab. 26:142–152.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mosconi L, Pupi A and De Leon MJ: Brain
glucose hypometabolism and oxidative stress in preclinical
Alzheimer's disease. Ann N Y Acad Sci 1147. 180–195. 2008.
View Article : Google Scholar
|
|
49
|
Nakanishi H and Wu Z: Microglia-aging:
Roles of microglial lysosome- and mitochondria-derived reactive
oxygen species in brain aging. Behav Brain Res. 201:1–7. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yoshida Y, Izumi H, Ise T, Uramoto H,
Torigoe T, Ishiguchi H, Murakami T, Tanabe M, Nakayama Y, Itoh H,
et al: Human mitochondrial transcription factor A binds
preferentially to oxidatively damaged DNA. Biochem Biophys Res
Commun. 295:945–951. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jeng JY, Yeh TS, Lee JW, Lin SH, Fong TH
and Hsieh RH: Maintenance of mitochondrial DNA copy number and
expression are essential for preservation of mitochondrial function
and cell growth. J Cell Biochem. 103:347–357. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Toki N, Kagami S, Kurita T, Kawagoe T,
Matsuura Y, Hachisuga T, Matsuyama A, Hashimoto H, Izumi H and
Kohno K: Expression of mitochondrial transcription factor A in
endometrial carcinomas: Clinicopathologic correlations and
prognostic significance. Virchows Arch. 456:387–393. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kang D, Kim SH and Hamasaki N:
Mitochondrial transcription factor A (TFAM): Roles in maintenance
of mtDNA and cellular functions. Mitochondrion. 7:39–44. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lee AL, Ogle WO and Sapolsky RM: Stress
and depression: Possible links to neuron death in the hippocampus.
Bipolar Disord. 4:117–128. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gałecki P, Florkowski A, Mrowicka M,
Pietras T and Gałecka E: Calcium ions, glutaminate acid,
hypothalamic-pituitary-adrenal axis, calcium dependent ATP-ase as
causes of oxidative damage in depression patients (part II). Pol
Merkur Lekarski. 24:72–75. 2008.(In Polish). PubMed/NCBI
|
|
56
|
Magistretti PJ and Pellerin L: Cellular
mechanisms of brain energy metabolism and their relevance to
functional brain imaging. Philos Trans R Soc Lond B Biol Sci.
354:1155–1163. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hasselbalch SG, Knudsen GM, Jakobsen J,
Hageman LP, Holm S and Paulson OB: Brain metabolism during
short-term starvation in humans. J Cereb Blood Flow Metab.
14:125–131. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
White H and Venkatesh B: Clinical review:
Ketones and brain injury. Crit Care. 15:2192011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Rowland NE: Food or fluid restriction in
common laboratory animals: Balancing welfare considerations with
scientific inquiry. Comp Med. 57:149–160. 2007.PubMed/NCBI
|
|
60
|
Kay GW, Verbeek MM, Furlong JM, Willemsen
MA and Palmer DN: Neuropeptide changes and neuroactive amino acids
in CSF from humans and sheep with neuronal ceroid lipofuscinoses
(NCLs, Batten disease). Neurochem Int. 55:783–788. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ni Y, Su M, Lin J, Wang X, Qiu Y, Zhao A,
Chen T and Jia W: Metabolic profiling reveals disorder of amino
acid metabolism in four brain regions from a rat model of chronic
unpredictable mild stress. FEBS Lett. 582:2627–2636. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Xu HB, Zhang RF, Luo D, Zhou Y, Wang Y,
Fang L, Li WJ, Mu J, Zhang L, Zhang Y and Xie P: Comparative
proteomic analysis of plasma from major depressive patients:
Identification of proteins associated with lipid metabolism and
immunoregulation. Int J Neuropsychopharmacol. 15:1413–1425. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Shao WH, Chen JJ, Fan SH, Lei Y, Xu HB,
Zhou J, Cheng PF, Yang YT, Rao CL, Wu B, et al: Combined
metabolomics and proteomics analysis of major depression in an
animal model: Perturbed energy metabolism in the chronic mild
stressed rat cerebellum. OMICS. 19:383–392. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kantrowitz JT and Javitt DC:
N-methyl-d-aspartate (NMDA) receptor dysfunction or dysregulation:
The final common pathway on the road to schizophrenia? Brain Res
Bull. 83:108–121. 2010. View Article : Google Scholar : PubMed/NCBI
|