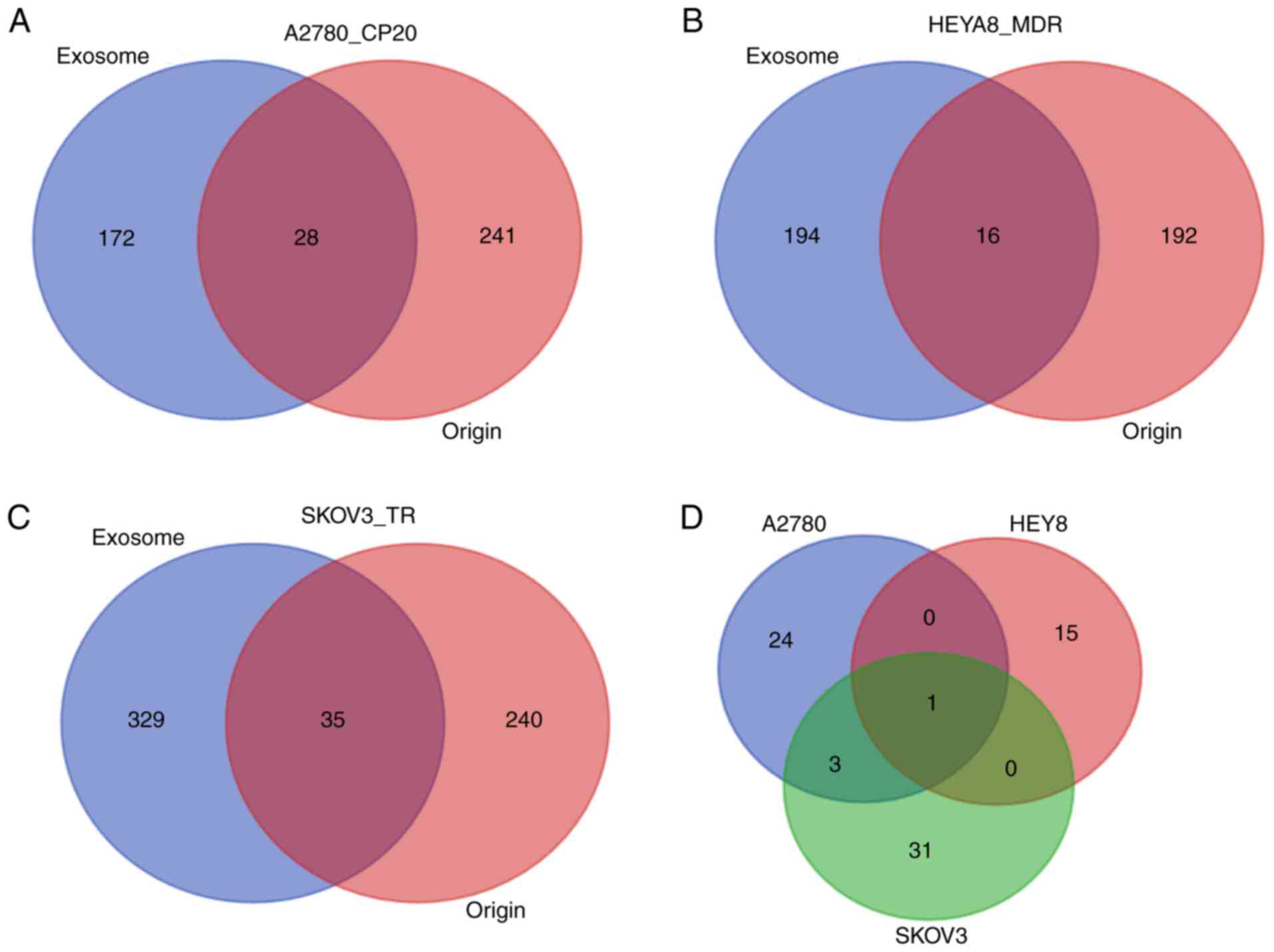
| A2780_CP20 | GO:0045944~positive
regulation of transcription from RNA polymerase II promoter |
1.24×10−7 | CCNT2, RNASEL,
RAI1, THRA, PPP2R5B, EDN1, CASK, NFKB1, CITED4, CBFB, SOHLH2,
CXCL10, EPCAM, TMEM173, OSR1, HSF2, RARA, FGF1, FGF2, ARHGEF2,
MTA2, STRN3, SOX12, MED12, DMRT2, FOSB, PTPRN, HMGA2, LILRB1,
INHBA, ZMIZ1, CD81, TFAP2B, SPDEF, SMARCA4, PEG3, CRTC2, TFAP4,
ONECUT1, CREM, ONECUT2, SOX2, PML, C14ORF166, NFIX, SRF, CALCOCO1,
ATF2, STAT6, PLAGL1, NPAS2, TCF20, CHD7, HOXA5, BCL11B, BCL11A,
GBX2, NFAT5, KDM3A, TRAF6, NFATC3, ESRRA, IL6, NOS1, CREB3, KLF12,
RFX4, TBX5, ARID3A, NR4A2, SMYD3, NR4A1, ARID3B, TEAD2, TEAD3, HGF,
DDX5, CELA1, USF1, FOXP1, BRCA1, HOXB4, MEF2D, CDH13, DLX2, NR1I3,
HIF3A, IRF2, NFIB, SLC9A1 |
|
|
GO:0060291~long-term synaptic
potentiation |
1.61×10−5 | SLC8A3, GFAP, STX4,
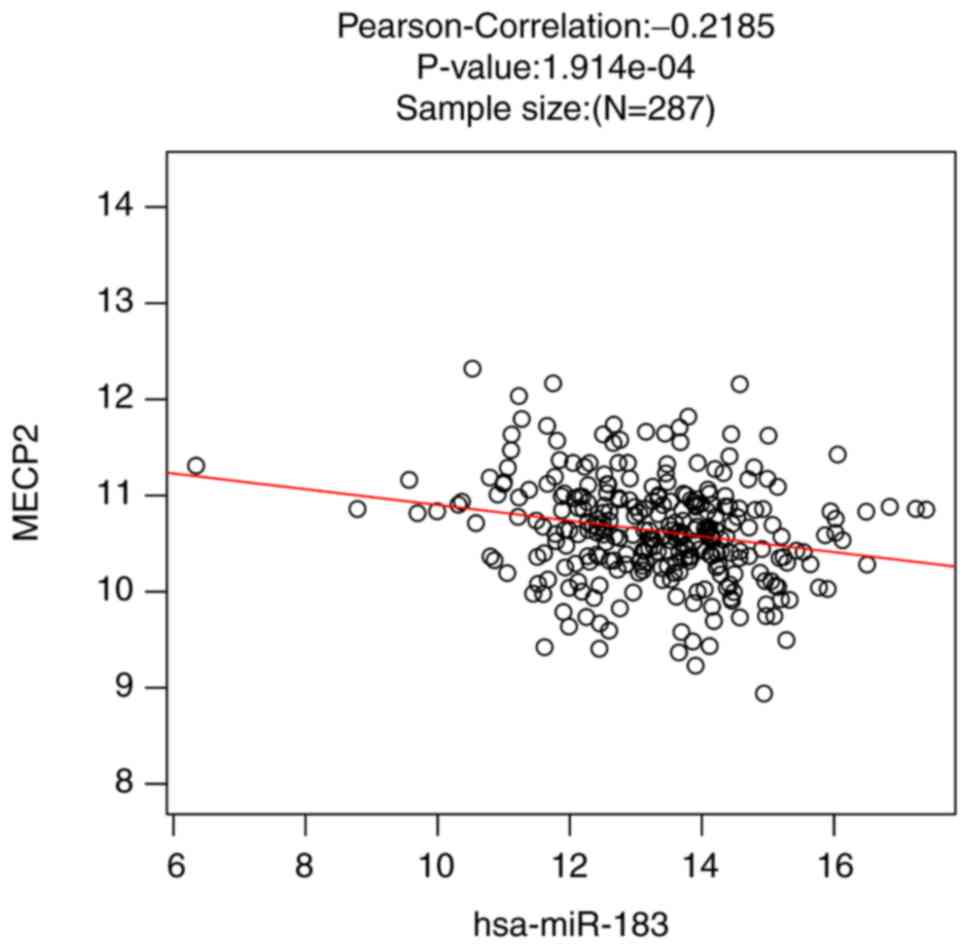
PLK2, SLC24A2, MECP2, NLGN1, GRIN2A, SNAP47, SHANK3, RGS14 |
|
|
GO:0000187~activation of MAPK
activity |
3.87×10−4 | IRAK1, IQGAP3, HGF,
KIT, GRM1, NTRK3, PEA15, PAK3, CD81, GNG3, FGF1, TRAF6, FRS2, FGF2,
MAP3K11, DUSP6 |
|
| GO:0007165~signal
transduction |
4.17×10−4 | COPS2, FGF18,
PPP2R5B, PPP2R5D, IQGAP3, NFKB1, CD53, CXCL10, TNFRSF11B, ANK1,
UNC5A, DLG4, PITPNC1, RARA, FAS, FGF1, MX1, FGF2, LTB, AKT3, PILRA,
IRAK1, MAGI2, LTBR, LIMK1, RASIP1, TLE1, CLIC1, HMGA2, CDS1, IL21,
DEPDC4, PLAUR, LILRB1, GAPVD1, IGSF1, GRN, NRGN, LALBA, ERBB4,
CREM, CCL8, NOSTRIN, GAST, KIT, CALCOCO1, RASAL2, STAT6, IGF1R,
DGKG, BCL11B, BCL11A, PSTPIP1, NFAT5, CC2D1A, HLA-DOA, DKKL1,
IL1RAPL1, INPP5B, TRAF4, DTNA, EBP, ABR, NR4A2, DPYSL5, NR4A1,
CD300C, ECM1, CCL11, LAT, RPS6KA6, NR1I3, ADCY9, GRIA2, CXCL14,
PNOC, BRE, ADRA1A, HIVEP2, CD79B, RIT1, ANTXR1, GRB7, ABCC8, PLAU,
ARHGAP10, BCAR3 |
|
|
GO:0006357~regulation of transcription
from RNA polymerase II promoter |
1.40×10−3 | ENY2, TSHZ3,
HTATIP2, THRA, NFKB1, CITED4, CBFB, ATF2, HOXC6, STAT6, PLAGL1,
HSF2, WDR77, CAMK2D, FOXD2, NFATC3, CHD3, KLF7, BRD2, ZMYM2, KLF12,
SOX12, MED11, FOSB, TEAD3, DDX5, UBN1, USF1, ECM1, BRCA1, FOXP1,
RBBP8, INHBA, SMARCE1, TIAL1, HIF3A, CKS2, KDM4C, SMARCA4 |
|
| GO:0032958~inositol
phosphate biosynthetic process |
2.44×10−3 | PPIP5K1, IPPK,
FGF2, IP6K3 |
|
| GO:1903507~negative
regulation of nucleic acid-templated transcription |
2.56×10−3 | HOXC6, COPS2,
SSX4B, PRMT5, SSX6, SSX4, SIAH2, SSX3, SSX1 |
|
| GO:0000122~negative
regulation of transcription from RNA polymerase II promoter |
3.72×10−3 | COPS2, EDN1, NFKB1,
OSR1, RARA, ATF7IP, GSC, RREB1, STRN3, MTA2, MECP2, ZHX3, TLE1,
FOSB, HMGA2, PKIA, RBBP8, KDM2B, TIMELESS, SPDEF, TFAP2B, TGIF1,
PEG3, SMARCA4, TSHZ3, SOX2, TRIB3, NFIX, TAGLN3, ATF2, STAT6,
ANKRD33, BCL11A, CC2D1A, BCOR, TRAF6, TCF25, ESRRA, KLF12, PTPN2,
NR4A2, WHSC1, SMYD2, CELA1, DDX5, EHMT2, FOXP1, DLX2, HOXB4,
SAP130, ATF7, IRF2, HDAC8, NCOR2, NFIB |
|
| GO:0043547~positive
regulation of GTPase activity |
7.06×10−3 | FGF18, ARFGAP3,
RALGPS1, ERBB4, CCL8, PLEKHG4B, RASGEF1C, KIT, MCF2L, RASAL2,
PLEKHG3, AGAP6, GARNL3, DLG4, CAMK2D, RAPGEF5, FGF1, FRS2, INPP5B,
FGF2, FBXO8, ARHGDIA, AGAP4, GIT1, ARHGEF2, IL5, ABR, IL2RA,
LAMTOR1, ARHGEF5, SPTBN4, GRIN2A, DOCK3, RGS14, CCL11, NCAM1, LAT,
GAPVD1, GNAQ, GFRA1, SH3BGRL3, RGS9, ARHGAP10, BCAR3 |
|
|
GO:0006351~transcription,
DNA-templated |
8.28×10−3 | MORF4L1, THRA,
MAF1, CITED4, HOXC6, SFSWAP, MIER3, TCEANC2, RARA, ZNF646, MECP2,
MED11, ZHX3, PTPRN, SPOCD1, PA2G4, KDM2B, KDM2A, SCYL1, TIMELESS,
ASCC3, PRDM6, SPDEF, TGIF1, DNTTIP1, EIF2AK2, SMARCA4, TSHZ3,
CRTC2, ERBB4, C14ORF166, ZNF618, OTP, PLAGL1, TCF20, SSX6, SSX4,
BCOR, TCF25, SSX3, THAP11, MN1, SSX1, ZNF423, ZBTB48, YEATS4, KAT8,
KLF7, ZMYM2, ESRRA, KLF8, KLF12, NR4A2, NR4A1, TEAD2, WHSC1, TEAD3,
SMYD2, UIMC1, FOXP1, BRCA1, HOXB4, NR1I3, SAP130, PHF1, ATF7,
HDAC8, NCOR2, ZNF410, CCNT2, E2F3, YLPM1, ZBTB37, ZNF678, BZW1,
SSX4B, OSR1, DEDD2, ZBTB22, ATF7IP, DMRT2, TLE1, ZNF335, HOXC10,
HIPK1, NAB2, ZMIZ1, CARM1, PEG3, SCML4, ZNF808, ZBTB10, CREM, PML,
TRIB3, NFIX, CALCOCO1, ZNF32, STAT6, HIC2, NPAS2, CHD7, BCL11A,
POU2F1, KDM3A, HBP1, CC2D1A, GTF3C2, FOXD2, BAZ2B, CHD5, CHD3,
BRD2, CREB3, HMBOX1, AFF3, ATXN1, MEF2D, JMJD6, IFT57, HIF3A,
KDM4C, ZBTB2, MESP2, NFIB |
|
|
GO:0018108~peptidyl-tyrosine
phosphorylation |
1.28×10−2 | FGF18, ZMYM2, IL5,
ERBB4, EFEMP1, ABI2, KIT, EPHB3, CDC37, NTRK3, EPHA5, SCYL1, CLK3,
EIF2AK2, FGF1, FGF2 |
|
| GO:0000082~G1/S
transition of mitotic cell cycle |
1.61×10−2 | INHBA, CUL2,
CDKN1A, CUL5, EIF4E, PLK2, CDKN2D, IQGAP3, CAMK2D, ORC5, MARK4,
RBBP8 |
|
| GO:0008285~negative
regulation of cell proliferation |
2.34×10−2 | TFAP4, ERBB4, PML,
COPS8, SRF, CUL2, CUL5, BCL11B, CDKN2D, RARA, FGF2, CHD5, NOX4,
IL6, MAGI2, NF2, PTPN2, TBX5, SMYD2, TMEM115, NOTCH2, INHBA, CDH13,
PPM1D, CDKN1A, NME1, BTG1, TFAP2B, MYO16, ADRA1A, EIF2AK2 |
|
| GO:0001666~response
to hypoxia |
3.30×10−2 | MUC1, KCNMA1, NOX4,
ATP1B1, NOS1, PML, MECP2, NR4A2, EGLN2, AGER, USF1, SRF, PKM,
CAMK2D, CD24, PLAU |
| HEYA8_MDR | GO:0045944~positive
regulation of transcription from RNA polymerase II promoter |
2.38×10−8 | CCNT2, HLF, ZNF292,
THRA, ARID4A, PPP2R5B, ARID4B, ZEB2, FOXO3, JAG1, PAX2, CBFA2T2,
CBFB, PROP1, SERPINE1, H2AFZ, RARB, PITX1, IL1A, BRD8, CYR61,
DAB2IP, HYAL2, RARG, SOX11, SOX12, TP53, IL25, PTPRN, ELL3, GRHL1,
GRHL2, PPARGC1B, AHR, MYCN, SENP2, ADRB2, ZMIZ2, JUN, ZNF746,
ACVR1, CAMTA1, CREM, TFE3, PML, SOX4, NFIX, MYBL2, CHD8, CHD7,
POU2F2, PPP3CB, HOXA10, NFAT5, PPP1R12A, PPP3CA, NKX2-2, BCL9,
PIK3R2, ESRRA, FOXL2, IKZF2, KLF13, MAML1, TEAD3, SMAD1, SIRT1,
CDH13, ATF3, EBF3, TRPS1, NEUROD1, NHLH2, ZNF462, IRF4, BMP7,
NR5A2, SETD3, APBB1, NFIB, BMPR1A |
|
| GO:0045892~negative
regulation of transcription, DNA-templated |
5.68×10−6 | ELF2, THRA, ARID4A,
PDGFB, TSG101, CREM, BTRC, PML, PAX2, CBFA2T2, HIC2, CHD8, TSC22D4,
PRAMEF2, EED, PER2, PEX14, BCL6, LOXL3, NR2F2, DEDD2, PITX1, BAHD1,
FOXL2, DAB2IP, KLF10, TP53, MECP2, UBE2I, BASP1, SPEN, SIRT1,
ADIPOQ, PPARGC1B, AHR, FOXN3, GZF1, ATXN1, PRICKLE1, JUN, USP47,
ZNF746, HDAC9, BMP7, RASD1 |
|
| GO:0043065~positive
regulation of apoptotic process |
1.70×10−5 | SNCA, SPINK2, SOX4,
PAWR, FOXO3, MAP3K5, MTCH1, PLEKHG5, RHOB, BCL6, RARB, UNC5C,
WNT10B, DAB2IP, FOXL2, RARG, ABR, NF1, TP53, BAD, ARHGEF9, SIRT1,
BCL2L11, NTRK3, IGF2R, BNIP3L, SMPD1, NEUROD1, DCUN1D3, BMP7,
APBB1 |
|
| GO:0000122~negative
regulation of transcription from RNA polymerase II promoter |
3.04×10−3 | JDP2, BACH2, MTDH,
CPEB3, SNCA, NFIX, ZEB2, PAWR, FOXO3, SUFU, CHD8, TSC22D3, TCERG1,
PROP1, EED, PER2, BCL6, RARB, NR2F2, EPO, DNMT3A, SATB1, FOXL2,
DAB2IP, ESRRA, WNT10B, RARG, RFX5, SOX11, KLF10, TP53, MECP2,
UBE2I, HES6, SPEN, SNAI1, SIRT1, GZF1, ATF3, TRPS1, DLL4, MNT,
TGIF2, PTCH1, ZNF746, HDAC9, HDAC8, NFIB |
|
| GO:0001666~response
to hypoxia |
3.18×10−3 | MUC1, AHCY, BECN1,
NF1, PML, MECP2, EGLN2, BAD, CXCL12, ADIPOQ, DDIT4, EDNRA, CASP3,
CAMK2D, HSD11B2, THBS1, EPO |
|
|
GO:0000186~activation of MAPKK
activity |
3.32×10−3 | MAP3K5, CRKL,
GNAI2, PLCG1, MAP3K3, GRM1, MAP3K12, ADAM9 |
|
| GO:0045893~positive
regulation of transcription, DNA-templated |
5.49×10−3 | SUPT3H, ELF2,
PDGFB, BTRC, TFE3, SOX4, AFAP1L2, CDH1, FOXO3, PAX2, CAMKK2, CHD8,
PPP3CB, NR2F2, EPO, IRAK1, FOXL2, SOX11, TP53, MECP2, SNAI1, FOXN3,
AHR, MYCN, JUN, USP21, NEUROD1, PTCH1, COL1A1, IRF4, NR5A2, NEK4,
BMP7, APBB1, SETD3, ACVR1 |
| SKOV3_TR | GO:0008284~positive
regulation of cell proliferation |
5.42×10−3 | FGFR2, AVPR2, PGF,
CNTFR, PRDX3, CUL3, IGF1R, TNFRSF11A, CNOT6L, ILK, RARB, NRG1,
FGFBP1, EPO, FN1, STAMBP, SHMT2, CAPNS1, RELA, SOX11, MECP2, DLL1,
NTRK3, CRKL, S100B, EREG, BNC1, DHPS, GRK5, CARM1 |
|
| GO:0007050~cell
cycle arrest |
6.77×10−3 | ING4, CDK6, TCF7L2,
TP73, DDIT3, CDKN1C, CUL3, PPP1R9B, PPM1G, CUL2, CDKN1A, ILK,
APBB1 |
|
|
GO:0097193~intrinsic apoptotic signaling
pathway |
2.47×10−2 | CUL3, CUL2, CDKN1A,
BBC3, CLU |
|
| GO:0043065~positive
regulation of apoptotic process |
3.41×10−2 | ING4, ING3, APH1A,
TFAP4, CLU, SNCA, ARHGEF9, ITSN1, BCL2L11, TP73, NTRK3, S100B,
HOXA5, RIPK2, FAM162A, RARB, APBB1, FGD4, EIF2B5 |