Introduction
Lung cancer is the leading cause of
cancer-associated mortality in males and females worldwide
(1). Lung cancer can be
pathologically subdivided into two principal types: Small cell lung
cancer and non-small cell lung cancer (NSCLC) (2). NSCLC is the most common type of lung
cancer and accounts for ~80% of all lung cancer cases (3). Numerous risk factors of NSCLC have
been identified, including long-term tobacco smoking, specific gene
mutations and exposure to radon gas, asbestos and other types of
environmental pollutants (4–6).
Notable developments in the diagnosis and therapy of NSCLC have
been made; however, the overall survival of patients with NSCLC
remains unsatisfactory (7). The
5-year survival rate of patients diagnosed with NSCLC at stages I
and IV is 67 and 1%, respectively (8). Tumour recurrence and metastasis are
mainly responsible for the poor therapeutic outcomes of patients
with NSCLC (9,10). Therefore, further identification of
the mechanisms underlying NSCLC occurrence and development is
important for the development of novel therapeutic strategies.
microRNAs (miRNAs/miRs) are a group of
single-stranded, highly conserved and small non-coding RNA
molecules that serve important roles in the oncogenesis of NSCLC
(11). miRNAs can negatively
regulate target gene expression by directly binding to the
3′-untranslated regions (UTRs) of target genes, which lead to the
degradation or translational suppression of targeted messenger RNAs
(mRNAs) (12). At present,
>1,000 miRNAs have been validated, and these miRNAs can regulate
~60% of all human genes (13).
Therefore, miRNAs are involved in the regulation of various
biological activities, including cell proliferation, cycle,
apoptosis, differentiation, metabolism and metastasis (14–16).
Numerous miRNAs are dysregulated in NSCLC; their dysregulation has
been associated with the pathogenesis and development of NSCLC
(17–19); aberrantly expressed miRNAs may
serve as tumour suppressors or oncogenes, depending on the
biological roles of their target genes (20). miRNAs may be developed as novel
therapeutic targets in the diagnosis and treatment of patients with
NSCLC.
miR-629-5p (miR-629) is upregulated in various types
of human cancers (21–23) and serves oncogenic roles in
carcinogenesis and the progression of cancer; however, the
expression profile, biological roles and associated mechanisms of
miR-629 in NSCLC remain unclear. In the present study, miR-629
expression was detected in NSCLC tissues and cell lines, the
effects of miR-629 in NSCLC cells were also investigated. The
mechanisms underlying the oncogenic roles of miR-629 in NSCLC cells
were also determined.
Materials and methods
Patients and tissue specimens
A total of 51 pairs of NSCLC and adjacent normal
tissues were obtained from patients with NSCLC (32 males, 19
females; age range, 43–69 years) who underwent surgical resection
at the Shanghai Ninth People's Hospital (Shanghai, China) between
June 2014 and January 2017. Patients with NSCLC did not receive
preoperative radiotherapy and chemotherapy. All patients with NSCLC
were divided into miR-629 high or low expression groups based on
the median value of miR-629 expression. TNM staging system was used
for the staging of NSCC (24).
Following resection, all tissue specimens were immediately frozen
in liquid nitrogen and stored at −80°C until further RNA isolation.
The present study was approved by the Ethics Committee of the
Shanghai Ninth People's Hospital. Written informed consent was also
obtained from the patients enrolled.
Cell culture and transfection
The non-tumorigenic bronchial epithelium cell line
BEAS-2B was obtained from the American Type Culture Collection
(Manassas, VA, USA) and cultured in LHC-9 medium with 10% fetal
bovine serum (FBS; both obtained from Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). A total of four human NSCLC
cell lines, including SK-MES-1, A549, H460 and SPC-A1 were
purchased from the Institute of Biochemistry and Cell Biology of
the Chinese Academy of Sciences (Shanghai, China) and were cultured
in Dulbecco's Modified Eagle's medium (DMEM) containing 10% FBS,
100 IU/mm penicillin and 100 µg/mm streptomycin (all from Gibco;
Thermo Fisher Scientific, Inc.). Cells were cultured at 37°C in a
humidified atmosphere with 5% CO2.
miR-629 inhibitor and control miRNA (NC inhibitor)
were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China).
The miR-629 inhibitor sequence was 5′-ACCCAAAUGCAACCCUCUUGA-3′ and
the NC inhibitor sequence was 5′-ACUACUGAGUGACAGUAGA-3′. To restore
the expression of RUNX3, a pCMV-RUNX3 plasmid was obtained from
RiboBio (Guangzhou, China); empty pCMV plasmids served as the
control. To knock down the expression of endogenous RUNX3, small
interfering RNA (siRNA) against the expression of RUNX3 (RUNX3
siRNA) and a negative control siRNA (NC siRNA) were purchased from
OriGene Technologies, Inc. (Beijing, China). The RUNX3 siRNA
sequence was 5′-TGACGAGAACTACTCCGCT-3′ and the NC siRNA sequence
was 5′-UUCUCCGAACGUGUCACGUTT-3′. Cells were seeded into 6-well
plates one day prior to transfection. Transient transfection was
conducted using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols. The
concentration of plasmids, miRNAs and siRNAs used was 4 µg, 100
pmol, and 100 pmol, respectively. Following 8 h of transfection,
cell culture medium was replenished with fresh DMEM containing 10%
FBS.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue specimens or
cells using TRIzol® reagent (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocols and was subjected
to complementary DNA (cDNA) synthesis using a TaqMan®
MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The temperature protocol for reverse
transcription was: 16°C for 30 min, 42°C for 30 min and 85°C for 5
min. The expression levels of miR-629 were determined using a
TaqMan MicroRNA Assay kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and was normalized by U6. The temperature
protocol for the reaction was: 50°C for 2 min, 95°C for 10 min; 40
cycles of denaturation at 95°C for 15 sec; and annealing/extension
at 60°C for 60 sec. To measure the mRNA expression levels of RUNX3,
RT was conducted using a PrimeScript™ RT Reagent kit, followed by
qPCR with a SYBR Green qPCR Master Mix (both from Takara
Biotechnology Co., Ltd., Dalian, China) by using an ABI 7500
thermocycler (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The temperature protocol for reverse transcription was: 37°C for 15
min and 85°C for 5 sec. The cycling conditions for qPCR were: 5 min
at 95°C, followed by 40 cycles of 95°C for 30 sec and 65°C for 45
sec. GAPDH served as an internal control for the mRNA expression of
RUNX3. The relative expression levels of miR-629 and RUNX3 were
calculated using the 2−∆ΔCq method (25). The primers were designed as
follows: miR-629, 5′-ACTTGTCCTATAGAAGCACAAC-3′ (forward) and
5′-ACTTGTCCTATAGAAGCACAAC-3′ (reverse); U6,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and
5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); RUNX3,
5′-GATGGCAGGCAATGACGA-3′ (forward) and 5′-TGCTGAAGTGGCTTGTGGT-3′
(reverse); and GAPDH, 5′-AGCCTTCTCCATGGTGGTGAA-3′ (forward) and
5′-ATCACCATCTTCCAGGAGCGA-3′ (reverse). Each assay was repeated
three times.
MTT assay
The viability of NSCLC cells was determined using an
MTT assay. For this assay, transfected cells were harvested and
seeded into 96-well plates with an initial density of 3,000 cells
in each well. At 0, 24, 48 and 72 h post-inoculation, a total of 20
µl MTT solution (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) was added into each well. Following incubation for another
4 h at 37°C, the culture medium was discarded, and 200 µl dimethyl
sulfoxide (Sigma-Aldrich; Merck KGaA) was used to dissolve the
purple formazan. The optical density of each well was measured at a
wavelength of 490 nm using an automatic multi-well
spectrophotometer (BioTek Instruments, Inc., Winooski, VT,
USA).
In vitro invasion assay
Transfected cells were collected, washed with PBS
and suspended into FBS-free DMEM. A total of 1×105 cells
in 200 µl FBS-free DMEM was added into the upper compartment of the
24-well Transwell chambers that were precoated with Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA); 600 µl DMEM supplemented
with 10% FBS was used as a chemoattractant in the lower
compartments. After 24 h of incubation at 37°C, the non-invaded
cells were carefully removed using a cotton swab. The invaded cells
were then fixed with 100% methanol at 37°C for 30 min, stained with
0.5% crystal violet at 37°C for 30 min, washed with PBS and
air-dried. The invaded cells were imaged with an IX71 inverted
microscope (magnification, 200×; Olympus Corporation, Tokyo, Japan)
and quantified by counting the number of invaded cells in five
randomly selected fields.
Bioinformatic prediction and
luciferase reporter assay
To investigate the mechanisms underlying the roles
of miR-629 in NSCLC, bioinformatic analysis was performed to
predict the putative targets of miR-629 using TargetScan (release
7.2; March 2018; http://www.targetscan.org/) and microRNA.org (http://www.microrna.org/). The 3′-UTR of RUNX3
containing the wild-type (Wt) or mutant (Mut) binding sequences for
miR-629 was generated by Shanghai GenePharma Co., Ltd., and cloned
into the pGL3 luciferase vector (XbaI and HpaI; Promega
Corporation, Madison, WI, USA). The Wt and Mut luciferase plasmids
were defined as Wt-RUNX3-3′-UTR and Mut-RUNX3-3′-UTR, respectively.
For this assay, miR-629 inhibitor or NC inhibitor, together with
Wt-RUNX3-3′-UTR or Mut-RUNX3-3′-UTR, was transfected into cells
using Lipofectamine 2000, according to the manufacturer's
protocols. After 48 h of post-transfection, luciferase activities
were determined using a Dual-Luciferase Reporter Assay System
(Promega Corporation) in accordance with the manufacturer's
protocols. Firefly luciferase activity was normalized to that of
Renilla luciferase activity.
Protein extraction and western blot
analysis
Total protein was isolated from the tissue specimens
or cells using radioimmunoprecipitation assay buffer, and the
protein concentration was determined using a bicinchoninic acid
assay kit (both from Nanjing KeyGen Biotech Co., Ltd., Nanjing,
China). Equal amounts of protein per lane (30 µg) were loaded and
separated using 10% SDS-PAGE. The PVDF membranes were then blocked
in 5% fat-free dry milk in Tris-buffered saline containing 0.1%
Tween-20 (TBST) at room temperature for 2 h and incubated overnight
at 4°C with primary antibodies. The primary antibodies included:
Mouse anti-human monoclonal RUNX3 antibody (1:1,000; ab135248;
Abcam, Cambridge, UK) and mouse anti-human monoclonal GAPDH
antibody (1:1,000; ab9482; Abcam). After washing four times with
TBST, the membranes were incubated with goat anti-mouse IgG
horseradish peroxidase-conjugated secondary antibody (1:5,000;
ab6789; Abcam) at room temperature for 2 h. Protein signals were
visualised using Pierce® ECL Plus Western Blotting
Substrate (Pierce; Thermo Fisher Scientific, Inc.). GAPDH served as
an endogenous control for the normalisation of expression. Protein
expression was quantified using Quantity One software version 4.62
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Each assay was
repeated three times.
Statistical analysis
SPSS version 13.0 software (SPSS, Inc., Chicago, IL,
USA) was applied for all statistical analysis. All data are
presented as the mean ± standard deviation from at least three
independent experiments. The association between miR-629 and the
clinicopathological characteristics of NSCLC patients was
determined using a χ2 test. The differences between
groups were analyzed with a Student's t-test or one-way analysis of
variance (ANOVA) plus multiple comparisons. Student-Newman-Keuls
test was applied as the post-hoc test following ANOVA. P<0.05
was considered to indicate a statistically significant
difference.
Results
miR-629 expression is upregulated in
NSCLC tissues and cells
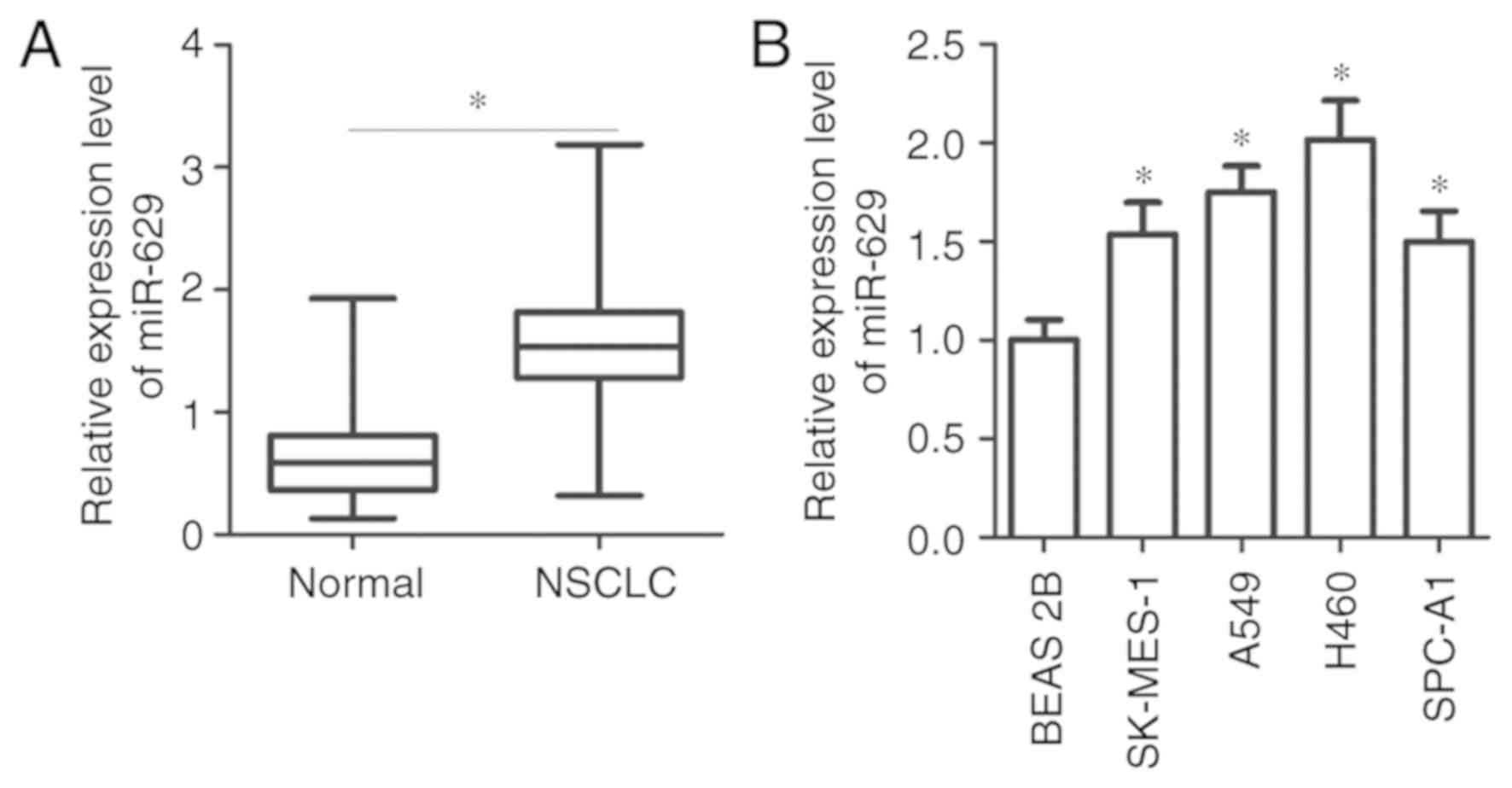
RT-qPCR analysis was performed to detect miR-629
expression in 51 pairs of NSCLC and adjacent normal tissues. The
results revealed that miR-629 expression was significantly
upregulated in NSCLC tissues compared with in adjacent normal
tissues (P<0.05; Fig. 1A). In
addition, the association between miR-629 expression and the
clinicopathological characteristics of patients with NSCLC was
investigated. All patients with NSCLC were divided into miR-629
high or low expression groups based on the median value of miR-629
expression. As presented in Table
I, high miR-629 expression levels were associated with tumour
size (P=0.015), clinical stage (P=0.007) and lymph node metastasis
(P=0.003). Furthermore, miR-629 expression was determined in NSCLC
cell lines. The data of RT-qPCR analysis revealed that the
expression levels of miR-629 were higher in all four tested NSCLC
cell lines, including SK-MES-1, A549, H460 and SPC-A1, compared
with the BEAS-2B cell line (P<0.05; Fig. 1B). These findings suggested that
miR-629 upregulation may be associated with NSCLC progression.
 | Table I.Association between miR-629 and the
clinicopathologic characteristics of non-small cell lung
cancer. |
Table I.
Association between miR-629 and the
clinicopathologic characteristics of non-small cell lung
cancer.
| Clinicopathological
characteristics | High miR-629
expression | Low miR-629
expression | P-value |
|---|
| Sex |
|
| 0.329 |
|
Male | 18 | 14 |
|
|
Female | 8 | 11 |
|
| Age (years) |
|
| 0.475 |
|
<50 | 6 | 8 |
|
|
≥50 | 20 | 17 |
|
| Tumor size
(cm) |
|
| 0.015 |
|
<3 | 11 | 19 |
|
| ≥3 | 15 | 6 |
|
|
Differentiation |
|
| 0.322 |
|
Moderate-well | 12 | 15 |
|
|
Poor | 14 | 10 |
|
| Clinical stage |
|
| 0.007 |
|
I–II | 9 | 18 |
|
|
III–IV | 17 | 7 |
|
| Lymph node
metastasis |
|
| 0.003 |
|
Negative | 10 | 20 |
|
|
Positive | 16 | 5 |
|
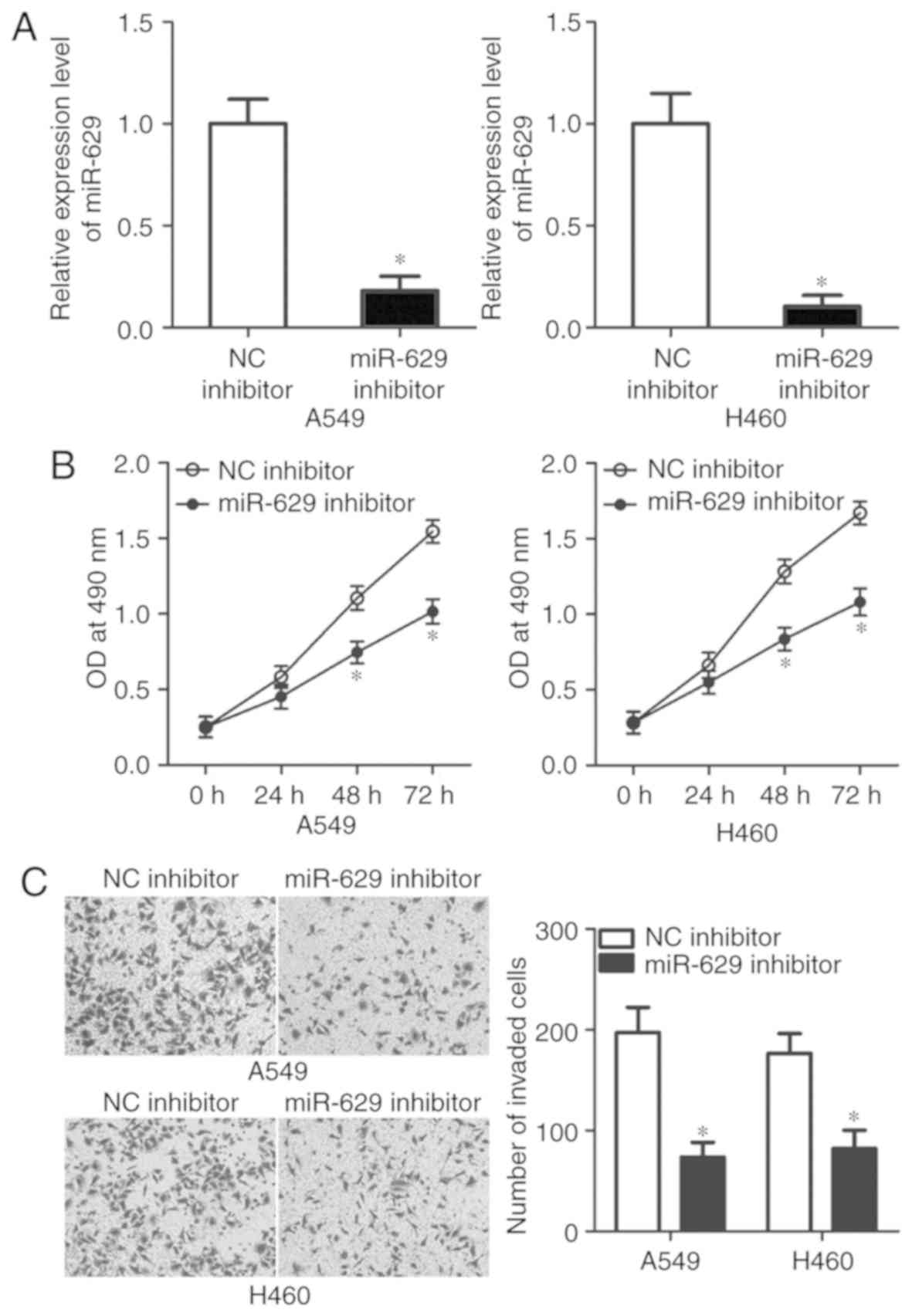
miR-629 downregulation suppresses A549
and H460 cell viability and invasion
To explore the detailed roles of miR-629 in NSCLC,
A549 and H460 cell lines were selected in the subsequent
experiments as these two cell lines exhibited higher miR-629
expression levels compared with SK-MES-1 and SPC-A1. miR-629
inhibitor was used to knock down miR-629 expression in A549 and
H460 cells (P<0.05; Fig. 2A).
An MTT assay was performed to investigate the effects of miR-629 on
NSCLC cell viability. The data revealed that miR-629 inhibition
suppressed the viability of A549 and H460 cells at 48 and 72 h
compared with the control (P<0.05; Fig. 2B). The effects of miR-629 knockdown
on the invasive ability of NSCLC cells were analysed in
vitro via an invasion assay. As presented in Fig. 2C, the invasive potential of A549
and H460 cells was significantly decreased by miR-629 inhibitor
compared with in the NC inhibitor groups (P<0.05). These results
suggested that miR-629 may serve an oncogenic role in the
development of NSCLC.
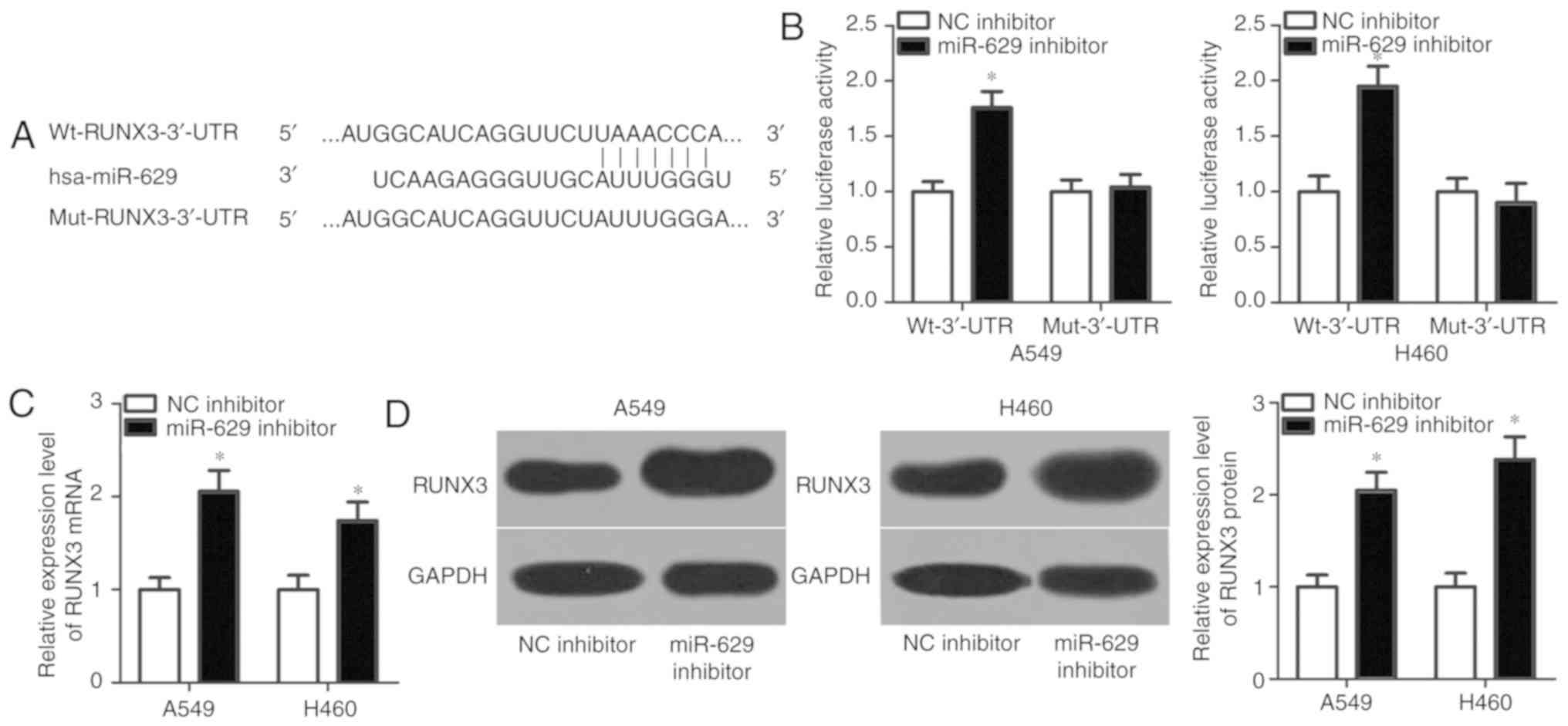
miR-629 directly targets RUNX3 in
NSCLC cells
To understand the molecular mechanisms underlying
the oncogenic roles of miR-629 in the progression of NSCLC,
bioinformatic analysis was conducted to predict the putative
targets of miR-629. RUNX3, a tumour suppressor in NSCLC (26–30),
was determined to contain a potential binding site for miR-629 in
its 3′-UTR (Fig. 3A). Luciferase
reporter assays were performed to examine the association between
miR-629 and RUNX3 in NSCLC. miR-629 downregulation significantly
increased the luciferase activity of the plasmid with wild-type
3′-UTR in A549 and H460 cells compared with the control
(P<0.05); however, the luciferase activities were notably
unaffected in the plasmid carrying mutant 3′-UTR of RUNX3 (Fig. 3B). To further investigate the
interaction between miR-629 and RUNX3 in NSCLC, RT-qPCR and western
blot analyses were employed to respectively measure the expression
levels of RUNX3 mRNA and protein in A549 and H460 cells transfected
with miR-629 inhibitor or NC inhibitor. Transfection with miR-629
inhibitor significantly upregulated the expression of RUNX3 mRNA
(P<0.05; Fig. 3C) and protein
(P<0.05; Fig. 3D) in A549 and
H460 cells compared with the control. These results suggested that
RUNX3 is a direct target of miR-629 in NSCLC cells.
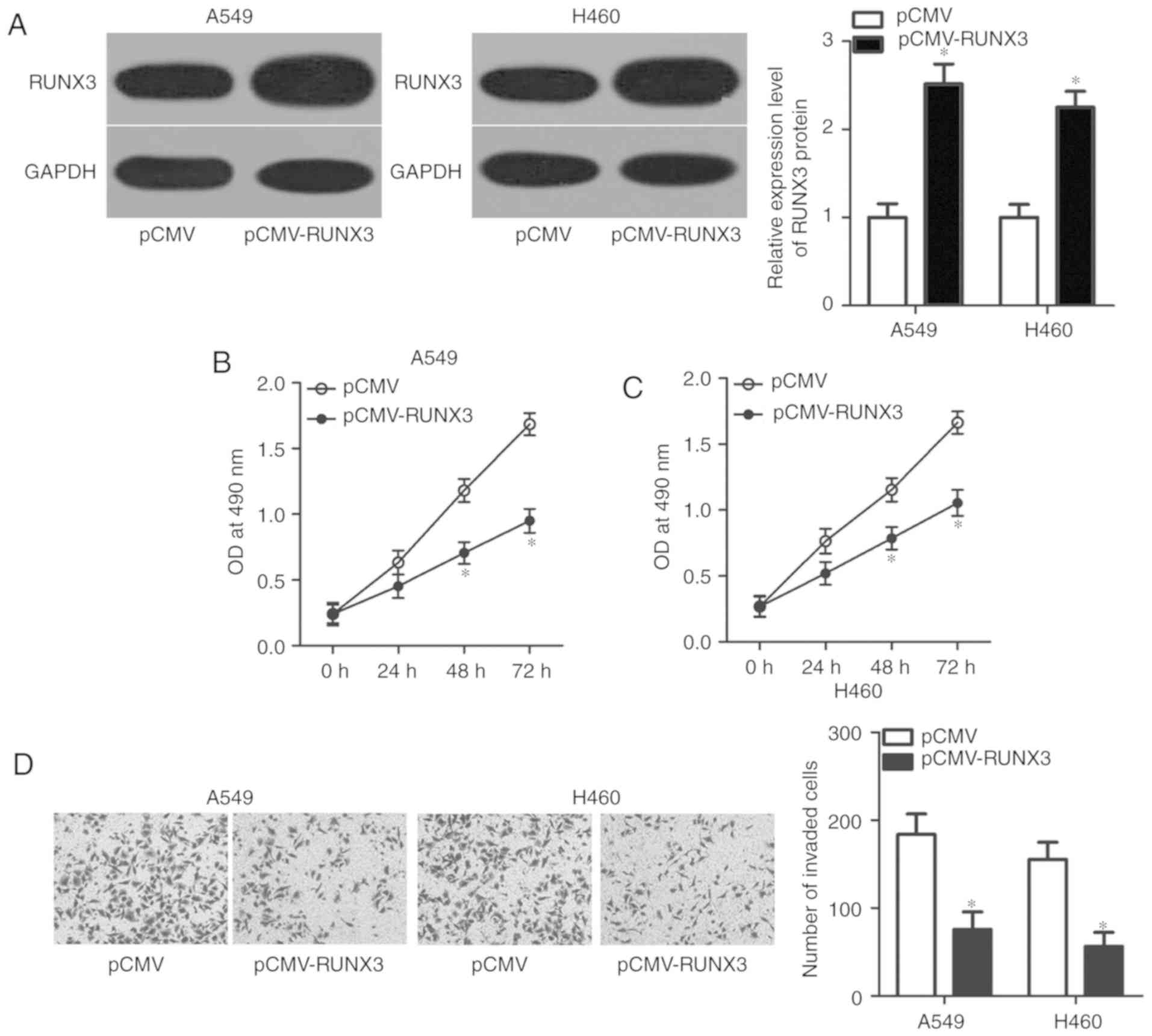
RUNX3 upregulation suppresses A549 and
H460 cell viability and invasion
The roles of RUNX3 in NSCLC cells were investigated
in the present study. A549 and H460 cells were transfected with
pCMV-RUNX3 or pCMV; RUNX3 protein expression was significantly
upregulated in A549 and H460 cells following transfection with
pCMV-RUNX3 compared with the control (P<0.05; Fig. 4A). Subsequent MTT and invasion
assays revealed that, similar to miR-629 knockdown, RUNX3
upregulation significantly suppressed the viability at 48 and 72 h,
(P<0.05; Fig. 4B and C) and
invasion (P<0.05; Fig. 4D) of
A549 and H460 cells compared with the control. These results
further demonstrated that RUNX3 is a functional downstream target
of miR-629 in NSCLC.
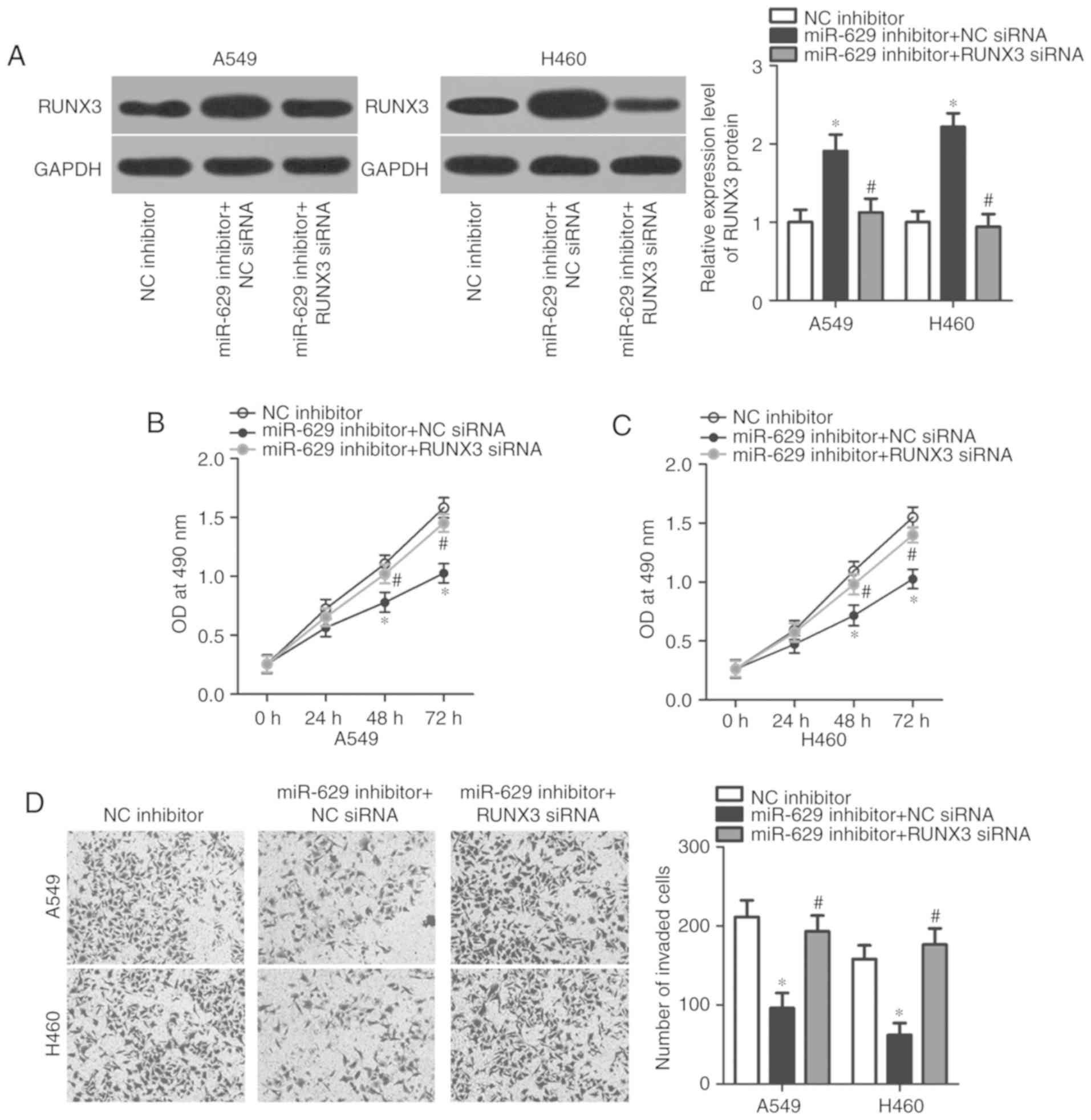
RUNX3 knockdown reverses the
phenotypes induced by miR-629 inhibition in A549 and H460
cells
Considering that RUNX3 was validated as a direct
target of miR-629 in NSCLC cells, whether RUNX3 upregulation is
required for the oncogenic roles of the miR-629 on NSCLC cells was
investigated in the present study. Rescue experiments were
performed by co-transfecting A549 and H460 cells with miR-629
inhibitor and RUNX3 siRNA or NC siRNA. Western blot analysis
demonstrated that co-transfection with RUNX3 siRNA significantly
downregulated the expression of RUNX3 in A549 and H460 cells
transfected with miR-629 inhibitor compared with cells exhibiting
miR-629 downregulation (P<0.05; Fig. 5A). MTT and in vitro invasion
assays revealed that RUNX3 knockdown rescued the effects of the
miR-629 inhibitor on A549 and H460 cell viability (P<0.05;
Fig. 5B and C) and invasion
(P<0.05; Fig. 5D). In summary,
our results suggested that miR-629 exerts oncogenic activity in
NSCLC cells by regulating RUNX3 expression.
Discussion
Dysregulated miRNAs directly modulate the biological
functions of NSCLC cells and contribute to the initiation and
progression of NSCLC (31–33); however, the specific roles and
underlying mechanisms of the dysregulated miRNAs in NSCLC require
further investigation. In the present study, it was reported that
miR-629 expression was significantly upregulated in NSCLC tissues
and cell lines. High miR-629 expression levels were highly
associated with the tumour size, clinical stage and lymph node
metastasis of patients with NSCLC. miR-629 inhibition suppressed
the viability and invasive ability of NSCLC cells. RUNX3 was
confirmed as a direct target gene of miR-629 in NSCLC cells. In
addition, RUNX3 overexpression exhibited similar effects of miR-629
inhibition in NSCLC cells. The rescue experiments demonstrated that
RUNX3 knockdown abrogated the effects of miR-629 downregulation in
NSCLC cells. In summary, miR-629 may exhibit oncogenic activity in
NSCLC by directly targeting RUNX3. Therefore, miR-629 serves a
pivotal role in NSCLC and may be an effective target for the
therapy of patients with this disease.
miR-629 dysregulation has been observed in numerous
types of human cancer (21–23).
For example, miR-629 expression is upregulated in breast cancer.
Patients with breast cancer and levels of high miR-629 exhibit
poorer overall survival and disease-free survival than those with
low miR-629 expression (21). In
addition, miR-629 was identified as an independent risk factor for
lung metastasis of breast cancer (21). miR-629 is also highly expressed in
clear cell renal cell carcinoma (22), and cervical (23), ovarian (34) and pancreatic cancers (35). These findings indicate that miR-629
is frequently upregulated in human cancers and suggest that miR-629
may be identified as a novel diagnostic and prognostic biomarker
for patients with these types of cancer.
miR-629 dysregulation is closely associated with the
malignant phenotype of cancers. For instance, miR-629
downregulation attenuates cell viability and migration of breast
cancer in vitro and decreases lung metastasis in vivo
(21). Jingushi et al
(22) reported that miR-629
downregulation inhibits cell migration and invasion of clear cell
renal cell carcinoma. Phuah et al (23) revealed that miR-629 knockdown
prohibits cell proliferation and promotes apoptosis, and thus
increases the sensitivity of cervical cancer cells to
1′S-1′-acetoxychavicol acetate. Shao et al (34) demonstrated that inhibiting
miR-suppressed inhibited cell metastasis and induced apoptosis in
ovarian cancer. Yan et al (35) reported that miR-629 inhibition
supresses proliferation, while increasing the apoptosis of
pancreatic cancer cells. These findings suggest that miR-629 may be
considered as a therapeutic target in the treatment of patients
with these specific types of cancer.
Numerous targets of miR-629 have been reported,
including leukaemia inhibitory factor receptor (21) in breast cancer, tripartite
motif-containing 33 (22) in clear
cell renal cell carcinoma, ras suppressor protein 1 (23) in cervical cancer, testis-specific
Y-like protein 5 (34) in ovarian
cancer and forkhead box O3 (35)
in pancreatic cancer. Identifying the targets of miR-629 in NSCLC
may improve understanding of the mechanisms underlying the
initiation and progression of NSCLC, which may facilitate the
identification of valuable therapeutic targets of patients with
NSCLC. RUNX3, which is located on chromosome 1p36, was identified
as a direct target gene of miR-629 in NSCLC in the present study.
RUNX3 was notably downregulated in NSCLC, which was reported in
patients with poorly differentiated NSCLC. Additionally, patients
with NSCLC and low RUNX3 expression levels demonstrated lower
five-year survival rates than those with high expression (26). RUNX3 serves crucial roles in the
oncogenesis and development of NSCLC by regulating cell
proliferation (27), invasion
(28), epithelial-mesenchymal
transition (29) and tumorigenesis
(30). In the present study, it
was demonstrated that miR-629 directly targeted RUNX3 to inhibit
the progression of NSCLC. Therefore, miR-629-based inhibition of
RUNX3 may be a promising therapeutic method to treat patients with
NSCLC.
In conclusion, miR-629 was overexpressed in NSCLC
tissues and cell lines. High miR-629 expression levels were closely
associated with tumour size, clinical stage and lymph node
metastasis in patients with NSCLC. miR-629 served oncogenic roles
in NSCLC by directly targeting RUNX3. The findings of the present
study may provide novel evidence for the potential of miR-629 as a
therapeutic target for NSCLC; however, the association between
miR-629 and the prognosis of patients with NSCLC was not
investigated. This limitation of our study may be resolved in
future experiments.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Experiment
research on Photodynamic therapy combined with Cisplatin treat
central type lung cancer (grant no. 20154Y0156).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
BZ and YC made substantial contributions to the
design of the present study and performed functional assays. The
authors read and approved the final draft of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shanghai Ninth People's Hospital (Shanghai, China),
and was performed in accordance the guidelines of the Ethics
Committee of Shanghai Ninth People's Hospital. Written informed
consent was obtained from all patients for the use of their
clinical tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nascimento AV, Bousbaa H, Ferreira D and
Sarmento B: Non-small cell lung carcinoma: An overview on targeted
therapy. Curr Drug Targets. 16:1448–1463. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Holdenrieder S and Stieber P: New
challenges for laboratory diagnostics in non-small cell lung
cancer. Cancer Biomark. 6:119–121. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ramnath N, Dilling TJ, Harris LJ, Kim AW,
Michaud GC, Balekian AA, Diekemper R, Detterbeck FC and Arenberg
DA: Treatment of stage III non-small cell lung cancer: Diagnosis
and management of lung cancer, 3rd ed: American College of Chest
Physicians evidence-based clinical practice guidelines. Chest. 143
(Suppl 5):e314S–e340S. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ding X, Yang Y, Sun Y, Xu W, Su B and Zhou
X: MicroRNA-585 acts as a tumor suppressor in non-small-cell lung
cancer by targeting hSMG-1. Clin Transl Oncol. 19:546–552. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu X and Chen J: Advances in diagnosis
and treatment and whole process management of anaplastic lymphoma
kinase (ALK)-positive non-small cell lung cancer. Zhonghua Zhong
Liu Za Zhi. 37:1–4. 2015.(In Chinese). PubMed/NCBI
|
|
7
|
Rosell R and Karachaliou N: Lung cancer:
Maintenance therapy and precision medicine in NSCLC. Nat Rev Clin
Oncol. 10:549–550. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schnabel PA and Junker K: Neuroendocrine
tumors of the lungs. From small cell lung carcinoma to diffuse
idiopathic pulmonary neuroendocrine cell hyperplasia. Pathologe.
35:557–564. 2014.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Z, Song Y, Liu L, Hou N, An X, Zhan D,
Li Y, Zhou L, Li P, Yu L, et al: miR-199a impairs autophagy and
induces cardiac hypertrophy through mTOR activation. Cell Death
Differ. 24:1205–1213. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mao M, Wu Z and Chen J: MicroRNA-187-5p
suppresses cancer cell progression in non-small cell lung cancer
(NSCLC) through down-regulation of CYP1B1. Biochem Biophys Res
Commun. 478:649–655. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Moretti F, D'Antona P, Finardi E, Barbetta
M, Dominioni L, Poli A, Gini E, Noonan DM, Imperatori A, Rotolo N,
et al: Systematic review and critique of circulating miRNAs as
biomarkers of stage I–II non-small cell lung cancer. Oncotarget.
8:94980–94996. 2017.PubMed/NCBI
|
|
12
|
Gompelmann D, Eberhardt R and Herth FJ:
Advanced malignant lung disease: What the specialist can offer.
Respiration. 82:111–123. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Donadeu FX, Schauer SN and Sontakke SD:
Involvement of miRNAs in ovarian follicular and luteal development.
J Endocrinol. 215:323–334. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liwak U, Faye MD and Holcik M: Translation
control in apoptosis. Exp Oncol. 34:218–230. 2012.PubMed/NCBI
|
|
16
|
Rutnam ZJ and Yang BB: The involvement of
microRNAs in malignant transformation. Histol Histopathol.
27:1263–1270. 2012.PubMed/NCBI
|
|
17
|
Wang N and Zhang T: Down-regulation of
microRNA-135 promotes sensitivity of non-small cell lung cancer to
gefitinib by targeting TRIM16. Oncol Res. 26:1005–1014. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jin JJ, Liu YH, Si JM, Ni R and Wang J:
Overexpression of miR-1290 contributes to cell proliferation and
invasion of non small cell lung cancer by targeting interferon
regulatory factor 2. Int J Biochem Cell Biol. 95:113–120. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao X, Li S, Li W, Wang G, Zhao W, Han J,
Diao C, Wang X and Zhang M: MicroRNA-539 suppresses tumor cell
growth by targeting the WNT8B gene in non-small cell lung cancer. J
Cell Biochem. 120:2017.
|
|
20
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang J, Song C, Tang H, Zhang C, Tang J,
Li X, Chen B and Xie X: miR-629-3p may serve as a novel biomarker
and potential therapeutic target for lung metastases of
triple-negative breast cancer. Breast Cancer Res. 19:722017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jingushi K, Ueda Y, Kitae K, Hase H, Egawa
H, Ohshio I, Kawakami R, Kashiwagi Y, Tsukada Y, Kobayashi T, et
al: miR-629 targets TRIM33 to promote TGFbeta/Smad signaling and
metastatic phenotypes in ccRCC. Mol Cancer Res. 13:565–574. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Phuah NH, Azmi MN, Awang K and Nagoor NH:
Suppression of microRNA-629 enhances sensitivity of cervical cancer
cells to 1′S-1′-acetoxychavicol acetate via regulating RSU1. Onco
Targets Ther. 10:1695–1705. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hattori A, Takamochi K, Oh S and Suzuki K:
New revisions and current issues in the eighth edition of the TNM
classification for non-small cell lung cancer. Jpn J Clin Oncol.
49:3–11. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Araki K, Osaki M, Nagahama Y, Hiramatsu T,
Nakamura H, Ohgi S and Ito H: Expression of RUNX3 protein in human
lung adenocarcinoma: Implications for tumor progression and
prognosis. Cancer Sci. 96:227–231. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Torshabi M, Faramarzi MA, Tabatabaei Yazdi
M, Ostad SN and Gharemani MH: Runx3 expression inhibits
proliferation and distinctly alters mRNA expression of bax in AGS
and A549 cancer cells. Iran J Pharm Res. 10:355–361.
2011.PubMed/NCBI
|
|
28
|
Wang Y, Li Y, Wu B, Shi C and Li C:
MicroRNA-661 promotes non-small cell lung cancer progression by
directly targeting RUNX3. Mol Med Rep. 16:2113–2120. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee JM, Shin JO, Cho KW, Hosoya A, Cho SW,
Lee YS, Ryoo HM, Bae SC and Jung HS: Runx3 is a crucial regulator
of alveolar differentiation and lung tumorigenesis in mice.
Differentiation. 81:261–268. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee KS, Lee YS, Lee JM, Ito K, Cinghu S,
Kim JH, Jang JW, Li YH, Goh YM, Chi XZ, et al: Runx3 is required
for the differentiation of lung epithelial cells and suppression of
lung cancer. Oncogene. 29:3349–3361. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cortinovis D, Monica V, Pietrantonio F,
Ceresoli GL, La Spina CM and Wannesson L: MicroRNAs in non-small
cell lung cancer: Current status and future therapeutic promises.
Curr Pharm Des. 20:3982–3990. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Boeri M, Pastorino U and Sozzi G: Role of
microRNAs in lung cancer: microRNA signatures in cancer prognosis.
Cancer J. 18:268–274. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vannini I, Fanini F and Fabbri M:
MicroRNAs as lung cancer biomarkers and key players in lung
carcinogenesis. Clin Biochem. 46:918–925. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shao L, Shen Z, Qian H, Zhou S and Chen Y:
Knockdown of miR-629 inhibits ovarian cancer malignant behaviors by
targeting testis-specific Y-like protein 5. DNA Cell Biol.
36:1108–1116. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yan H, Li Q, Wu J, Hu W, Jiang J, Shi L,
Yang X, Zhu D, Ji M and Wu C: MiR-629 promotes human pancreatic
cancer progression by targeting FOXO3. Cell Death Dis. 8:e31542017.
View Article : Google Scholar : PubMed/NCBI
|