Introduction
Toll-like receptor (TLR)-mediated production of
proinflammatory cytokines is necessary to eliminate invading
pathogens. However, uncontrolled TLR activation can result in many
diseases, including diabetes (1),
obesity (2), cancer (3) and root resorption (4). Lipopolysaccharide (LPS) triggers
innate immunity and activates multiple signaling pathways in
macrophages, which results in the production of inflammatory
cytokines (5,6). These activities are essential for the
control and maintenance of balanced inflammatory reactions.
MicroRNAs (miRNAs/miRs), a class of short non-coding
RNAs that are 19–24 nucleotides in length, can bind to the
3′-untranslated region (UTR) of target genes, resulting in target
mRNA degradation and translation inhibition (7). A number of miRNAs associated with
LPS-induced inflammatory responses have been identified in
macrophages, including miR-146a (8), miR-71 (9), miR-709 (10), miR-181a (11) and miR-27a (12). However, there are many
uncharacterized miRNAs that need to be explored with respect to
their function in the inflammatory response.
Phosphatase and tensin homolog (PTEN) is involved in
the tumorigenesis of many malignancies (13). PTEN is a lipid phosphatase that can
dephosphorylate phosphoinositol (3–5)-trisphosphate at position 3, thus
antagonizing phosphoinositide 3-kinase (PI3K) signaling. PTEN is
involved in various cellular processes, including proliferation,
cellular architecture and survival (14). A previous study suggested that this
protein may have an important role in inflammatory responses
(15). Agrawal et al
(16) reported that overexpression
of PTEN in dendritic cells from elderly individuals significantly
increases LPS-induced secretion of tumor necrosis factor (TNF)-α
and interleukin (IL)-6. In addition, Cao et al (17) demonstrated that PTEN was a positive
mediator of macrophage inflammatory responses.
The aim of the present study was to investigate and
characterize miRNAs that are differentially expressed in
macrophages in response to LPS stimulation. The results indicated
that miR-494-3p, which can target the 3′-UTR of the PTEN gene, was
involved in LPS-induced immune responses. In addition, the
mechanisms associated with this observation were determined.
Exploring the role of miR-494-3p in LPS-stimulated macrophages,
including RAW264.7 cells, may aid in the development of
anti-miR-494-3p as novel therapeutic agents for inflammatory
diseases.
Materials and methods
Affymetrix microarray data
The GSE43300 array dataset, based on the GPL7723
platform, was downloaded from the Gene Expression Omnibus (GEO;
http://www.ncbi.nlm.nih.gov/geo/)
database. These data were deposited by Chen et al (18). A total of two LPS-treated cell
groups and two negative control groups were included in the present
study. Raw data and annotation files were used for subsequent
analysis.
Data preprocessing
The R statistical software (version 3.5.0) program
in Bioconductor (version 3.8; http://www.bioconductor.org/) was used, as it provides
a robust multiarray average algorithm to preprocess raw expression
data. A total of 579 miRNA expression values were obtained and the
limma package (19) in
Bioconductor was used to analyze differential miRNA expression
between the LPS-treated and control groups. The t-test in the limma
package was used to calculate P-values for differential miRNA
expression. Log2-fold change ≥1.2 and P<0.05 were
used as the cut-off criteria. The pheatmap package (version 1.0.12)
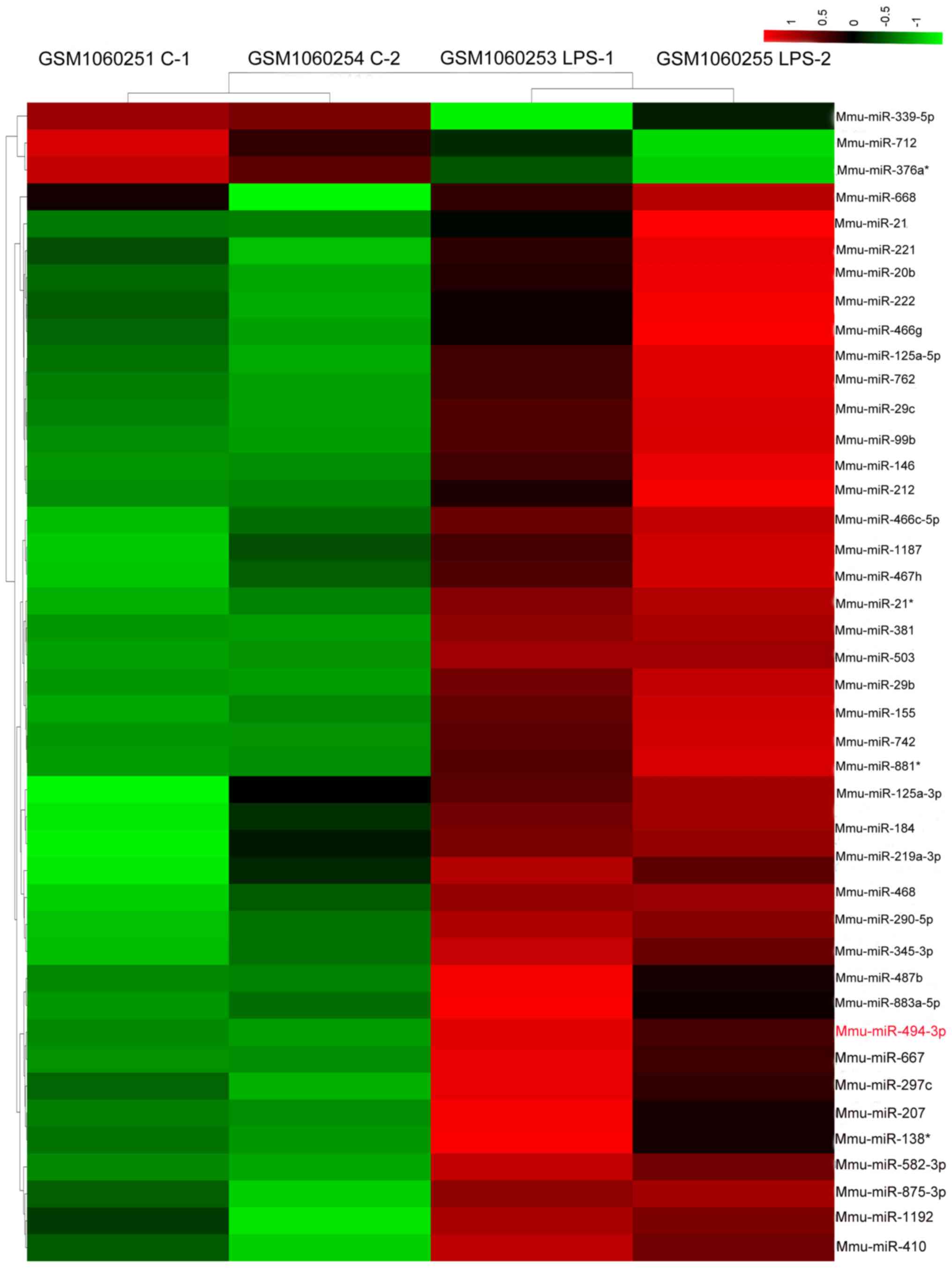
was used to generate the heat map presented in Fig. 1.
Cell lines and transfection
The RAW264.7 murine macrophage leukemia cell line
and 293T cell line were purchased from the American Type Culture
Collection (Manassas, VA, USA) and cultured in RPMI-1640 medium
supplemented with 10% fetal bovine serum (Biological Industries
USA, Cromwell, CT, USA) and antibiotics (100 U/ml penicillin and
100 µg/ml streptomycin) at 37°C with 5% CO2.
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) was used to transfect cells
with miR-494-3p mimics (5′-UGAAACAUACACGGGAAACCUC-3′; 20 µM),
miR-494-3p inhibitor (5′-GAGGUUUCCCGUGUAUGUUUCA-3′; 20 µM),
negative control (5′-UUCUCCGAACGUGUCACGUTT-3′; 20 µM), inhibitor
negative control (5′-CAGUACUUUUGUGUAGUACAA-3′; 20 µM), short
interfering RNA (siRNA)-PTEN (sense, 5′-CAAAUCCAGAGGCUAGCAGUU-3′
and antisense, 5′-CUGCUAGCCUCUGGAUUUGTT-3′; 20 µM), siRNA negative
control (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′; 20 µM; all from Shanghai GenePharma
Co., Ltd., Shanghai China) and pmiRGLO Dual-Luciferase miRNA Target
Expression Vector plasmids (500 ng/µl; Promega Corporation,
Madison, WI, USA), according to the manufacturer's instructions.
Following transfection for 48 h, cells were continuously stimulated
with LPS at a concentration of 1 µg/ml for 24 h at 37°C in the
presence of 5% CO2. Total RNA and protein was
subsequently extracted for reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) and western blotting
analysis.
RNA extraction and RT-qPCR
RAW264.7 cells were seeded in 6-well plates and
incubated overnight, following which cells were treated with
various concentrations of LPS (0, 1, 2, 3 and 6 µg/ml) for 24 h at
37°C in the presence of 5% CO2. The expression levels of
IL-1β and TNF were detected by RT-qPCR. TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to isolate
total RNA from RAW264.7 cells. cDNA was synthesized using a
PrimeScript™ RT reagent kit (RR037A; Takara Biotechnology Co.,
Ltd., Dalian, China), according to the manufacturer's protocols.
qPCR was performed using the SYBR-Green qPCR Master Mix (2X
RealStar Green Power Mixture; A311-05; GenStar, Beijing, China) on
an FTC-3000 thermal cycler (Funglyn Biotech Inc., Richmond Hill,
ON, Canada). qPCR was performed as follows: 95°C for 10 min,
followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. U6 and
GAPDH were used as the internal reference genes. The
2−ΔΔCq method was used to calculate the relative
expression of genes (20). All
primers used in the study are listed in Table I. The RNA extraction and RT-qPCR
experiments were repeated three times.
 | Table I.Primers used for reverse
transcription-quantitative polymerase chain reaction and luciferase
reporter construction. |
Table I.
Primers used for reverse
transcription-quantitative polymerase chain reaction and luciferase
reporter construction.
| Gene | Sequence |
|---|
| U6 |
F:5′-CTCGCTTCGGCAGCACA-3′ |
|
|
R:5′-AACGCTTCACGAATTTGCGT-3′ |
| miR-494-3p |
F:5′-ATCCAGTGCGTGTCGTG-3′ |
|
|
R:5′-TGCTTAGCTTATCAGACTG-3′ |
| GAPDH |
F:5′-GGTTGTCTCCTGCGACTTCA-3′ |
|
|
R:5′-GGTGGTCCAGGGTTTCTTACT-3′ |
| TNF-α |
F:5′-TCTTCTCATTCCTGCTTGTGG-3′ |
|
|
R:5′-GGTCTGGGCCATAGAACTGA-3′ |
| IL-1β |
F:5′-AGTTGACGGACCCCAAAAG-3′ |
|
|
R:5′-AGCTGGATGCTCTCATCAGG-3′ |
| PTEN |
F:5′-AAATGCGTACCTACCTTGCC-3′ |
|
|
R:5′-TGTTGTTAGCCCACCAGAA-3′ |
|
pmiRGLO/PTEN-WT-3′UTR |
F:5′-CAGGGTTTTGATTTTGAATGTTTCAC-3′ |
|
|
R:5′-TCGAGTGAAACATTCAAAATCAAAACCCTGAGCT-3′ |
|
pmiRGLO/PTEN-MUT-3′UTR |
F:5′-CAGCCTTTGATTTACAAACTTCAC-3′ |
|
|
R:5′-TCGAGTGAAGTTTGTAAATCAAAGGCTGAGCT-3′ |
Western blot analysis
Radioimmunoprecipitation assay buffer supplemented
with protease and phosphatase inhibitors (Beyotime Institute of
Biotechnology, Shanghai, China) was used to lyse RAW264.7 cells.
Protein concentration was determined using a bicinchoninic acid kit
(E162-01; GenStar). The nuclear protein was obtained using a
Cytoplasmic and Nuclear Extract kit (KGP1100; Nanjing KeyGen
Biotech Co., Ltd., Nanjing, China). Protein (20 µg/lane) was then
added to 5X loading buffer, separated by 15% SDS-PAGE and
electrophoretically transferred to a polyvinylidene difluoride
membrane (EMD Millipore, Billerica, MA, USA). The membrane was were
blocked with 5% non-fat milk at room temperature for 1 h and
incubated with primary antibodies overnight at 4°C: β-actin
(1:1,000; cat. no. 4970; Cell Signaling Technology, Inc., Danvers,
MA, USA); lamina (1:1,000; 10298-1-AP; ProteinTech Group, Inc.,
Chicago, IL, USA); IL-1β (1:1,000; cat. no. 31202; Cell Signaling
Technology, Inc.); TNF-α (1:2,000; 60291-1-Ig; ProteinTech Group,
Inc.); PTEN (1:1,000; cat. no. 9552; Cell Signaling Technology,
Inc.); AKT serine/threonine kinase 1 (Akt1; 1:1,000; cat. no. 2967;
Cell Signaling Technology, Inc.), phosphorylated (p)-Akt1 (1:1,000;
cat. no. 9018; Cell Signaling Technology, Inc.) and p65 (1:1,500;
ab16502; Abcam, Cambridge, UK). The membranes were subsequently
incubated for 1 h at room temperature with donkey anti-rabbit
immunoglobulin G secondary antibodies (1:2000; cat. no.
711-005-152; Jackson ImmunoResearch, West Grove, PA, USA). The
protein bands were visualized using Immobilon™ chemiluminescent
substrate (WBKLS0050; EMD Millipore). β-actin and lamina were used
as the loading controls. Blots were scanned using a Bio-Rad
ChemiDoc™ XRS (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and
the band densities were quantified using Quantity One®
analysis software version 4.6.2 (Bio-Rad Laboratories, Inc.).
Construction of luciferase reporter
plasmid and dual luciferase assay
Putative miRNA target genes were predicted by
application of TargetScan 7.2 (http://www.targetscan.org/mmu_72/), picTar (https://pictar.mdc-berlin.de/), PITA version 6
(https://genie.weismann.ac.il/pubs/mir07/mir07_data.html)
and miRanda version 3.3a (http://www.microrna.org/microrna/home.do). Venn
diagram analysis (Venny; version 2.1; http://bioinfogp.cnb.csic.es/tools/venny/) was used to
identify the intersection between the four target predictions; PTEN
was identified as a candidate mRNA associated with miR-494-3p. To
validate that miR-494-3p can target PTEN, a reporter plasmid
containing the 3′-UTR sequence of PTEN was generated. Primers were
designed based on the National Center for Biotechnology Information
reference sequence NM_008960.2, and specific primers were used for
the amplification of wild-type and mutant 3′-UTR sequences from
PTEN mRNA (Table I). The wild-type
and mutated 3′-UTR fragment were cloned downstream of the
luciferase reporter gene of the pmiRGLO-reporter vector, using the
XhoI and SacI restriction sites. One day prior to
transfection, 293T cells were seeded into a 96-well plate at a
density of 1×104 cells/well. Then, cells were
co-transfected with miR-494-3p mimics and PTEN luciferase reporter
constructs using Lipofectamine 3000. Blank pmiRGLO dual-luciferase
was used as a positive control. After 24 h, a
Dual-Luciferase® Reporter Assay system (Promega
Corporation) was used to measure reporter activity. The ratio of
the Renilla fluorescence value to the firefly fluorescence
value was calculated.
Confocal microscopy
To determine the effect of miR-494-3p on the protein
level of PTEN, immunocytochemistry was performed using the PTEN
antibody. Cells were seeded 24 h prior to treatment. Following
transfection with miR-494-3p mimics or negative controls for 48 h,
RAW264.7 cells were stimulated with LPS (1 µg/ml) for 24 h at 37°C
with 5% CO2, and PTEN expression was determined. The
transfected RAW264.7 cell lines were fixed with 4% formaldehyde for
30 min at room temperature, then incubated with 0.5% Triton X-100
for 10 min at room temperature. Cells were washed three times with
PBS and blocked with goat serum (5%; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for 1 h at room temperature. The cells were
incubated with anti-PTEN antibody (1:100) at 4°C overnight, washed
three times with PBS, and then incubated with goat anti-rabbit
secondary antibody (Cy3 AffiniPure Goat Anti-Rabbit Immunoglobulin
G; 1:1,000; E031640-01, EarthOx Life Sciences, Milbrae, CA, USA)
for 1 h at 37°C in the dark. The cells were incubated with DAPI
(1:10,000; cat. no. 4083; Cell Signaling Technology, Inc.) for 5
min at room temperature. Images of PTEN expression were acquired by
confocal microscopy (magnification, ×10; A1; Nikon Corporation,
Tokyo, Japan).
Statistical analysis
SPSS software version 13.0 (SPSS, Inc., Chicago, IL,
USA) was used to perform statistical analyses. Data are presented
as the mean ± standard deviation of at least three independent
experiments. One-way analysis of variance followed by Bonferroni
post hoc test was used to compare the means of multiple groups.
Student's t-test was used to assess differences between two groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Data processing and analysis of
differential miRNA expression
Following preprocessing of the raw expression data
from the GSE43300 array dataset, 44 upregulated and three
downregulated miRNAs were identified in the LPS-treated samples
compared with in the control samples. The number of upregulated
genes was higher than the number of downregulated genes. A
hierarchical heatmap of the 47 differentially expressed miRNAs is
shown in Fig. 1. Notably,
miR-494-3p was upregulated by LPS in RAW264.7 cells.
LPS stimulation regulates miR-494-3p
expression in murine macrophages
It was previously demonstrated that RAW264.7 cells
can release inflammatory cytokines in response to LPS stimulation
(21). To verify this, RAW264.7
cells were treated with different concentrations of LPS, and IL-1β
and TNF-α expression were measured. The IL-1β and TNF-α mRNA and
protein expression levels were increased following LPS stimulation,
with peak expression occurring with 1 µg/ml LPS (Fig. 2A and B). To determine whether LPS
treatment would affect miR-494-3p expression in RAW264.7 cells,
RT-qPCR was performed. As shown in Fig. 2C, peak expression of miR-494-3p
occurred following stimulation with 1 µg/ml LPS. A previous study
also selected 1 µg/ml LPS to induce inflammatory responses in
RAW264.7 cells (22). The
expression miR-494-3p, IL-1β and TNF-α following LPS treatment (1
µg/ml) was upregulated; however, the expression of all the
molecules notably decreased following stimulation with >2 µg/ml
LPS compared with 1 µg/ml LPS. This may be due to high
concentrations of LPS inducing apoptosis in cells instead of
inflammatory responses (23).
Further investigation into the underlying mechanisms is required.
The results indicated that miR-494-3p expression may be closely
associated with LPS-induced inflammatory responses in RAW264.7
cells.
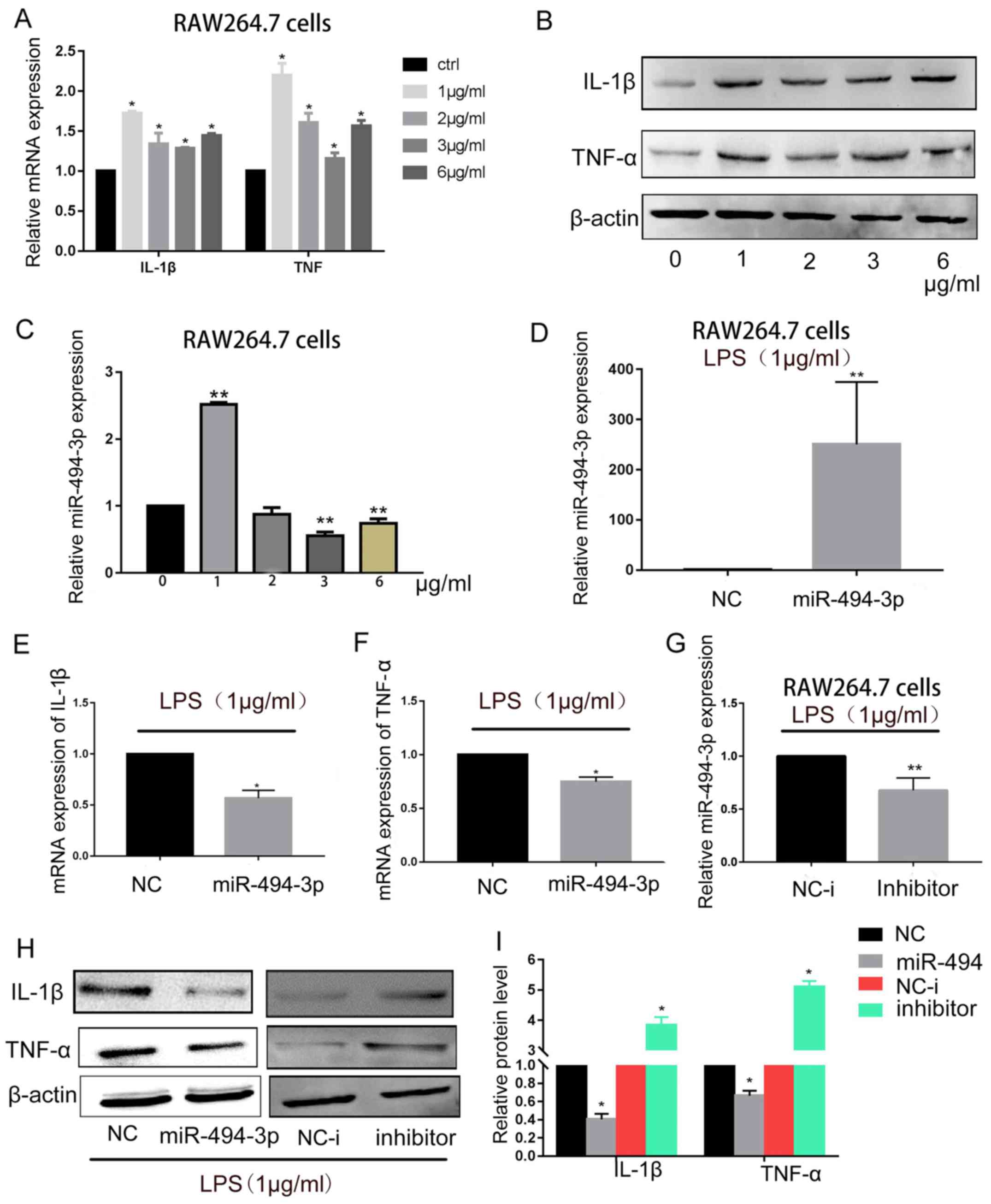 | Figure 2.miR-494-3p inhibits inflammatory
responses in LPS-treated RAW264.7 cells. RAW264.7 cells were
treated with 1–6 µg/m1 LPS for 24 h. (A) RT-qPCR and (B) western
blotting of IL-1β and TNF-α were performed. (C) RT-qPCR was used to
determine miR-494-3p expression levels. *P<0.05, **P<0.01 vs.
0 µg/ml LPS. (D) Next, RAW264.7 cells were transfected with 50 nM
miR-494-3p mimics or NC. After 24 h, cells were treated with 1
µg/ml LPS for 24 h. Transfection efficiency was confirmed by
RT-qPCR. (E and F) IL-1β and TNF-α mRNA expression levels were
detected using RT-qPCR. (G) RAW264.7 cells were transfected with 50
nM miR-494-3p inhibitor or inhibitor NC. After 24 h, cells were
treated with 1 µg/ml LPS for 24 h. Transfection efficiency was
confirmed by RT-qPCR. (H) IL-1β and TNF-α protein expression levels
were detected using western blotting. (I) Quantification of protein
expression. Data are presented as the mean ± standard deviation of
at least three independent experiments. *P<0.05, **P<0.01 vs.
NC. IL, interleukin; LPS, lipopolysaccharide; miR, microRNA; NC,
negative control; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; TNF, tumor necrosis factor. |
miR-494-3p regulates LPS-induced
inflammation
To explore the regulatory function of miR-494-3p
during LPS-induced inflammation, RAW264.7 cells were transfected
with miR-494-3p mimics or negative control (Fig. 2D), and RT-qPCR was performed to
measure IL-1β and TNF-α mRNA levels. Overexpression of miR-494-3p
downregulated IL-1β and TNF-α expression levels in LPS-treated
RAW264.7 cells (Fig. 2E and F). To
further confirm these results, an inhibitor was used to suppress
miR-494-3p expression (Fig. 2G),
and the results demonstrated that the protein expression levels of
IL-1β and TNF-α were increased in RAW264.7 cells (Fig. 2H and I). These results suggested
that miR-494-3p may regulate proinflammatory cytokine production
via a negative feedback loop in RAW264.7 cells.
miR-494-3p targets the 3′-UTR of PTEN
and regulates its expression
Next, to explore the molecular function of
miR-494-3p in suppressing inflammatory responses, bioinformatics
analysis was used to identify potential target genes. Putative
miRNA target genes were identified by employing TargetScan, picTar,
PITA and miRanda. The miRNA databases predicted a seed match for
miR-494-3p in the 3′-UTR region of PTEN (Fig. 3A and B). Sequences from the
predicted miR-494-3p target site in the 3′-UTR of PTEN, and mutant
variants, were successfully cloned into the pmiRGLO vector.
RAW264.7 cells were co-transfected with miR-494-3p mimics and the
pmiRGLO vector. Transfection with miR-494-3p mimics inhibited the
luciferase activity of pmiRGLO-PTEN-wild-type, but not that of
pmiRGLO-PTEN-mutant (Fig. 3C).
This indicated that miR-494-3p bound specifically to the PTEN
3′-UTR. Confocal microscopy and western blotting was used to
determine whether miR-494-3p inhibited the protein expression
levels of PTEN in RAW264.7 cells. PTEN protein expression levels
were decreased in RAW264.7 cells transfected with miR-494-3p
mimics. Conversely, it was increased with a miR-494-3p inhibitor
(Fig. 3D).
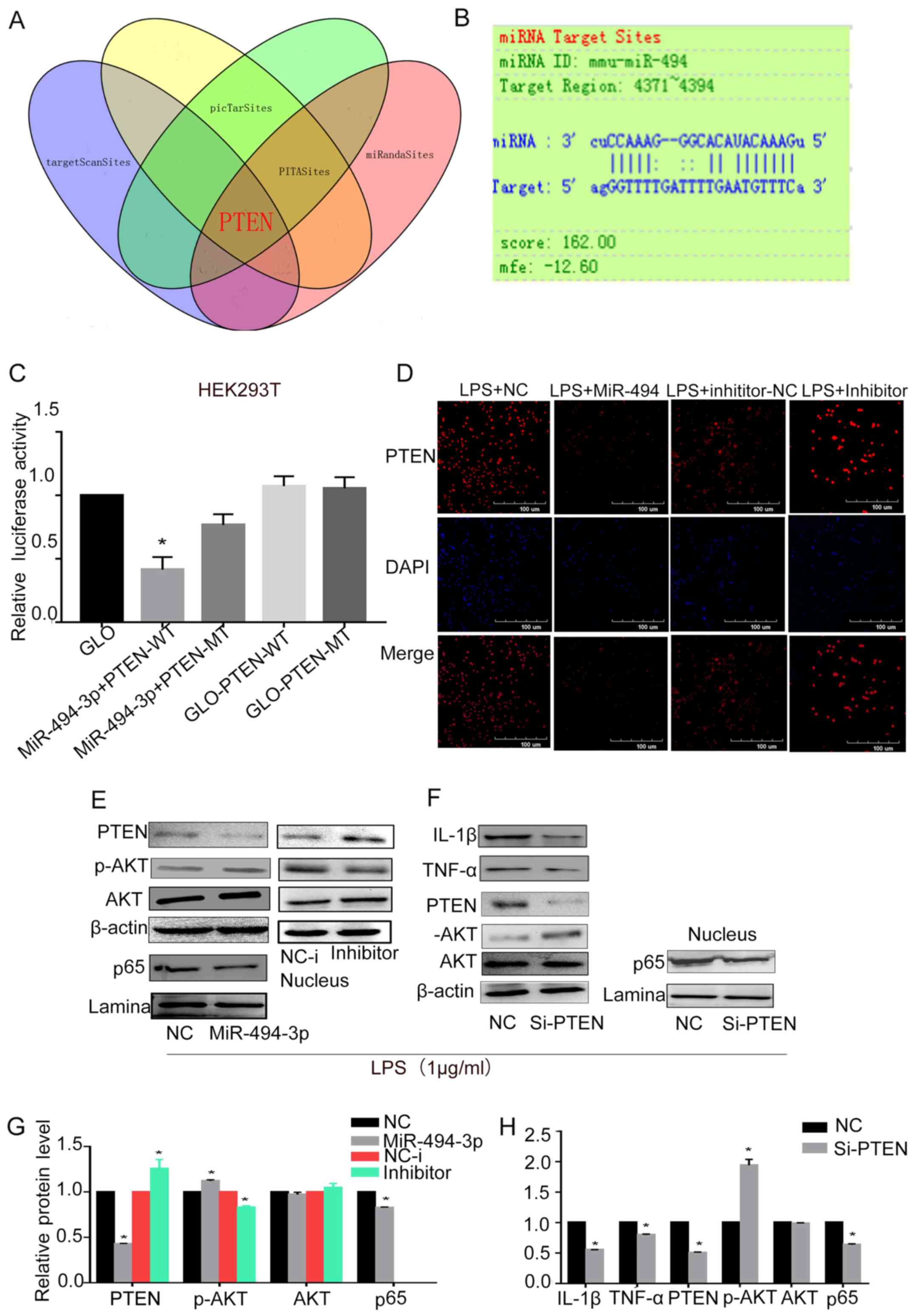 | Figure 3.miR-494-3p regulates PTEN/Akt/NF-κB
signaling. (A) miR-494-3p target prediction using four online
tools. (B) Alignment of miR-494-3p and the 3′-UTR of PTEN. (C)
Luciferase activity in 293T cells co-transfected with a reporter
vector containing the 3′-UTR of PTEN and miR-494-3p mimics.
RAW264.7 cells were transfected with miR-494-3p mimics, miR-494-3p
inhibitor, NC or NC-i. After 24 h, cells were treated with 1 µg/ml
LPS for 24 h. *P<0.05 vs. GLO-PTEN-WT. (D) PTEN expression was
detected by confocal microscopy. (E) Western blot analysis was used
to determine the protein expression levels of PTEN, Akt1, p-Akt1
and p65. (F) RAW264.7 cells were transfected with PTEN siRNA or
control siRNA. After 48 h, cells were treated with 1 µg/ml LPS for
24 h. Western blot analysis was used to determine the protein
expression levels of IL-1β, TNF-α, PTEN, Akt1, p-Akt1 and p65. (G
and H) Quantification of the protein expression presented in E and
F. Data are presented as the mean ± standard deviation of at least
three independent experiments. *P<0.05 (miR-494-3p) vs. nc;
*P<0.05 (inhibitor) vs. NC-i; *P<0.05 (si-PTEN) vs. NC. Akt1,
AKT serine/threonine kinase 1; IL, interleukin; LPS,
lipopolysaccharide; miR, microRNA; MT, mutant; NC, negative
control; NC-i, inhibitor NC; p, phosphorylated; PTEN, phosphatase
and tensin homolog; siRNA, small interfering RNA; TNF, tumor
necrosis factor; UTR, untranslated region; WT, wild-type. |
miR-494-3p targets PTEN and suppresses
LPS-induced nuclear factor (NF)-κB signaling
TLR4, a member of the TLR family, interacts all four
Toll-interleukin receptor domain-containing adaptor proteins to
induce inflammatory responses; stimulation of TLR activates the
NF-κB pathway (24). Based on a
previous study, Akt1 activation is enhanced in PTEN-/-cells
(25). This suggests that PTEN
might augment TLR4-induced inflammatory responses by suppressing
Akt1 activation (17). In
addition, NF-κB pathways have been demonstrated to enhance
LPS-induced inflammatory responses (26). Using miR-494-3p or negative control
mimics to transfect RAW264.7 cells, it was revealed that miR-494-3p
enhanced LPS-induced p-Akt expression, and suppressed the nuclear
expression of its downstream molecule, p65 (Fig. 3E and G), which may induce decreased
levels of the inflammatory mediators, IL-1β and TNF-α. Taken
together, these results indicated that miR-494-3p enhanced Akt1
phosphorylation and suppressed the NF-κB pathway.
PTEN silencing inhibits LPS-induced
inflammatory responses
To determine the role of PTEN in miR-494-3p-mediated
inhibition of LPS-induced inflammatory responses, RAW264.7 cells
were transfected with PTEN siRNA or control siRNA. Western blotting
was performed to measure the siRNA efficiency; PTEN siRNA
significantly reduced the expression of PTEN (Fig. 3F). Akt1 phosphorylation was
upregulated in RAW264.7 cells following transfection with PTEN
siRNA compared with control siRNA (Fig. 3F and H). The protein expression
levels of proinflammatory cytokines and p65 were also measured
(Fig. 3F). The results suggested
that miR494-3p targets PTEN and suppresses LPS-induced inflammation
via the Akt/NF-κB pathway.
Discussion
LPS binds to TLR4, activates NF-κB signaling
pathways, and induces the expression and release of inflammatory
factors, including IL-1β and TNF-α (27,28).
The production of these, and other proinflammatory cytokines,
results in a positive feedback loop to promote inflammation
(29). In the present study, miRNA
expression data from the GEO database were analyzed, and 44
upregulated miRNAs were identified in LPS-treated RAW264.7 cells
compared with in control cells. RT-qPCR was used to validate the
differential expression of miR-494-3p. The results indicated that
miR-494-3p may have a role in regulation of inflammatory
responses.
There is increasing research focusing on the
critical role of miRNAs in innate immunity (30,31).
For example, miR-718 was reported to repress proinflammatory
cytokine production by targeting PTEN (32). In the present study, it was
demonstrated that miR-494-3p may suppress LPS-induced inflammation
in RAW264.7 cells. Notably, it was confirmed that PTEN is a target
of miR-494-3p. Previous studies suggested that PTEN expression is
tightly regulated and that several miRNAs, including miR-214, −21
and −217, downregulated the expression of PTEN (33–35).
Another study demonstrated that miR-494-3p can target PTEN in human
glioblastoma cells, and regulate cellular proliferation and
apoptosis through the Akt signaling pathway (36).
PTEN is a phosphatidylinositol phosphate phosphatase
that is specific for the 3-position of the inositol ring.
Phosphatidylinositol (3,4,5)-trisphosphate can be dephosphorylated
by PTEN, resulting in inactivation of Akt1 (37). Consistent with the results from
other studies, the results in the present study demonstrated that
inhibition of PTEN enhanced LPS-induced Akt1 activation (38,39).
In the early stage of immune responses, Akt1
suppresses NF-κB signaling and downregulates the expression of
proinflammatory cytokines (40).
Inhibition of PI3K/Akt can enhance TNF-α expression by increasing
the transcriptional activity of p65 in LPS-treated human monocytes
(39). The results in the present
study indicated that inhibition of PTEN expression suppressed p65
translocation to the nucleus and decreased expression of
NF-κB-dependent proinflammatory cytokines. The transcription
factor, p65, is important for regulating inflammatory processes
(41). It can translocate to the
nucleus where it binds target DNA sequences to promote gene
transcription. p65 activation triggers the transcription of many
proinflammatory cytokines, including IL-1β and TNF-α, after LPS
stimulation in RAW264.7 cells (42). It is also involved in many
biological processes, and its function is tightly regulated by many
proteins, including Akt1 (39).
Taken together, the results demonstrated that PTEN inhibition may
lead to the downregulation of p65 expression, leading to
suppression of inflammatory pathways.
miR-494-3p is involved in many biological processes.
It modulates the progression of Parkinson's disease in both in
vitro and in vivo models by targeting Sirtuin 3
(43). miR-494-3p can also act as
an oncogene by modulating NOTCH1 and PTEN/PI3K/Akt signaling, and
stimulating growth and metastasis of A549 cells (44). However, to the best of our
knowledge, there are no studies on the functional role of
miR-494-3p in regulating inflammatory responses. In the present
study, miR-494-3p overexpression reduced IL-1β and TNF-α
production, whereas downregulation of miR-494-3p enhanced the
production IL-1β and TNF-α. Further investigation revealed that
miR-494-3p downregulated PTEN expression, and miR-494-3p-mediated
suppression of IL-1β and TNF-α was associated with increased p-Akt1
expression and decreased p65 expression. These data indicated that
miR-494-3p regulated inflammation by decreasing p65 translocation
to the nucleus. This study also demonstrated that miR-494-3p was
associated with suppressing immune responses by downregulating PTEN
in RAW264.7 cells. In the future, additional experiments will be
conducted to explore the relationship between miR-494-3p and NF-κB
signaling pathways. In addition, further studies are needed to
explore the detailed mechanisms through which miR-494-3p regulates
inflammatory responses. Thus, miR-494-3p may serve as a novel
marker of inflammatory responses.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81070870).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SZ performed the research and wrote the paper. KH
completed the data preprocessing and heatmap generation. JC and ZJ
made significant contributions towards the design of the present
study. WZ made substantial contributions to the acquisition of
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Xie W and Du L: Diabetes is an
inflammatory disease: Evidence from traditional Chinese medicines.
Diabetes Obes Metab. 13:289–301. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Johnson AR, Milner JJ and Makowski L: The
inflammation highway: Metabolism accelerates inflammatory traffic
in obesity. Immunol Rev. 249:218–238. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vendramini-Costa DB and Carvalho JE:
Molecular link mechanisms between inflammation and cancer. Curr
Pharm Des. 18:3831–3852. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin YP, Love RM, Friedlander LT, Shang HF
and Pai MH: Expression of Toll-like receptors 2 and 4 and the
OPG-RANKL-RANK system in inflammatory external root resorption and
external cervical resorption. Int Endod J. 46:971–981. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ingalls RR, Heine H, Lien E, Yoshimura A
and Golenbock D: Lipopolysaccharide recognition, CD14, and
lipopolysaccharide receptors. Infect Dis Clin North Am. 13341–353.
(vii)1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Su GL, Simmons RL and Wang SC:
Lipopolysaccharide binding protein participation in cellular
activation by LPS. Crit Rev Immunol. 15:201–214. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang C, Liu XJ, QunZhou, Xie J, Ma TT,
Meng XM and Li J: MiR-146a modulates macrophage polarization by
inhibiting Notch1 pathway in RAW264.7 macrophages. Int
Immunopharmacol. 32:46–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zheng Y, Guo X, He W, Shao Z, Zhang X,
Yang J, Shen Y, Luo X and Cao J: Effects of Echinococcus
multilocularis miR-71 mimics on murine macrophage RAW264.7 cells.
Int Immunopharmacol. 34:259–262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li M, Chen H, Chen L, Chen Y, Liu X and Mo
D: miR-709 modulates LPS-induced inflammatory response through
targeting GSK-3β. Int Immunopharmacol. 36:333–338. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie W, Li Z, Li M, Xu N and Zhang Y:
miR-181a and inflammation: miRNA homeostasis response to
inflammatory stimuli in vivo. Biochem Biophys Res Commun.
430:647–652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng Y, Du L, Jiao H, Zhu H, Xu K, Guo S,
Shi Q, Zhao T, Pang F, Jia X and Wang F: Mmu-miR-27a-5p-dependent
upregulation of MCPIP1 inhibits the inflammatory response in
LPS-induced RAW264.7 macrophage cells. Biomed Res Int.
2015:6076922015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Furnari FB, Lin H, Huang HS and Cavenee
WK: Growth suppression of glioma cells by PTEN requires a
functional phosphatase catalytic domain. Proc Natl Acad Sci USA.
94:12479–12484. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Myers MP, Pass I, Batty IH, Van der Kaay
J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP and Tonks NK: The
lipid phosphatase activity of PTEN is critical for its tumor
suppressor function. Proc Natl Acad Sci USA. 95:13513–13518. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu D, Hattori H, Jo H, Jia Y, Subramanian
KK, Loison F, You J, Le Y, Honczarenko M, Silberstein L and Luo HR:
Deactivation of phosphatidylinositol 3,4,5-trisphosphate/Akt
signaling mediates neutrophil spontaneous death. Proc Natl Acad Sci
USA. 103:14836–14841. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Agrawal A, Agrawal S, Cao JN, Su H, Osann
K and Gupta S: Altered innate immune functioning of dendritic cells
in elderly humans: A role of phosphoinositide 3-kinase-signaling
pathway. J Immunol. 178:6912–6922. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cao X, Wei G, Fang H, Guo J, Weinstein M,
Marsh CB, Ostrowski MC and Tridandapani S: The inositol
3-phosphatase PTEN negatively regulates Fc gamma receptor
signaling, but supports Toll-like receptor 4 signaling in murine
peritoneal macrophages. J Immunol. 172:4851–4857. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen Y, Liu W, Sun T, Huang Y, Wang Y, Deb
DK, Yoon D, Kong J, Thadhani R and Li YC: 1,25-Dihydroxyvitamin D
promotes negative feedback regulation of TLR signaling via
targeting microRNA-155-SOCS1 in macrophages. J Immunol.
190:3687–3695. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu J, Zhang Y, Wu G, Xiao Z, Zhou H and
Yu X: Inhibitory effects of oligochitosan on TNF-α, IL-1β and
nitric oxide production in lipopolysaccharide-induced RAW264.7
cells. Mol Med Rep. 11:729–733. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He Y, Sun X, Huang C, Long XR, Lin X,
Zhang L, Lv XW and Li J: MiR-146a regulates IL-6 production in
lipopolysaccharide-induced RAW264.7 macrophage cells by inhibiting
Notch1. Inflammation. 37:71–82. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ding Y, Wang L, Zhao Q, Wu Z and Kong L:
MicroRNA-93 inhibits chondrocyte apoptosis and inflammation in
osteoarthritis by targeting the TLR4/NF-κB signaling pathway. Int J
Mol Med. 43:779–790. 2019.PubMed/NCBI
|
|
24
|
Shi H, Wang XL, Quan HF, Yan L, Pei XY,
Wang R and Peng XD: Effects of betaine on LPS-stimulated activation
of microglial M1/M2 phenotypes by suppressing TLR4/NF-κB pathways
in N9 cells. Molecules. 24:E3672019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu YL, Yan Y, Webster C, Shao L, Lensing
SY, Ni H, Feng W, Colorado N, Pathak R, Xiang Z, et al: Timing of
the loss of Pten protein determines disease severity in a mouse
model of myeloid malignancy. Blood. 127:1912–1922. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ko W, Sohn JH, Jang JH, Ahn JS, Kang DG,
Lee HS, Kim JS, Kim YC and Oh H: Inhibitory effects of
alternaramide on inflammatory mediator expression through
TLR4-MyD88-mediated inhibition of NF-кB and MAPK pathway signaling
in lipopolysaccharide-stimulated RAW264.7 and BV2 cells. Chem Biol
Interact. 244:16–26. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
da Silveira Cruz-Machado S, Carvalho-Sousa
CE, Tamura EK, Pinato L, Cecon E, Fernandes PA, de Avellar MC,
Ferreira ZS and Markus RP: TLR4 and CD14 receptors expressed in rat
pineal gland trigger NFKB pathway. J Pineal Res. 49:183–192.
2010.PubMed/NCBI
|
|
28
|
He W, Qu T, Yu Q, Wang Z, Lv H, Zhang J,
Zhao X and Wang P: LPS induces IL-8 expression through TLR4, MyD88,
NF-kappaB and MAPK pathways in human dental pulp stem cells. Int
Endod J. 46:128–136. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sieve I, Ricke-Hoch M, Kasten M, Battmer
K, Stapel B, Falk CS, Leisegang MS, Haverich A, Scherr M and
Hilfiker-Kleiner D: A positive feedback loop between IL-1β, LPS and
NEU1 may promote atherosclerosis by enhancing a pro-inflammatory
state in monocytes and macrophages. Vascul Pharmacol.
103-105:16–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Niu Y, Mo D, Qin L, Wang C, Li A, Zhao X,
Wang X, Xiao S, Wang Q, Xie Y, et al: Lipopolysaccharide-induced
miR-1224 negatively regulates tumour necrosis factor-α gene
expression by modulating Sp1. Immunology. 133:8–20. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
O'Connell RM, Rao DS, Chaudhuri AA and
Baltimore D: Physiological and pathological roles for microRNAs in
the immune system. Nat Rev Immunol. 10:111–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kalantari P, Harandi OF, Agarwal S, Rus F,
Kurt-Jones EA, Fitzgerald KA, Caffrey DR and Golenbock DT: miR-718
represses proinflammatory cytokine production through targeting
phosphatase and tensin homolog (PTEN). J Biol Chem. 292:5634–5644.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fang Y, Qiu J, Jiang ZB, Xu SR, Zhou ZH
and He RL: Increased serum levels of miR-214 in patients with PCa
with bone metastasis may serve as a potential biomarker by
targeting PTEN. Oncol Lett. 17:398–405. 2019.PubMed/NCBI
|
|
34
|
Du G, Cao D and Meng L: miR-21 inhibitor
suppresses cell proliferation and colony formation through
regulating the PTEN/AKT pathway and improves paclitaxel sensitivity
in cervical cancer cells. Mol Med Rep. 15:2713–2719. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nie X, Fan J, Li H, Yin Z, Zhao Y, Dai B,
Dong N, Chen C and Wang DW: miR-217 promotes cardiac hypertrophy
and dysfunction by targeting PTEN. Mol Ther Nucleic Acids.
12:254–266. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li XT, Wang HZ, Wu ZW, Yang TQ, Zhao ZH,
Chen GL, Xie XS, Li B, Wei YX, Huang YL, et al: miR-494-3p
regulates cellular proliferation, invasion, migration, and
apoptosis by PTEN/AKT signaling in human glioblastoma cells. Cell
Mol Neurobiol. 35:679–687. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bayascas JR and Alessi DR: Regulation of
Akt/PKB Ser473 phosphorylation. Mol Cell. 18:143–145. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Aksoy E, Vanden Berghe W, Detienne S,
Amraoui Z, Fitzgerald KA, Haegeman G, Goldman M and Willems F:
Inhibition of phosphoinositide 3-kinase enhances TRIF-dependent
NF-kappa B activation and IFN-beta synthesis downstream of
Toll-like receptor 3 and 4. Eur J Immunol. 35:2200–2209. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guha M and Mackman N: The
phosphatidylinositol 3-kinase-Akt pathway limits lipopolysaccharide
activation of signaling pathways and expression of inflammatory
mediators in human monocytic cells. J Biol Chem. 277:32124–32132.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee YG, Lee J, Byeon SE, Yoo DS, Kim MH,
Lee SY and Cho JY: Functional role of Akt in macrophage-mediated
innate immunity. Front Biosci (Landmark Ed). 16:517–530. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tak PP and Firestein GS: NF-kappaB: A key
role in inflammatory diseases. J Clin Invest. 107:7–11. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shin JS, Im HT and Lee KT: Saikosaponin B2
suppresses inflammatory responses through IKK/IκBα/NF-κB signaling
inactivation in LPS-induced RAW 264.7 macrophages. Inflammation.
42:342–353. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Geng L, Zhang T, Liu W and Chen Y:
miR-494-3p modulates the progression of in vitro and in vivo
Parkinson's disease models by targeting SIRT3. Neurosci Lett.
675:23–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Faversani A, Amatori S, Augello C, Colombo
F, Porretti L, Fanelli M, Ferrero S, Palleschi A, Pelicci PG,
Belloni E, et al: miR-494-3p is a novel tumor driver of lung
carcinogenesis. Oncotarget. 8:7231–7247. 2017. View Article : Google Scholar : PubMed/NCBI
|