Introduction
Globally, gastric cancer (GC) is the fifth leading
cause of cancer and the third leading cause of cancer-related
deaths, accounting for 5.7% of cases and 8.2% of deaths (1). Surgery remains the only curative
therapy for stomach cancer. Patients with advanced GC are not
eligible for surgery and are treated with chemotherapies, such as
mitomycin, cisplatin, and 5-fluorouracil (5-FU) (2). However, chemoresistance has
contributed to a poor 5-year survival rate in GC patients (3). Chemoresistance is a complex
biological problem, and is facilitated by a number of events,
including decreased drug uptake, increased drug efflux, changes in
the level and structure of intracellular targets, and increased
repair of DNA damage (3). The
specific mechanism that regulates chemoresistance in GC is not well
understood.
Recent studies have reported that transcription
factor activating enhancer-binding protein 2e (TFAP2E)
methylation may be involved in chemoresistance to 5-FU (4). Activating protein 2 (AP-2) is a
family of closely related transcription factors, including AP-2α,
AP-2β, AP-2γ, AP-2δ and AP-2ε. TFAP2E is highly homologous
to other members of the family in its DNA binding and dimerization
domains but less so in its N-terminal domain (5). TFAP2E is located on chromosome
1p34 and has 2 cytosine-phosphate-guanine (CpG) islands, which
underscore the potential for regulation of gene expression by CpG
methylation. Our previous study revealed that patients with GC also
had TFAP2E hypermethylation and were resistant to
fluorouracil-based chemotherapy (4). Methylation has a multifaceted link
with miRNAs, and exosomes are rich in miRNAs (6). Therefore, the aim of the present
study was to determine whether GC cells secreted exosomes and
whether exosomal miRNAs were involved in chemoresistance to
5-FU.
Exosomes are cell-derived vesicles that are present
in many eukaryotic fluids, including blood, urine, and in
conditioned medium of cell cultures (7). Studies have revealed that exosomes
can regulate tumor progression and chemosensitivity (8). Exosomes contain a large amount of
proteins, a rich variety of mRNAs and miRNAs (9). Previous studies have also shown that
miRNA gene promoters are frequent targets of aberrant DNA
methylation (6). DNA methylation
and miRNAs have been reported to be involved in the chemoresistance
of GC (10), glioma (11) and hepatocellular carcinoma
(12). Therefore, it was
hypothesized that TFAP2E methylation leads to chemoresistance to
5-FU by regulating exosomal miRNAs in GC.
As aforementioned, our previous study indicated a
potential role for TFAP2E hypermethylation in chemosensitivity,
which may be exploited to develop new treatment strategies for
patients with GC (4). In the
present study, drug-resistant human GC MGC-803/5-FU cells were
established and the mechanism underlying the development of GC
chemoresistance to 5-FU was determined. Our analysis of the
association between TFAP2E methylation and exosomal miRNAs
may increase the development of chemoresistance in GC.
Materials and methods
Antibodies and reagents
Commercially available antibodies and reagents were
as follows: Total exosome isolation reagent (cat. no. 4478359)
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
rabbit polyclonal anti-TFAP2E (1:15,000; cat. no. ab56295); rabbit
polyclonal anti-CD63 (1:20,000; cat. no. ab68418); rabbit
polyclonal anti-CD9 (1:50,000; cat. no. ab223052); rabbit
monoclonal anti-CD81 (1:2,000; cat. no. ab109201); Rabbit
polyclonal anti-GFP (1:1,000; cat. no. ab6556) (all from Abcam,
Cambridge, MA, USA), PKH26 Red Fluorescent Cell Linker Mini kit
(cat. no. MINI26) (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany),
Annexin V-FITC apoptosis detection kit (cat. no. AD10), Cell
Counting Kit-8 (CCK-8) (cat. no. CK04) (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan).
Cell culture and lentivirus
transfection
The GC cell line MGC-803 was obtained from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China).
Drug-resistant subline, MGC-803/5-FU was successfully established
from the parental MGC-803 cells as recently reported (13). MGC-803 was cultured as the control
group in some of the experiments. MGC-803/5-FU was maintained in
drug-free medium for 1 week prior to subsequent analysis, avoiding
toxic effects. Cells used for exosome isolation were cultured in
medium with 10% exosome-depleted serum.
TFAP2E was overexpressed via transfection of
lenti-TFAP2E (GeneChem, Inc., Shanghai, China). MGC-803/5-FU
were transfected with lenti-TFAP2E or lenti-NC at a
confluence of 30–50%, >95% of the cells were viable 12 h later.
The medium was then changed, the cells were incubated for a further
3 days, and passaged for further experiments. Transfection
efficacies were assessed via western blotting.
Cell cytotoxicity and apoptosis
assays
Cell cytotoxicity was determined by the CCK-8
according to the manufacturer's instructions. Cells were seeded in
96-well plates with or without the specific treatments, and
incubated for 48 h. Then, 10 µl of CCK-8 solution was added into
each well. Absorbance values were determined at 450 nm by a
microplate reader (Thermo Fisher Scientific, Inc., Waltham, MA,
USA).
An equal number of cells was seeded and treated with
or without the specific treatments. After 48 h, the cells were
collected and resuspended in binding buffer. Apoptotic cells were
double-stained by an Annexin V-FITC/PI or an Annexin V-APC/PI
Apoptosis Detection kit following the manufacturer's instructions
(Dojindo Molecular Technologies, Inc., Shanghai, China). Flow
analysis was performed using FACSCalibur flow cytometry (BD
Biosciences, Franklin Lakes, NJ, USA).
TFAP2E methylation analysis
All samples in this research were measured in
duplicate. Methylation-sensitive high-resolution melting analysis
(MSHRM) was performed with precision melt analysis software to
analyze the methylation level of TFAP2E in the cells. Methylation
standards (100, 75, 50, 25, 10, 1 and 0%) were established by
diluting unmethylated control DNA in a pool of 100% methylated one.
Primers were listed as follows: Forward,
5′-GTTTTATTTTAGAAGCGGTTTT-3′ and reverse,
5′-CGAACGCTTACCTACAATCA-3′. The amplicon was located at the second
CpG island of the TFAP2E gene in intron 3. The PCR mixture was
prepared in a final volume of 20 ml, containing 10 ml LightCycler
480 HRM Master Mix, 2 ml of 25 mmol/l MgCl2, 10 mmol/l
of primers, 50 ng DNA template, and ribonuclease-free
H2O. The cycling protocol for MS-HRM analysis was
initial denaturation at 95°C for 10 min followed by 50 cycles at
94°C for 10 sec, an annealing temperature for 15 sec, and extension
at 72°C for 10 sec, followed by an MS-HRM step of 95°C for 1 min,
40°C for 1 min, and continuous acquisition between 65°C and 97°C at
1 min acquisition/0.2°C. The standard curves with known methylation
ratios were obtained, and the methylation ratio of each sample via
these standard curves was measured.
Exosome isolation and analysis
Exosomes were isolated from the conditioned medium
by differential centrifugation. Briefly, the conditioned medium was
centrifuged at 300 × g for 10 min and then at 2,000 × g for 30 min
at 4°C to remove cells. Subsequently, this supernatant was
centrifuged at 10,000 × g for 30 min to remove cell debris,
followed by filtration through a 220-nm filter to remove particles
with a diameter >200 nm. Then, exosomes were pelleted by
ultracentrifugation at 100,000 × g for 70 min (14). MGC-803 in logarithmic phase was
transfected with the lentivirus encoding green fluorescent protein
(GFP), according to the manufacturer's instructions.
For exosome uptake research, they were labeled with
PKH26 Fluorescent Cell Linker kits (Merck KGaA) according to the
manufacturer's protocol. Then, exosomes were incubated with GC
cells and examined under a confocal microscope.
Microarray analysis
Exosome pellets from 10 ml supernatant of cells were
collected and homogenized in TRIzol (Thermo Scientific, Inc.).
Total RNA was extracted as aforementioned. The miRNA microarray
analysis was performed by Shanghai Biotechnology Corporation
(Shanghai, China). The Affymetrix GeneChip miRNA 2.0 Array contains
15,644 probe sets, including 1,105 human mature miRNAs. The raw
data were treated using the miRNA QCTool software (version 2.3.0;
Petros Eikon, Inc., Orangeville, ON, Canada).
Quantitative real-time PCR
Two differentially expressed miRNAs (hsa-miR-421 and
hsa-miR-106a-5p) identified by microarray in exosomes were selected
for further validation by quantitative real-time
reverse-transcription PCR (qRT-PCR). Total RNA and exosomes were
extracted from cells using TRIzol reagent (Shanghai Sangong
Pharmaceutical Co., Ltd., Shanghai, China). cDNA synthesis was
performed with 1 mg RNA using ReverAid™ First Strand cDNA Synthesis
kit (K1622; Fermentas; Thermo Fisher Scientific, Inc.). The
oligo-nucleotide primers used are listed in Table I. Equal amounts of each
reverse-transcription product (1 mg) were PCR amplified using
DreamTaq™ Green PCR Master Mix (2X) (K1081; Fermentas; Thermo
Fisher Scientific, Inc.) for 30 cycles consisting of 1 min at 95°C,
30 sec at 55°C, and 1 min 72°C. The amplified cDNA was run on 1%
agarose gels and visualized by UV light. Each experiment was
performed in duplicate and the results were standardized to
glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
 | Table I.Primers used for RT-PCR analysis of
gene expression. |
Table I.
Primers used for RT-PCR analysis of
gene expression.
| Gene symbol | Forward primer | Reverse primer | Accession no. |
|---|
| E2F1 |
5′-GGGGAGAAGTCACGCTATGA-3′ |
5′-CTCAGGGCACAGGAAAACAT-3′ | NM_005225.3 |
| MTOR |
5′-CGCTGTCATCCCTTTATCG-3′ |
5′-ATGCTCAAACACCTCCACC-3 | NM_004958.3 |
| STAT3 |
5′-ACCTGCAGCAATACCATTGAC-3′ |
5′-AAGGTGAGGGACTCAAACTGC-3 | NM_003150.3 |
| TFAP2E |
5′-CAGAGAGAAGTGGGCAGGAG-3′ |
5′-AGGACAGACAGCAACAGGACT-3′ | NM_178548.4 |
| miR-106a-5p |
5′-AAAAGTGCTTACAGTGCAGGTAG-3′ |
5′-GAAAAGTGCTTACAGTGCAGGT-3′ | – |
| miR-421 |
5′-TATGGTTGTTCTGCTCTCTGTGTC-3′ |
5′-CTCACTCACATCAACAGACATTAATT-3′ | – |
Western blot analysis
Total proteins of the isolated exosomes and cells
were extracted using a protein extraction kit, according to the
manufacturer's protocols. Briefly, the samples were washed with
phosphate-buffered saline and lysed in RIPA lysis buffer containing
phenylmethylsulfonyl fluoride. The protein concentration was
measured using a Bicinchoninic Acid protein assay kit (Sangon
Biotech Co., Ltd., Shanghai, China), and 2 mg/ml bovine serum
albumin (Gibco; Thermo Fisher Scientific, Inc.) was used as the
standard. Subsequently, 50 µg total protein was loaded in each
lane. Proteins were separated by 10% SDS-PAGE and transferred onto
a polyvinylidene fluoride membrane. The membranes were then blocked
with 5% non-fat milk at room temperature for 1 h, and then
incubated at 4°C overnight with a primary antibody. After washing,
the blots were incubated with Alexa Fluor-conjugated
goat-anti-rabbit antibody (1:10,000; cat. no. G-21234; Fermentas;
Thermo Fisher Scientific, Inc.) at 37°C for 1 h. Proteins were
visualized using an enhanced chemiluminescence kit, and images were
analyzed using a Bio-Rad gel imaging system (Bio-Rad Laboratories,
Inc., Hercules, CA, USA).
Statistical analysis
All data were expressed as the mean ± (standard
deviation) SD. Wherever appropriate, the data were subjected to
unpaired two-tailed Student's t-test. The differences between the
experimental groups were evaluated using one-way analysis of
variance (ANOVA) followed by a Bonferroni's post hoc test to allow
for multiple comparisons. P<0.05 was considered to indicate a
statistically significant difference.
Results
TFAP2E methylation is involved in
chemoresistance to 5-FU
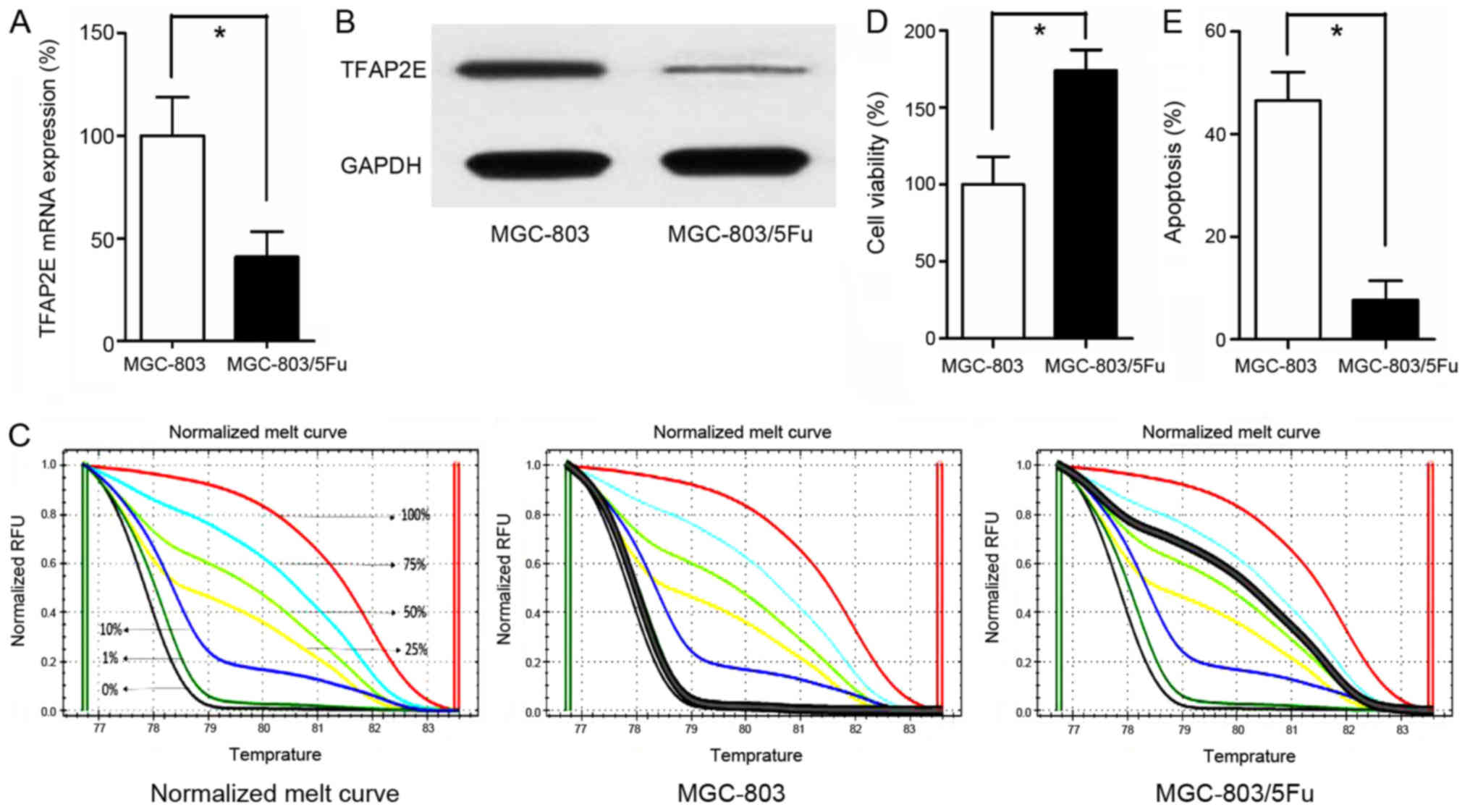
TFAP2E gene expression was significantly
lower in the MGC-803/5-FU group than in the MGC-803 group
(0.41-fold, P<0.001; Fig. 1A).
In addition to reduced gene expression, the TFAP2E protein level
was also assessed. As anticipated, the TFAP2E protein level was
higher in the MGC-803 group than in the MGC-803/5-FU group
(3.42-fold, P<0.001; Fig. 1B).
In addition to TFAP2E gene and protein expression level, the
methylation status of TFAP2E was detected using MSHRM.
First, the normalized melt curve for TFAP2E methylation
level was generated. Percentage of methylation (PM) values were
then calculated according to the following formula, which also
served as a surrogate measure of the methylation level of AP-2E in
our study (15):
PM=-d(ddt)Fluorescence value
at82°C-d(ddt)Fluorescence value at77.5°C+-d(ddt)Fluorescence value
at82°Cx100%
Our data revealed that the drug-resistant
MGC-803/5-FU cell line had a higher PM value (1.042±2.552%) than
the control group (66.037±11.201%) (P<0.001; Fig. 1C). Additionally, the cells were
treated with 10 µg/ml 5-FU for 48 h in 96-well plates. Cell
viability and apoptosis assays indicated that treatment with 5-FU
significantly decreased the viability and significantly increased
apoptosis of the MGC-803 group in comparison to the MGC-803/5-FU
group (viability, 100.00±17.944% vs. 173.857±13.558%, P<0.001;
apoptosis, 46.543±5.521% vs. 7.729±3.683%, P<0.001, MGC-803 vs.
MGC-803/5-FU; Fig. 1D and E).
qRT-PCR analysis of exosomal microRNAs
and target gene analysis
Increasing evidence indicates that tumor-derived
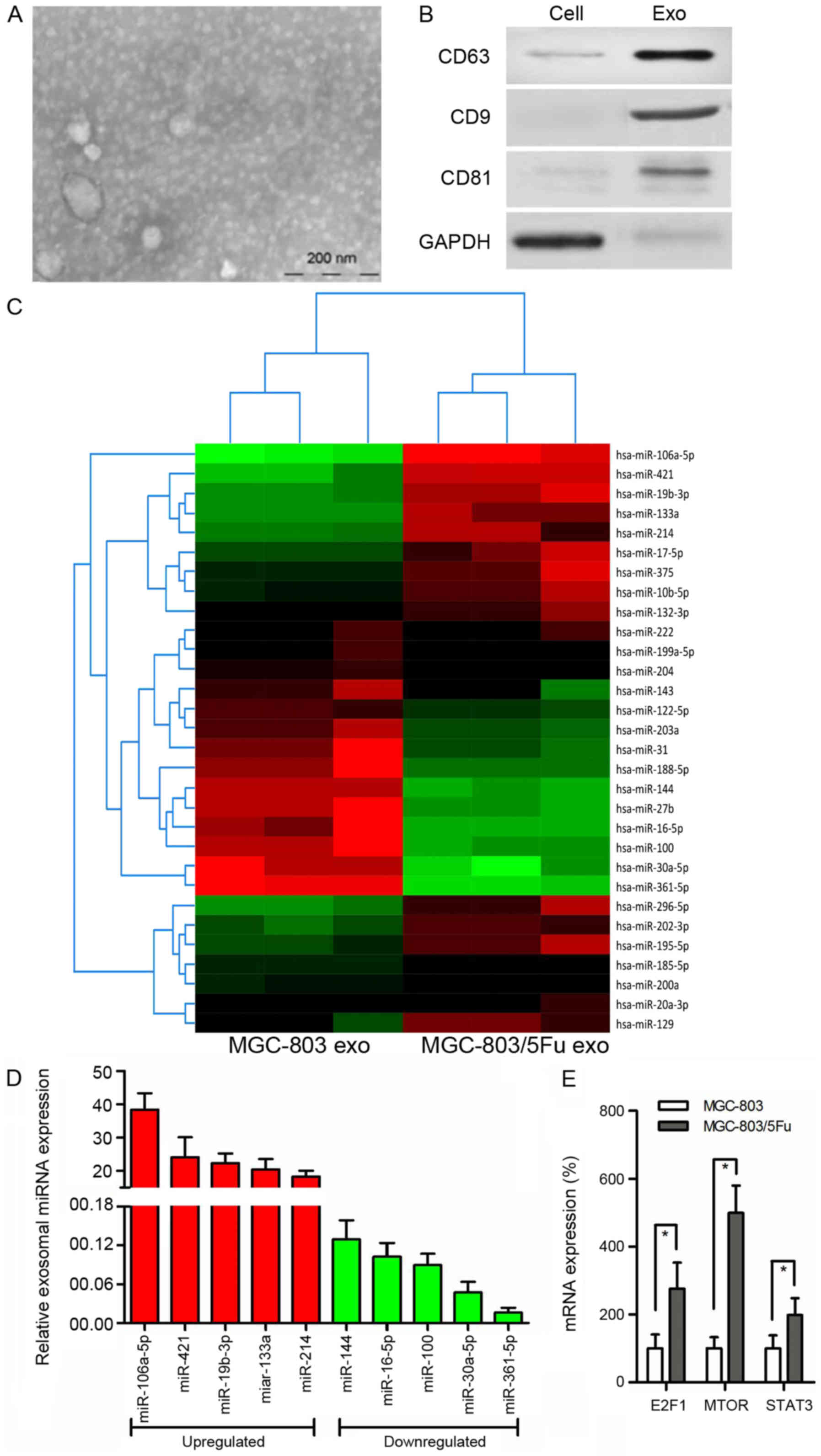
exosomes are involved in chemoresistance of GC. We used electron
microscopy and western blot analysis to characterize exosomes. The
purified exosomes were small round vesicles with diameters ranging
from 80 to 120 nm, and expressed the exosomal markers CD9, CD63 and
CD81 (Fig. 2A and B). To
investigate the functional role of exosomes in chemoresistance in
GC cells, miRNA microarray analysis was performed using RNA samples
extracted from MGC-803 exosomes and MGC-803/5-FU exosomes (Fig. 2C). Notably, exosomal miR-106a-5p,
miR-421, miR-19b-3p, miR-133a, and miR-214 were more highly
expressed, whereas miR-144, miR-16-5p, miR-100, miR-30a-5p, and
miR-361-5p were more reduced in MGC-803/5-FU exosomes in comparison
with MGC-803 exosomes (Fig. 2D).
TargetScan 7.1 (version 7.1; http://www.targetscan.org/vert_71/) was used to
predict the target genes for these 10 miRNAs. This analysis
predicted 2,119 genes as the target mRNAs, and KEGG pathway
analysis was performed. According to our data, 90 genes were
associated with cancer (P<0.001), and 24 of these genes were
associated with chemoresistance. Among these 24 genes, 10 genes
[CBLB (16), E2F1 (17), MET (18), SKP2 (19), FGFR3 (20), HIF1A (21), MTOR (22), MAPK1 (23), RHOA (24), and STAT3 (25)] have been revealed to be involved in
GC, and 7 of them [CBLB (16),
E2F1 (17), HIF1A (21), MTOR (22), MAPK1 (23), RHOA (24), and STAT3 (25)] have been reportedly associated with
5-FU. The relationship between methylation and drug resistance was
then explored. Only E2F1 (17),
MTOR (22) and STAT3 (26) have been reported to be associated
with methylation. E2F1 and STAT3 are targets of miR-106a-5p, while
MTOR is a target of miR-421. Therefore, their expression was
evaluated. There were higher levels of these 3 genes in the
MGC-803/5-FU group in comparison to the MGC-803 group (E2F1,
2.76-fold, P<0.001; MTOR, 4.99-fold, P<0.001; STAT3,
1.98-fold, P= 0.003) (Fig.
2E).
Exosomes mediate target gene
expression and chemoresistance to 5-FU
Exosomes from MGC-803/5-FU cells were obtained and
co-cultured with MGC-803 cells. It has been reported that exosomes
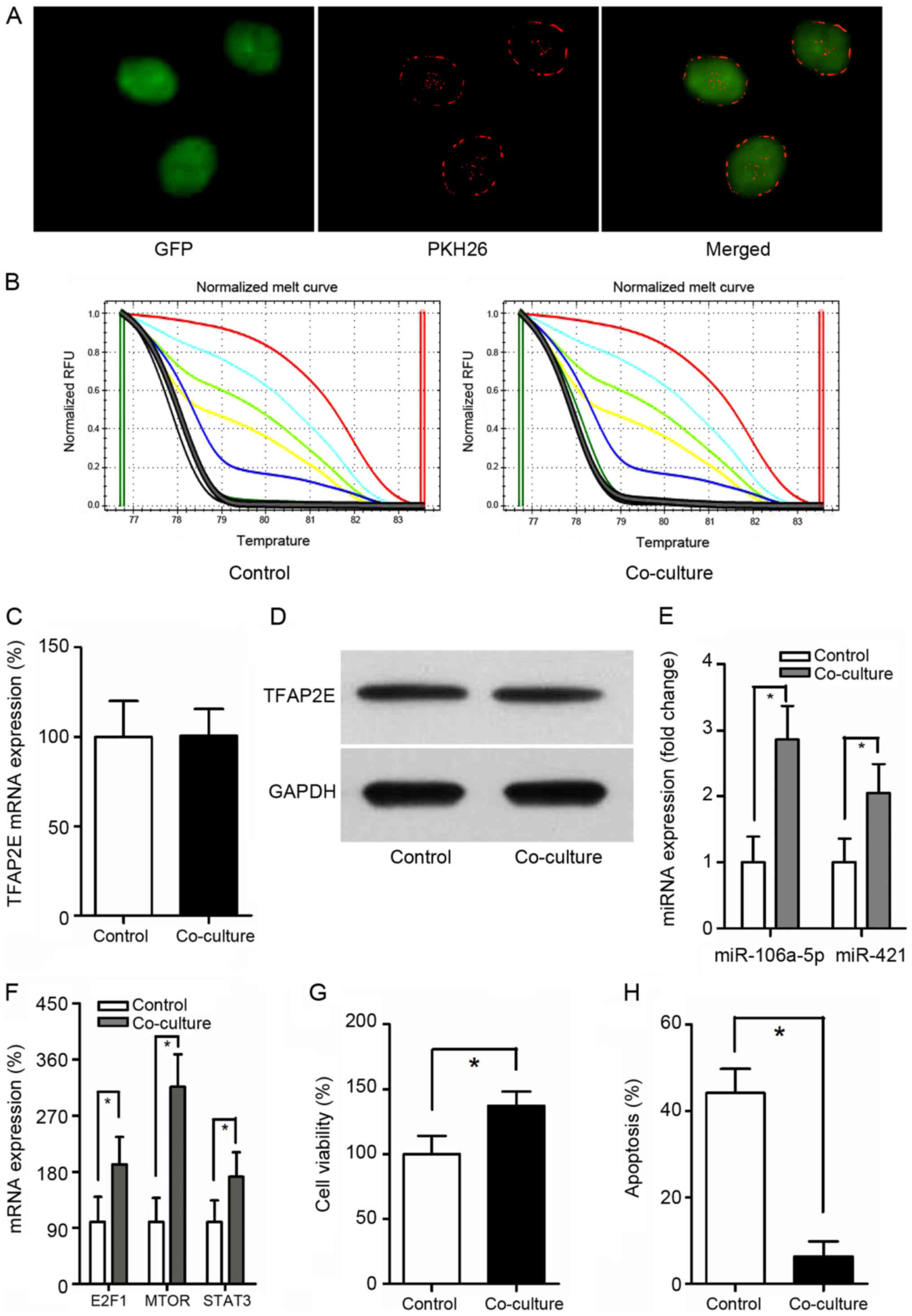
can be transferred from donor cells to recipient cells (27). To better observe the interaction,
GFP was employed and exposed to PKH26-labeled exosomes for 24 h.
When MGC-803-derived exosomes were labeled with PKH26 (red
fluorescent dye), a red staining on the cells could be observed by
fluorescence microscopy, suggesting exosome binding and
incorporation (Fig. 3A).
TFAP2E PM of the MGC-803 (control group) and the co-culture
group were assessed. Notably, there was no significant difference
between these 2 groups (P=0.549) (Fig.
3B). Similarly, no statistical significance in TFAP2E
gene and protein expression levels could be observed in the present
study (P=0.953, 0.771, respectively) (Fig. 3C and D), indicating that exosomes
may be downstream of and therefore, not directly involved in
TFAP2E methylation. The expression of miRNAs was then
assessed. The data revealed that the expression of miR-106a-5p and
miR-421 were increased with the treatment of MGC-803/5-FU exosomes
(miR-106a-5p: P<0.001; miR-421: P=0.001) (Fig. 3E). The expression of the predicted
target genes was also assessed. The target genes were higher in the
co-culture group than in the control group (E2F1, 1.92-fold,
P=0.004; MTOR, 3.16- fold, P<0.001; STAT3, 1.72- fold, P=0.007)
(Fig. 3F). Cell viability and
apoptosis assays indicated that treatment with 5-FU resulted in a
significant decrease of viability and a significant increase of
apoptosis in the MGC-803 group than in the co-culture group
(viability, 100.00±14.024% vs. 137.286±11.011%, P<0.001;
apoptosis, 44.129±5.588% vs. 6.300±3.492%, P<0.001, control vs.
co-culture) (Fig. 3G and H).
Demethylation reduces chemoresistance
by suppressing miRNA expression
MGC-803/5-FU was used as the control group for this
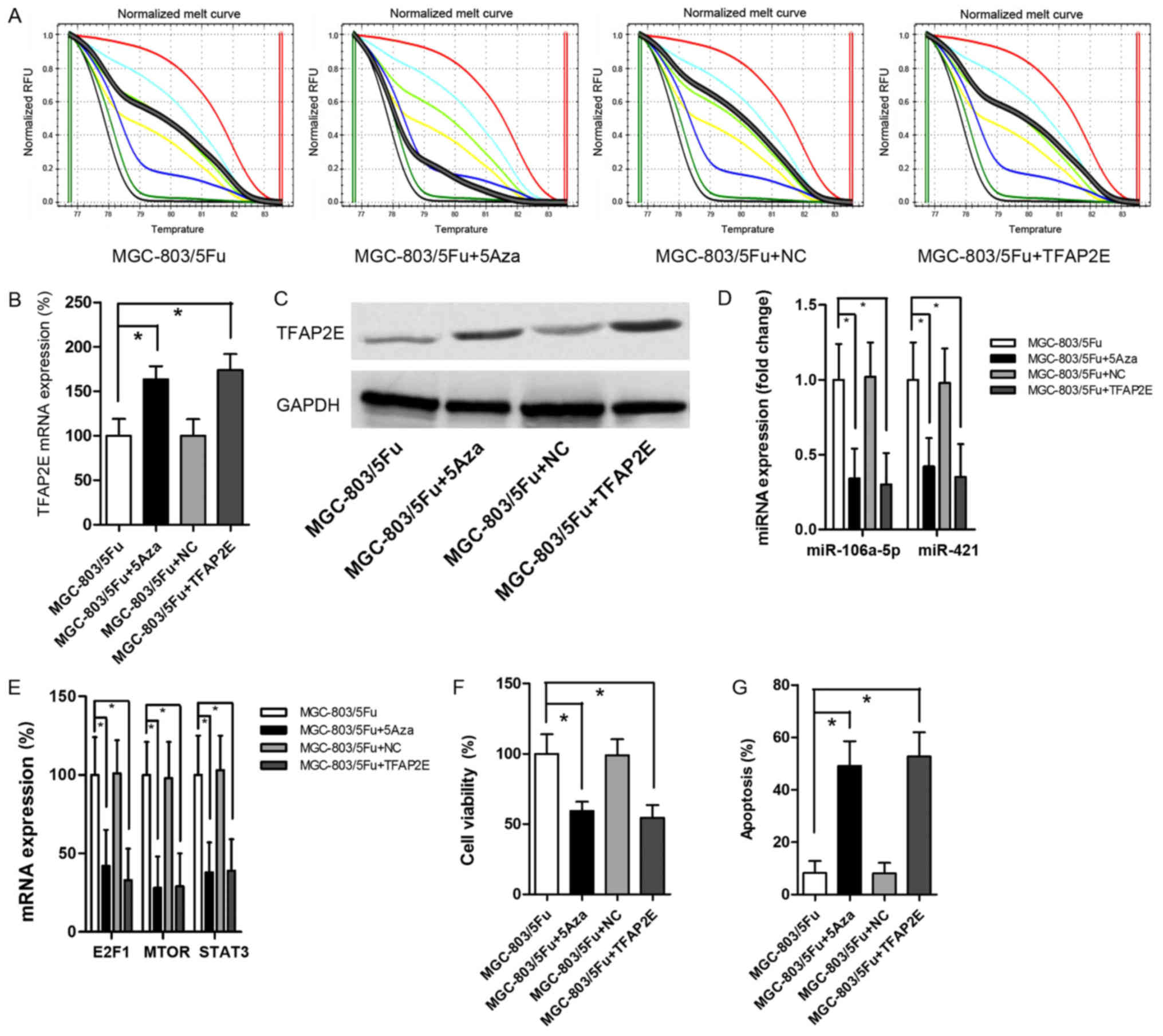
experiment. Cells were treated with 10 µM 5-Aza-2-deoxycytidine
(5Aza) to establish a baseline demethylation status as previously
reported (28). The data revealed
that 5Aza decreased TFAP2E PM significantly (P<0.001)
(Fig. 4A). After demethylation,
there were significant increases in TFAP2E mRNA expression
(1.62-fold) and protein expression (1.80-fold) in MGC-803/5-FU
cells (Fig. 4B and C).
Additionally, miR-106a-5p and miR-421 were significantly decreased
after 5Aza treatment (0.34-, 0.42-fold, respectively; P=0.001,
0.001, respectively; Fig. 4D). The
levels of the predicted target gene were also significantly
decreased (E2F1, 0.42-fold, P=0.001; MTOR, 0.28-fold, P<0.001;
STAT3, 0.38-fold, P<0.001; Fig.
4E). 5-FU chemoresistant tests indicated that 5Aza resulted in
a significant decrease in viability and a significant increase in
apoptosis (viability, 0.59-fold, P<0.001; apoptosis, 5.99-fold,
P<0.001; Fig. 4F and G).
MGC-803/5-FU cells were also transfected with a recombinant plasmid
to directly upregulate TFAP2E expression. The results
revealed a significant increase in TFAP2E expression after
transient transfection (Fig. 4B and
C). However, there was no obvious difference in TFAP2E
PM in comparison to the control group (Fig. 4A). Despite the low methylation
rate, high expression of TFAP2E significantly suppressed
miR-106a-5p, miR-421, the levels of predicted target genes, and
cell viability (Fig. 4D-F). These
results recapitulated the effects of 5Aza treatment.
Discussion
TFAP2E hypermethylation has been reported to
be associated with clinical no-response to chemotherapy in
colorectal cancer, and targeting of DKK4 may be an option to
overcome TFAP2E-mediated drug resistance (15). It has been reported that
5-FU-resistant gastric cancer (GC) cell lines MKN45 and MKN28 have
TFAP2E hypermethylation (29). In
the present study, whether TFAP2E methylation affected
chemoresistance to 5-FU in MGC-803 cells, was investigated. Our
previous study revealed that GC patients with TFAP2E
hypermethylation were resistant to fluorouracil-based chemotherapy
(4). However, the mechanism has
not been fully elucidated. In the present study, a drug-resistant
subline MGC-803/5-FU was successfully established, and high
TFAP2E PM, low TFAP2E gene and protein expression in
this subline were observed, which was consistent with previous
research. TFAP2E methylation resulted in reduction of its
gene expression. Additionally, the functional role of TFAP2E
methylation was investigated in chemoresistance to 5-FU in GC
cells.
Studies have demonstrated an increasingly important
role for exosomes in GC, as mediators of cell-to-cell crosstalk in
the tumor microenvironment (27).
Recent studies have indicated that exosomal miRNAs are associated
with GC chemoresistance by transferring a variety of RNAs. Zheng
et al (30), demonstrated
that exosomal transfer of tumor-associated macrophage-derived
miR-21 conferred DDP resistance to GC. However, 5-FU is the most
frequently used chemotherapy drug in the treatment of GC.
Therefore, miRNA microarray analysis was conducted and the target
mRNAs were predicted to explore the relationship between miRNAs and
chemoresistance to 5-FU.
In our study, 2,119 genes were predicted as target
mRNAs. Notably, only 3 genes were previously reported to be
associated with chemoresistance to 5-FU and gene methylation in GC.
E2F1, a transcription factor, plays a crucial role in the control
of cell cycle and tumor suppression and is also a target of the
transforming proteins of small DNA tumor viruses. Tahara et
al (17), have reported that
overexpression of E2F1 promoted the development of MDR in GC,
suggesting that E2F1 may represent a viable target for GC therapy.
Mammalian target of rapamycin (MTOR) regulates cell growth, cell
proliferation, cell motility, cell survival, and transcription. It
has been reported that MTOR promoted the development of GC, and its
inhibitor interacted with 5-FU in a synergistic manner in scirrhous
GC cells by the activation of apoptosis signals (31). Signal transducer and activator of
transcription 3 (STAT3) mediates the expression of a variety of
genes in response to cell stimuli, and thus plays a key role in
many cellular processes such as cell growth and apoptosis. Notably,
STAT3 is overactivated in GC stem-like cells (32). Therefore, these 3 mRNAs have been
implicated in GC tumorigenesis.
However, the regulatory relationship between
TFAP2E and exosomal miRNAs is unclear. Notably, it was
revealed that MGC-803 cells co-cultured with exosomes derived from
MGC-803/5-FU had increased expression of miR-106a-5p and miR-421.
Additionally, predicted target mRNAs were increased. Although
co-cultured cells appeared to be chemoresistant to 5-FU,
TFAP2E expression and its PM value were not significantly
different from the control, indicating that exosomal miRNAs could
not meditate methylation of TFAP2E. Therefore, it was
hypothesized that TFAP2E methylation may be upstream and
regulate the expression of exosomal miRNAs.
In order to assess our hypothesis, the methylation
status of MGC-803/5-FU cells was reduced using 5Aza, a commonly
used demethylation drug, and subsequently the expression of
miR-106a-5p, miR-421 and their predicted target mRNAs was assessed.
As anticipated, TFAP2E expression was increased and its PM
value was significantly decreased with 5Aza treatment. However,
5Aza significantly suppressed the expression of the aforementioned
miRNAs and their predicted target mRNAs, indicating that changes in
TFAP2E methylation may result in the decrease of miRNAs. The
ultimate result was the increased chemosensitivity of GC cells to
5-FU. These data were also consistent with our previous hypothesis.
Additionally, results of a TFAP2E rescue experiment using
transfection of a TFAP2E- expressing plasmid supported our
hypothesis. Although increased TFAP2E expression did not affect its
methylation status, the results indicated that hypermethylation may
act as an upstream regulator of its expression.
There were some limitations to the present study.
Inhibitors and mimics of miRNAs were not used in our study, and our
analysis methods used to predict target genes may have filtered out
some important or unreported genes. Additionally, other
5-FU-resistant GC cell lines were not used in the present study and
should be analyzed in future research. Future studies are also
required to more comprehensively assess the functional role of
E2F1, MTOR, and STAT3 in regulating chemoresistance in GC
cells.
In summary, our data indicated that chemoresistance
to 5-FU in GC cells may be facilitated by TFAP2E
hypermethylation-induced release of miRNA-containing exosomes.
These results indicated that targeting of miRNAs may be a viable
therapeutic strategy for GC patients.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Suzhou
Project (grant no. kjxw2018015).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
XH conceived and designed the study. SJ and ZJ
acquired the data. WX analyzed and interpreted the data. LW and LD
performed statistical analyses. SJ and WX drafted and revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Venerito M, Vasapolli R, Rokkas T and
Malfertheiner P: Gastric cancer: Epidemiology, prevention, and
therapy. Helicobacter. 23 (Suppl 1):e125182018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marin JJ, Al-Abdulla R, Lozano E, Briz O,
Bujanda L, Banales JM and Macias RI: Mechanisms of resistance to
chemotherapy in gastric cancer. Anticancer Agents Med Chem.
16:318–334. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun J, Du N, Li J, Zhou J, Tao G, Sun S
and He J: Transcription factor AP2epsilon: A potential predictor of
chemoresistance in patients with gastric cancer. Technol Cancer Res
Treat. 15:285–295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tummala R, Romano RA, Fuchs E and Sinha S:
Molecular cloning and characterization of AP-2 epsilon, a fifth
member of the AP-2 family. Gene. 321:93–102. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chhabra R: miRNA and methylation: A
multifaceted liaison. Chembiochem. 16:195–203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Keller S, Sanderson MP, Stoeck A and
Altevogt P. Exosomes: From biogenesis and secretion to biological
function. Immunol Lett. 107:102–108. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brinton LT, Sloane HS, Kester M and Kelly
KA: Formation and role of exosomes in cancer. Cell Mol Life Sci.
72:659–671. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van Niel G, Porto-Carreiro I, Simoes S and
Raposo G: Exosomes: A common pathway for a specialized function. J
Biochem. 140:13–21. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Toiyama Y, Okugawa Y and Goel A: DNA
methylation and microRNA biomarkers for noninvasive detection of
gastric and colorectal cancer. Biochem Biophys Res Commun.
455:43–57. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao B, Bian EB and Li J and Li J: New
advances of microRNAs in glioma stem cells, with special emphasis
on aberrant methylation of microRNAs. J Cell Physiol.
229:1141–1147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Anwar SL and Lehmann U: DNA methylation,
microRNAs, and their crosstalk as potential biomarkers in
hepatocellular carcinoma. World J Gastroenterol. 20:7894–7913.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang H, Zeng L, Wang J, Zhang X, Ruan Q,
Wang J, Cui S and Yang D: Reversal of 5-fluorouracil resistance by
EGCG is mediate by inactivation of TFAP2A/VEGF signaling pathway
and down-regulation of MDR-1 and P-gp expression in gastric cancer.
Oncotarget. 8:82842–82853. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeringer E, Barta T, Li M and Vlassov AV:
Strategies for isolation of exosomes. Cold Spring Harb Protoc.
2015:319–323. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Giovannetti E, Codacci-Pisanelli G and
Peters GJ: TFAP2E-DKK4 and chemoresistance in colorectal cancer. N
Engl J Med. 366:9662012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Feng D, Ma Y, Liu J, Xu L, Zhang Y, Qu J,
Liu Y and Qu X: Cbl-b enhances sensitivity to 5-fluorouracil via
EGFR- and mitochondria-mediated pathways in gastric cancer cells.
Int J Mol Sci. 14:24399–24411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tahara M, Ochiai A, Fujimoto J, Boku N,
Yasui W, Ohtsu A, Tahara E and Yoshida S: Expression of thymidylate
synthase, thymidine phosphorylase, dihydropyrimidine dehydrogenase,
E2F-1, Bak, Bcl-X, and Bcl-2, and clinical outcomes for gastric
cancer patients treated with bolus 5-fluorouracil. Oncol Rep.
11:9–15. 2004.PubMed/NCBI
|
|
18
|
Peng Z, Li Z, Gao J, Lu M, Gong J, Tang
ET, Oliner KS, Hei YJ, Zhou H and Shen L: Tumor MET expression and
gene amplification in chinese patients with locally advanced or
metastatic gastric or gastroesophageal junction cancer. Mol Cancer
Ther. 14:2634–2641. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wei Z, Jiang X, Liu F, Qiao H, Zhou B,
Zhai B, Zhang L, Zhang X, Han L, Jiang H, et al: Downregulation of
Skp2 inhibits the growth and metastasis of gastric cancer cells in
vitro and in vivo. Tumour Biol. 34:181–192. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Piro G, Carbone C, Cataldo I, Di
Nicolantonio F, Giacopuzzi S, Aprile G, Simionato F, Boschi F,
Zanotto M, Mina MM, et al: An FGFR3 autocrine loop sustains
acquired resistance to trastuzumab in gastric cancer patients. Clin
Cancer Res. 22:6164–6175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen F, Zhuang M, Zhong C, Peng J, Wang X,
Li J, Chen Z and Huang Y: Baicalein reverses hypoxia-induced 5-FU
resistance in gastric cancer AGS cells through suppression of
glycolysis and the PTEN/Akt/HIF-1α signaling pathway. Oncol Rep.
33:457–463. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun Y, Jiang Y, Huang J, Chen H, Liao Y
and Yang Z: CISD2 enhances the chemosensitivity of gastric cancer
through the enhancement of 5-FU-induced apoptosis and the
inhibition of autophagy by AKT/mTOR pathway. Cancer Med.
6:2331–2346. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiong HL, Zhou SW, Sun AH, He Y, Li J and
Yuan X: MicroRNA197 reverses the drug resistance of
fluorouracilinduced SGC7901 cells by targeting mitogenactivated
protein kinase 1. Mol Med Rep. 12:5019–5025. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kang WK, Lee I and Park C:
Characterization of RhoA-mediated chemoresistance in gastric cancer
cells. Cancer Res Treat. 37:251–256. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu GY and Tang XJ: Troxerutin (TXN)
potentiated 5-Fluorouracil (5-Fu) treatment of human gastric cancer
through suppressing STAT3/NF-κB and Bcl-2 signaling pathways.
Biomed Pharmacother. 92:95–107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
To KF, Chan MW, Leung WK, Ng EK, Yu J, Bai
AH, Lo AW, Chu SH, Tong JH, Lo KW, et al: Constitutional activation
of IL-6-mediated JAK/STAT pathway through hypermethylation of
SOCS-1 in human gastric cancer cell line. Br J Cancer.
91:1335–1341. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang X, Yuan X, Shi H, Wu L, Qian H and
Xu W: Exosomes in cancer: Small particle, big player. J Hematol
Oncol. 8:332015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Seelan RS, Mukhopadhyay P, Pisano MM and
Greene RM: Effects of 5-Aza-2′-deoxycytidine (decitabine) on gene
expression. Drug Metab Rev. 50:193–207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hong Y, Zhang J, Zhuang M, Li W, Wu P, Li
R, Hu N, Bian B, Song Z and Wu F: Efficacy of decitabine-loaded
gelatinases-stimuli nanoparticles in overcoming cancer drug
resistance is mediated via its enhanced demethylating activity to
transcription factor AP-2 epsilon. Oncotarget. 8:114495–114505.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie
G, Ma Y and Shen L: Exosomal transfer of tumor-associated
macrophage-derived miR-21 confers cisplatin resistance in gastric
cancer cells. J Exp Clin Cancer Res. 36:532017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Matsuzaki T, Yashiro M, Kaizaki R, Yasuda
K, Doi Y, Sawada T, Ohira M and Hirakawa K: Synergistic
antiproliferative effect of mTOR inhibitors in combination with
5-fluorouracil in scirrhous gastric cancer. Cancer Sci.
100:2402–2410. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hajimoradi M, Mohammad Hassan Z, Ebrahimi
M, Soleimani M, Bakhshi M, Firouzi J and Samani FS: STAT3 is
overactivated in gastric cancer stem-like cells. Cell J.
17:617–628. PubMed/NCBI
|