Introduction
Liver fibrosis is an inevitable pathological event
of cirrhosis that develops in a background of chronic liver disease
induced by various pathogenic factors, including viral infection,
poisoning, parasite infection, alcoholic hepatitis and fatty liver.
The continuous and repeated hepatocyte inflammation or necrosis
leads to a specific repair response in the body, frequently
resulting in fibrous tissue hyperplasia, insufficient fiber
degradation, extensive deposition of extracellular matrix (ECM) in
the liver and development of liver fibrosis. Hepatic stellate cells
(HSCs) are the major cells responsible for ECM synthesis in the
liver, and their activation can create an imbalance between
synthesis and degradation of ECM, with ensuing liver fibrosis
(1). In chronic liver injury, the
proliferation and phenotype of HSCs are altered. A switch occurs
from the static phenotype, which is rich in vitamin A, to the
activated myofibroblast-like phenotype, which is a hallmark of HSC
activation (2). Various clinical
investigations have indicated that liver fibrosis may be reduced or
reversed by fiber degradation. In addition, early cirrhosis may
also be reversed. The prognosis of chronic patients with liver
disease is markedly improved by inhibition, alleviation or reversal
of liver fibrosis. Thus, early diagnosis of liver fibrosis is
crucial for arresting its development and improving the clinical
outcome of patients with chronic liver disease (3).
The name syndecan-1 (SDC1) originates from the Greek
word ‘syndein’, which means the binding of cell microenvironment
components with the cytoskeleton. This protein regulates the
interaction between cells and the microenvironment, and acts as a
cell-surface co-receptor that participates in the regulation of a
series of physiological processes, such as differentiation and
development of tissues and organs, vascularization and tissue
regeneration (4). The SDC proteins
are removed from the cell surface by trypsin in cultured epithelial
cells. This causes the cells to lose their adhesion characteristics
and to exhibit pleiomorphic-like anchoring non-dependent growth.
The full-length cDNA sequence of SDC1 can be integrated into the
aforementioned cells in order to construct a stably expressing SDC1
cell line. The epithelial cell shape and growth characteristics can
recover in the presence of constitutive SDC1 expression (5,6).
However, whether SDC1 has an important role in the activation of
HSCs has not been previously reported. The aim of the present study
was to investigate the functional expression of the SDC1 protein in
mouse HSCs.
A number of previous studies have indicated that the
transforming growth factor-β1 (TGF-β1)/Smad3 signaling pathway
participates in the development of liver fibrosis (7,8). It
was recently demonstrated that the SDC family of proteins is an
important regulatory factor in the TGF-β1/Smad3 signaling pathway
(9). It remains unclear whether
this signaling pathway is also regulated by SDC1 during liver
fibrosis. The effects and possible mechanism underlying the role of
SDC1 in the development of liver fibrosis were investigated at the
cellular and molecular levels in the current study, hoping to
provide a new target for the prevention and treatment of this
disease.
The phenotypic transformation of myofibroblasts
through HSC activation is considered as the hallmark of liver
fibrosis. In the present study, mouse HSCs were isolated and
cultured in vitro. Freshly isolated HSCs are spontaneously
activated during in vitro culture. This model mimics the
process of liver injury. Thus, in the present study, the changes in
the mRNA and protein levels of α-smooth muscle actin (α-SMA), a
marker of HSC activation, SDC1 and TGF-β1/Smad3 signaling
pathway-related proteins were investigated during the spontaneous
activation of freshly isolated mouse HSCs that were cultured in
vitro. At specific time points (days 1, 3 and 7), reverse
transcription-quantitative PCR (RT-qPCR) analysis was used to
quantitatively measure the mRNA levels of the aforementioned
markers, whereas western blotting was used to quantitatively
analyze their corresponding protein levels. To further analyze the
effect of SDC1 on the activation of HSCs, SDC1 recombinant protein
was used to stimulate HSCs. Smad3-siRNA transfection resulted in
the activation of mouse HSCs, whereas SDC1 recombinant protein was
also used to stimulate HSCs and observe their activated status.
Materials and methods
Animals and HSC isolation
Male Kunming mice (8–10 weeks old, 22±2 g) were
provided by CHI Scientific, Inc. The mice were all raised under
standard laboratory conditions with free access to food and water,
and were allowed to acclimatize for 1 week. The experiments were
performed in accordance with the Guide for the Care and Use of
Laboratory Animals of China Jiaxing University, and approved by the
Ethics Committee of Animal Experimentation. All the laboratory
procedures were performed with the permission and under the
surveillance of the institutional ethics committee. Pronase E
(0.05%) and type IV collagenase (0.1%) were used to perfuse mouse
livers; the HSCs were isolated by Nycodenz density gradient
centrifugation at 4°C for 20 min (1,400 × g) and cultured. The cell
yield was measured by cell counting method, and the survival rate
of the cells was measured by Trypan blue staining (1 g/l) for 3 min
at 25°C. The isolated HSCs were seeded in DMEM containing 10% FBS
at 1×105/cm2, and cultured at 37°C in the
presence of 5% CO2. The HSCs were then collected for the
subsequent experiments.
Immunofluorescence staining
When the cells reached 80% confluence, the culture
medium was discarded. The cells were washed with warm PBS twice (10
min each time), fixed with 4% paraformaldehyde at room temperature
for 15 min, washed an additional two times with PBS (10 min each
time) and permeabilized with 0.1% Triton X-100 for 15 min at 4°C.
Excess Triton X-100 was removed by washing with PBS and the
cellular proteins were blocked with 4% BSA for 30 min at room
temperature. α-SMA (cat. no. MAB1420; R&D Systems, Inc.) and
desmin (cat. no. AF3844; R&D Systems, Inc.) primary antibodies
were diluted at 1:150 and incubated with the cells at 4°C
overnight. The cells were further washed with PBS three times (10
min each time). The secondary antibody used was a Cy3-conjugated
rabbit anti-goat immunoglobulin G (1:1,00; cat. no. BA1034; Wuhan
Boster Biological Technology, Ltd.). The secondary antibody was
added to the cells at 37°C for 1 h. The cells were washed with PBS
three times (10 min each time) and the cell nuclei were stained
with DAPI 1 µg/ml for 1 h at 25°C. Finally, the cells were observed
under a microscope.
RT-qPCR
Total RNA was extracted from cultured HSCs using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's recommendations. cDNA was
synthesized with the High-Capacity cDNA Reverse Transcription kit
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. SYBR Green Dye reagent was used to
quantify the products formed during qPCR. The amplification was
performed using 40 cycles of 95°C for 30 sec, 95°C for 5 sec and
60°C for 34 sec. The 2−ΔΔCq method (10) was used for normalization of raw
data using the housekeeping gene GAPDH. The experiments were
repeated at least three times. The sequences of the primers were as
follows: GAPDH, forward 5′-TGGCAAAGTGGAGATTGTT-3′ and reverse
5′-CTTCTGGGTGGCAGTGAT-3′; α-SMA, forward 5′-GCTGCTCCAGCTATGTGTGA-3′
and reverse 5′-GTTTTCCATGTCGTCCCAGT-3′; and SDC1, forward
5′-CAGCAGCAACACCGAGAC-3′; Smad3, forward 5′-CCTGGGCAAGTTCTCCAGAG-3′
and reverse 5′-CCATCGCTGGCATCTTCTGTG-3′.
Western blot analysis
RIPA lysis buffer (Wuhan Boster Biological
Technology, Ltd.) was used to extract total protein from HSCs, and
the protein concentration was measured using a UV spectrophotometer
following the bicinchoninic acid method. Proteins were separated on
a 10% SDS-PAGE gel and transferred to a PVDF membrane (EMD
Millipore). Subsequently, the membrane was blocked with 5% BSA
(Wuhan Boster Biological Technology, Ltd.) in TBS with Tween-20 for
1 h at room temperature and incubated overnight at 4°C with the
primary antibodies, including anti-SDC1 (1:1,000; cat. no.
SAB1305991; Sigma-Aldrich; Merck KGaA), α-SMA (1:500; cat. no.
MAB1420; R&D Systems, Inc.), Smad3 (1:1,000; cat. no. ab52903;
Abcam) and TGF-β1 (1:1,500; cat. no. SAB4502954; Sigma-Aldrich;
Merck KGaA). Following membrane washing, horseradish peroxidase
(HRP)-conjugated rabbit anti-goat IgG (1:2,000; cat. no. BA1060;
Wuhan Boster Biological Technology, Ltd.) or HRP-conjugated goat
polyclonal anti-rabbit IgG (1:2,000; cat no. BA1054; Wuhan Boster
Biological Technology, Ltd.) were added to the membrane for 2 h at
room temperature. The expression of the proteins was detected with
an ECL kit (cat. no. AR1170; Wuhan Boster Biological Technology,
Ltd.). GAPDH (1:1,000; cat. no. BM3896; Wuhan Boster Biological
Technology, Ltd.) was used as an internal reference, and the
densities of the western blot bands were analyzed using Image Lab
Software 5.2.1 (Bio-Rad Laboratories, Inc.)
Exogenous SDC1 stimulation
After 6 days of incubation, the isolated HSCs were
seeded in a 96-well plate at a density of 1×105
cells/ml. Following cell adherence, serum-free culture medium was
used to replace the medium after 24 h. The cells were divided into
three groups and incubated with recombinant SDC1 protein solution
at concentrations of 10 and 20 ng/ml in order to stimulate the
cells for 24 h. Subsequently, the cells were collected for α-SMA,
SMAD3 and TGF-β1 detection.
Small interfering RNA (siRNA)
targeting of mouse Smad3
siRNA targeting was performed for the cDNA sequence
of mouse Smad3. Guangzhou RiboBio Co., Ltd synthesized the
oligonucleotides. The sequences were as follows:
5′-CAGUUCUACCUCCAGUGUUdTdT-3′ (siRNA-1),
5′-CCAUGACAGUAGAUGGCUUdTdT-3′ (siRNA-2) and
5′-CGCAGAACGUGAACACCAAdTdT-3′ (siRNA-3). A negative control siRNA
(siRNAnc) was used as control (5′-UAUCACUGCGAUUACAUGCdTdT-3′). To
measure the knockdown efficacy of different siRNA oligonucleotides,
primary cultured mouse HSCs (2×105 cells/ml) were
transfected with 16 µg/ml siRNAs (siRNAnc, siRNA1, siRNA2 and
siRNA3) using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Following transfection, the cells were
cultured for 48 h and the inhibitory effects of the different siRNA
oligonucleotides were determined by measuring Smad3 mRNA using
RT-qPCR. The siRNA molecule with the highest inhibitory rate was
selected for subsequent experiments.
Statistical analysis
The statistical analysis of the data was performed
using SPSS 15.0 statistical software for Windows (SPSS, Inc.). The
results are presented as mean ± SEM. The data were analyzed by the
one-way ANOVA or repeated-measures ANOVA tests followed by Tukey's
post-hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Spontaneous activation of
mouse-isolated HSCs during culture
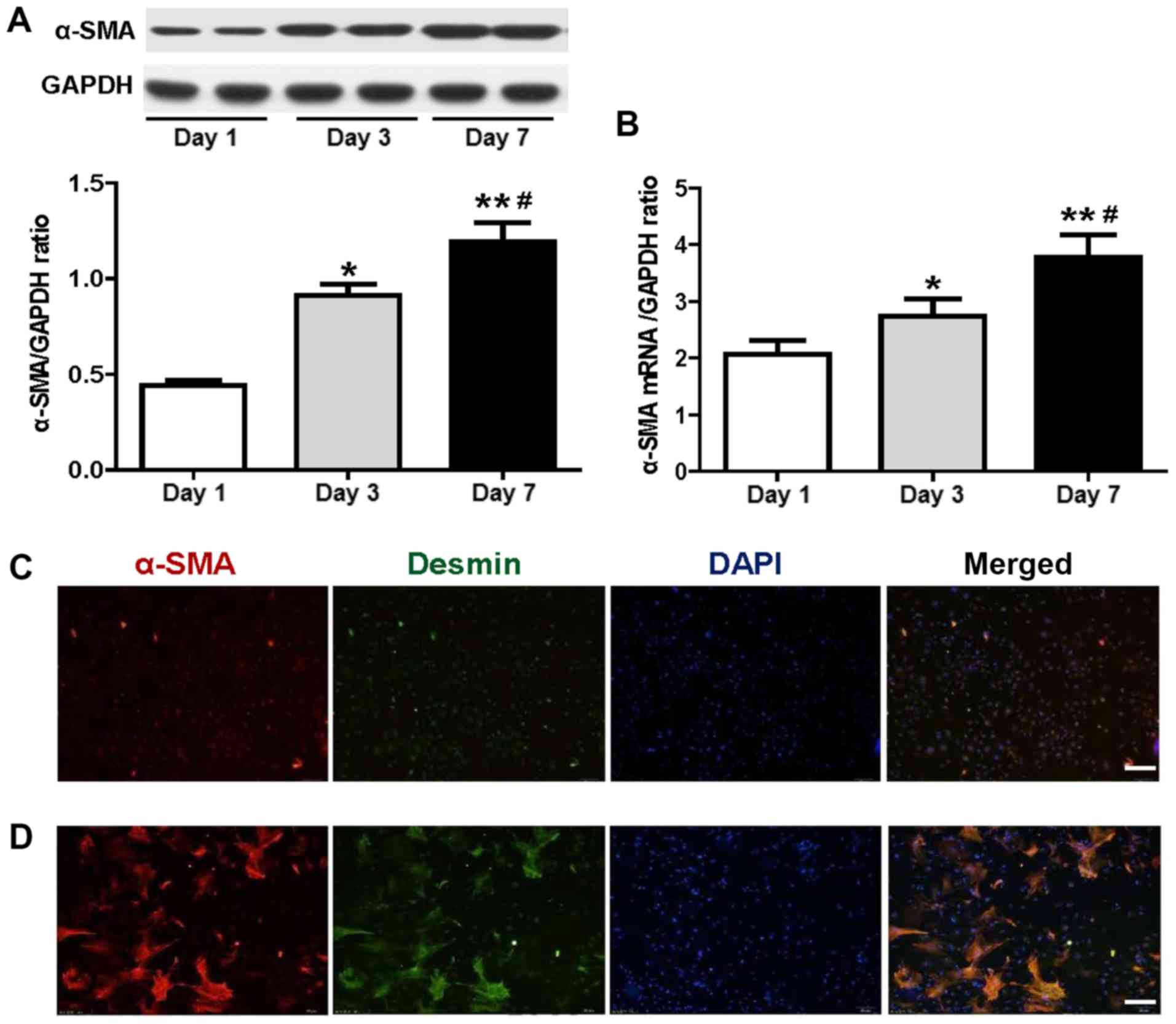
The results of the western blot analysis indicated
that the relative expression levels of α-SMA on days 3 and 7 were
increased compared with those of the cultured HSCs on day 1
(P<0.05; n=6; Fig. 1A). The
relative expression levels of α-SMA on day 7 were significantly
increased compared with those on day 3 (P<0.05; n=6; Fig. 1A). These data demonstrated that the
expression of α-SMA increased in a time-dependent manner. The
results of the RT-qPCR analysis were consistent with the
aforementioned data on the protein expression of α-SMA. The α-SMA
mRNA levels of the aforementioned groups exhibited significant
differences (P<0.05; n=6; Fig.
1B). Immunofluorescence staining indicated that, after 7 days
of HSC culture, α-SMA protein was highly expressed in myocytes that
were transformed into myofibroblasts (Fig. 1D). Double immunofluorescence
staining indicated cellular co-localization of α-SMA and muscle
markers (desmin) in HSCs cultured for 1 and 7 days (Fig. 1C and D, respectively). The
aforementioned experimental results highlighted that the isolated
HSCs had undergone transformation from static to activated cells
during in vitro culture.
Expression of SDC1 is downregulated
during HSC culture
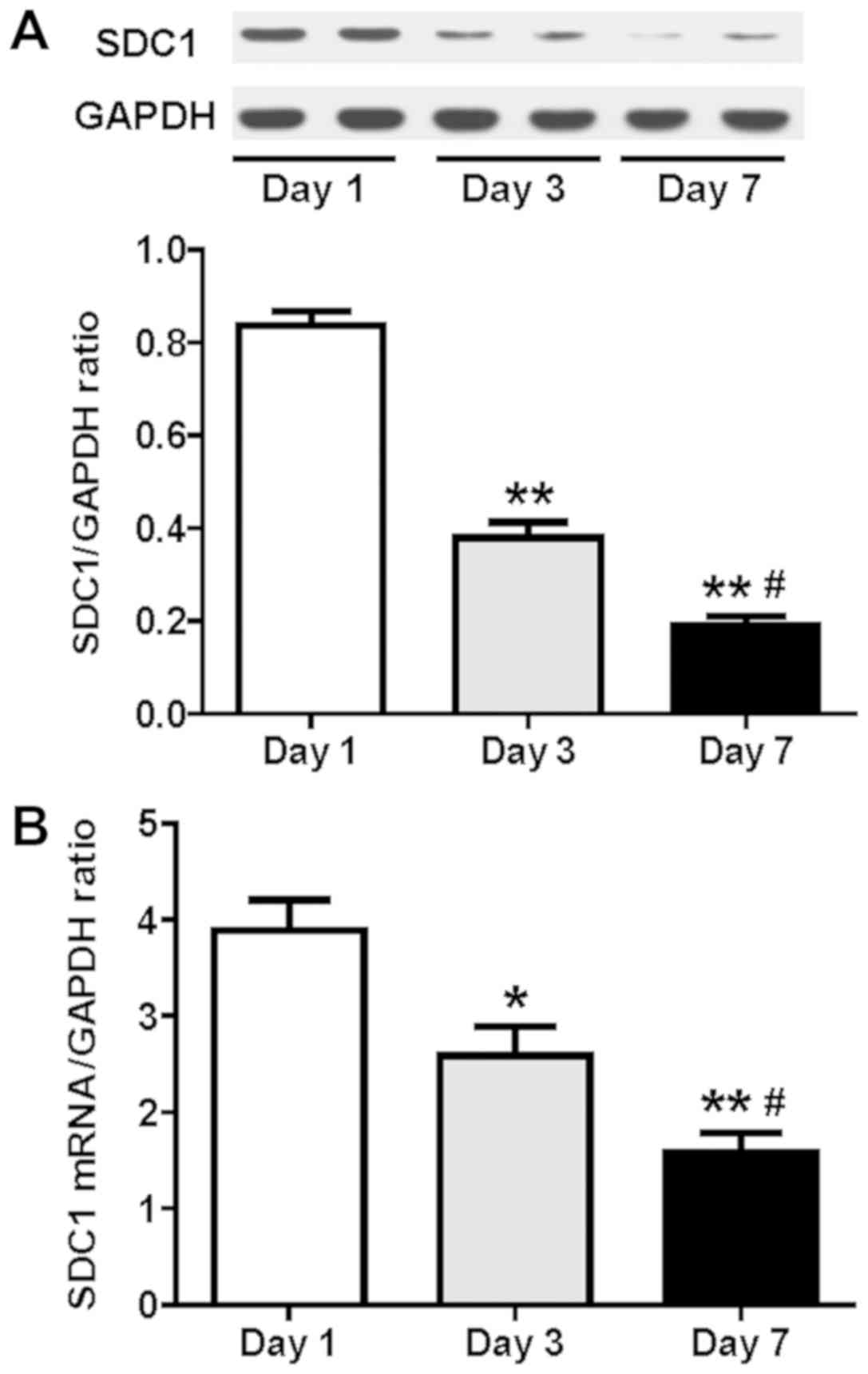
The results of the western blot analysis indicated
that the protein expression of the SDC1 was significantly
downregulated following culture of HSCs for 3 and 7 days compared
with that of the HSCs on day 1 (P<0.05; n=6; Fig. 2A). The relative expression levels
of the SDC1 protein on day 7 were also significantly decreased
compared with those of the cultured HSCs on day 3 (P<0.05; n=6;
Fig. 2A). The results of the SDC1
mRNA levels as detected by RT-qPCR were consistent with those of
the western blot assay (P<0.05; n=6; Fig. 2B). Upon activation of HSCs, the
expression levels of SDC1 exhibited a time-dependent decrease.
TGF-β1/Smad3 signaling pathway is
activated during HSC culture
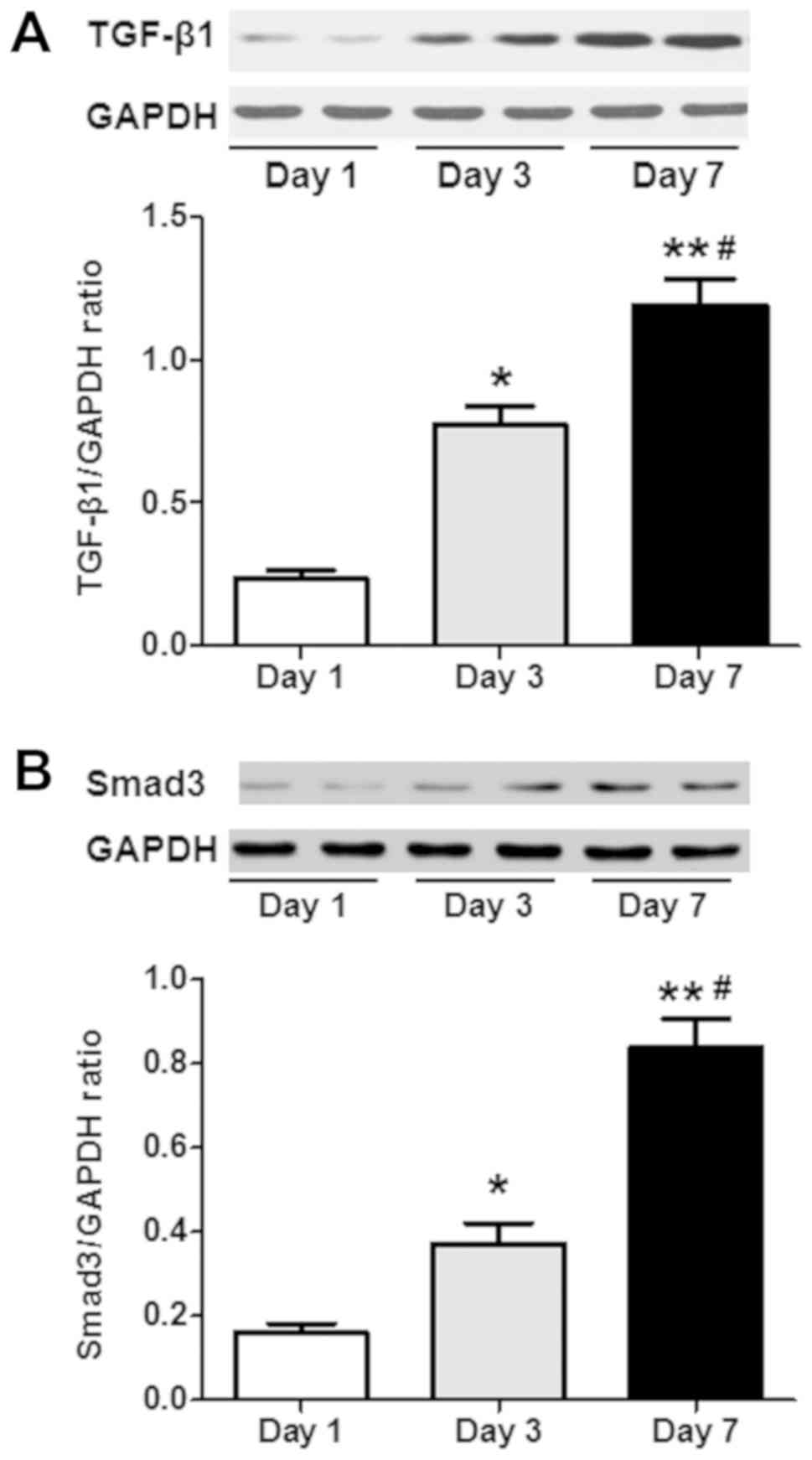
The results of the western blot analysis indicated
that HSCs isolated and cultured on day 1 exhibited upregulation of
the TGF-β1 and Smad3 proteins on days 3 and 7 (P<0.05; n=6;
Fig. 3). The relative expression
levels of the TGF-β1 and Smad3 proteins on day 7 were further
upregulated compared with those observed on day 3 (P<0.05; n=6;
Fig. 3). In the present study, the
TGF-β1/Smad3 signaling pathway was also activated during the HSC
culture.
SDC1 negatively regulates the
activation of HSCs and the TGF-β1/Smad3 signaling pathway during
HSC culture
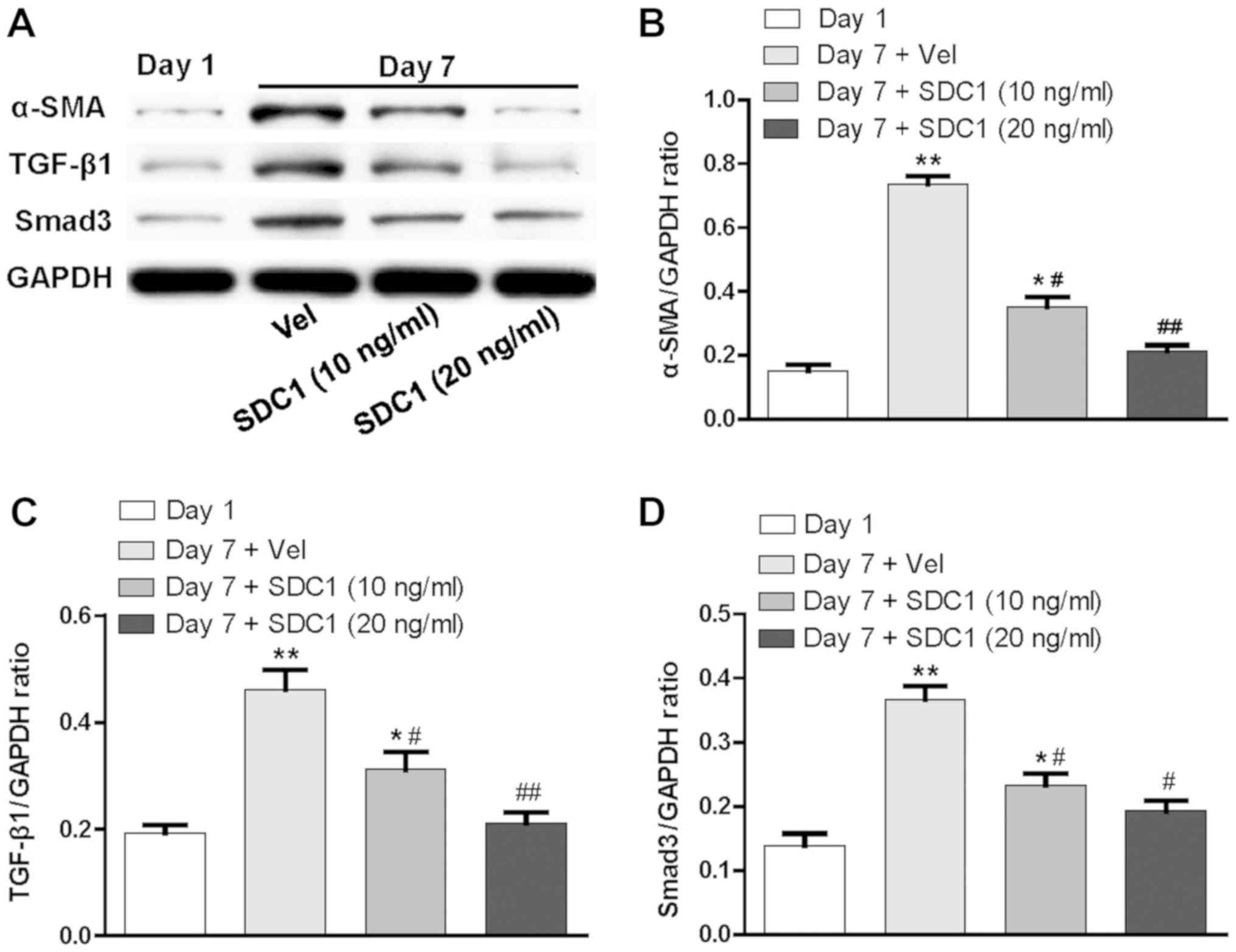
Having demonstrated that the protein expression
levels of α-SMA, TGF-β1 and Smad3 in HSCs cultured for 7 days were
significantly increased, recombinant SDC1 protein (10 and 20 ng/ml)
was added to stimulate HSCs for 24 h on day 6. The expression
levels of α-SMA, TGF-β1 and Smad3 were significantly downregulated
compared with those of the control group (P<0.05; n=6; Fig. 4). Thus, it was deduced that SDC1
negatively affects the activation of HSCs via the TGF-β1/Smad
signaling pathway.
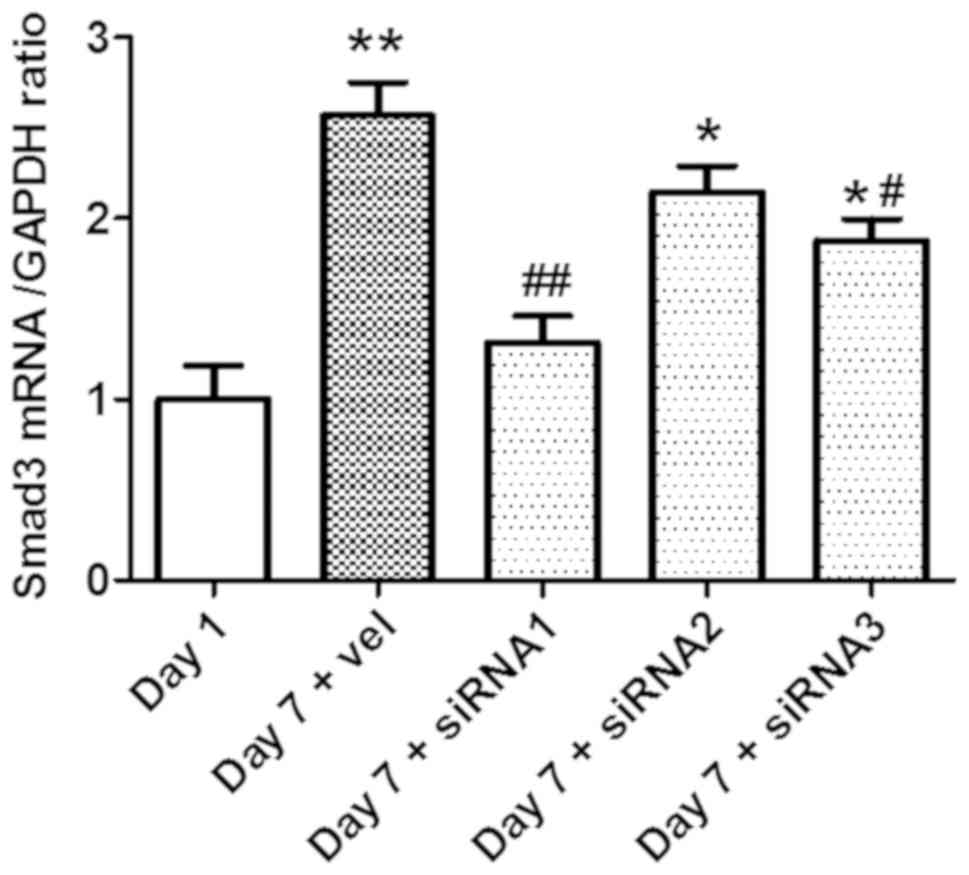
Smad3 expression is successfully
silenced by Smad3-siRNA
In order to select a siRNA oligonucleotide for
efficient knockdown of Smad3, three siRNA oligonucleotides
targeting mouse Smad3 were co-transfected to HSCs isolated from
mice. The transfection was performed after 7 days of initial HSC
culture, and the Smad3 mRNA expression levels were evaluated by
RT-qPCR. siRNA1 and siRNA3 achieved effective knockdown of the
Smad3 protein, with siRNA1 exhibiting the highest efficiency
(P<0.05; n=6; Fig. 5).
Therefore, siRNA1 was selected to knock down Smad3 in subsequent
experiment.
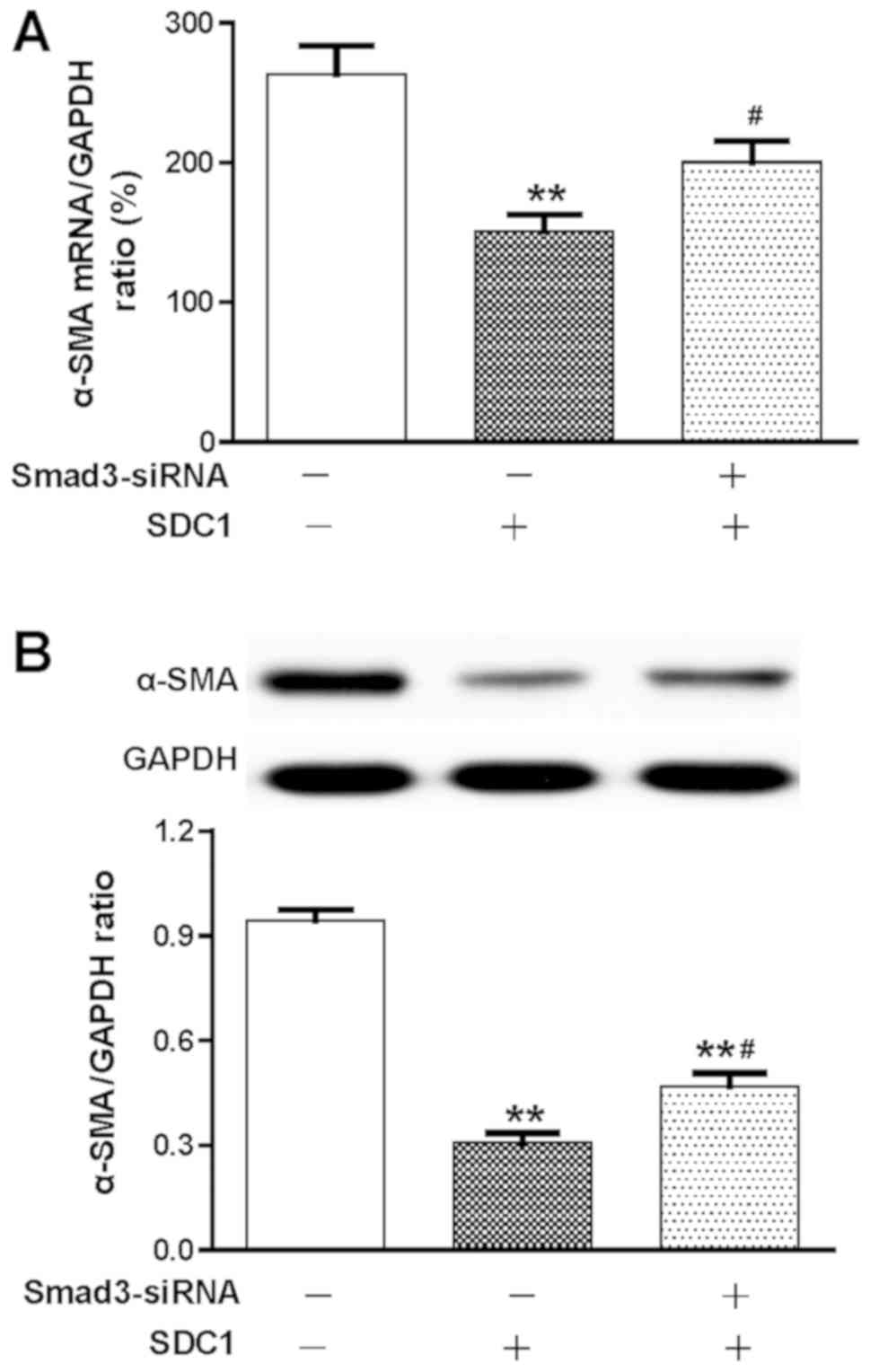
SDC1 negatively regulates the levels
of α-SMA upon HSC activation via the TGF-β1/Smad3 signaling
pathway
As shown in Fig. 6,
the expression levels of α-SMA in HSCs cultured for 7 days were
measured by RT-qPCR and western blot assays. SDC1 stimulation
significantly decreased the expression levels of α-SMA in HSCs
compared with those of the control groups (P<0.01; n=6; Fig. 6). Pretreatment with Smad3-siRNA
reduced the effects of SDC1, which increased the expression levels
of α-SMA (P<0.01; n=6; Fig. 6).
These data suggested that SDC1 markedly inhibits HSC activation,
which is likely mediated by activation of the TGF-β1/Smad3
signaling pathway.
Discussion
Liver fibrosis is an irreversible process caused by
progression of chronic liver disease of varying etiologies.
Prevention and early intervention to target fibrotic tissues is an
effective measure for stabilizing the disease and preventing
progression from liver fibrosis to cirrhosis, which can ultimately
prevent the development of liver cancer (11). Thus, the exploration of the
development of liver fibrosis is crucial for the prevention and
treatment of chronic liver diseases (12). HSCs belong to the family of hepatic
stromal cells, accounting for 5–8% of total liver cells. Static
HSCs are in a non-proliferative state and do not express α-SMA.
Upon liver injury, the phenotype of static HSCs is altered,
resulting in the formation of myofibroblasts. These cells have a
high proliferative rate and express α-SMA. α-SMA promotes the
transformation of HSCs into myofibroblasts and may be used as a
clinical diagnostic index for the initial stages of liver fibrosis
(13).
Freshly isolated HSCs can be spontaneously activated
during in vitro culture. The magnitude of activation is
parallel to the induction of liver injury. Thus, mouse HSCs were
used as a model in the present study. Western blot analysis and
immunofluorescence were used to detect the relevant mechanism
underlying HSC activation. The aim of these experiments was to
obtain crucial information on HSC activation that may prove useful
in the prevention and treatment of liver fibrosis. The results
indicated that the expression of α-SMA was increased in a
time-dependent manner in an HSC culture. Immunofluorescent staining
demonstrated that the expression levels of α-SMA were high on day
7, indicating that HSCs had differentiated to myofibroblasts,
transforming from a static to an activated state.
The development of liver fibrosis is regulated by
multiple growth factors, such as TGF, platelet-derived growth
factor and epidermal growth factor, which are essential regulatory
factors for HSC activation and proliferation (14,15).
The TGF-β1/Smad signaling pathway plays an important role in the
development of liver fibrosis, although its exact mechanism of
action remains unclear. Several studies to date have demonstrated
that the TGF-β1/Smad signaling pathway participates in the
activation of HSCs (16). The
results of the present study are in accordance with this
conclusion. The inhibition of the classic or the non-classic
TGF-β1/Smad signaling pathway may inhibit the proliferation of HSCs
and induce apoptosis. However, the exact mechanism through which
this process affects liver function is unclear. Furthermore, the
direct inhibition of this pathway may have several adverse effects.
Recent studies demonstrated that the SDC family of proteins
regulates the TGF-β1/Smad signaling pathway, thus playing an
important role in tissue differentiation, organ formation and liver
disease progression (17,18).
SDC1 is a surface transmembrane proteoglycan that
acts as a membrane adhesion receptor. SDC1 is an important
component of the intercellular region and of the plasma membrane
(19). SDC1 regulates the
interaction between cells and the microenvironment by covalent
binding to ligands, and participates in physiological processes,
such as vascularization and tissue regeneration. Its expression is
highly regulated and is cell type- and developmental stage-specific
(20). The cells employ integrin
and SDC1 receptors in order to mediate cell-ECM adhesion, cell
proliferation and differentiation, and tumor growth inhibition
(21). The present study
demonstrated that, during HSC culture in vitro, the SDC1
mRNA and protein expression levels decreased in a time-dependent
manner. Although it is reported that SDC1 is upregulated in
patients with liver cirrhosis (22), a recent study demonstrated that
SDC1 inhibits the early stages of liver fibrogenesis by interfering
with the action of TGF-β1 and upregulating matrix metalloproteinase
14 (23). There is a change in
SDC1 over time; its expression during the earlier stages of
fibrogenesis is higher in liver cells, whereas it starts to
decrease during the later stages (23). These results indicated that SDC1
may also participate in the regulation of HSC activation.
In the current study, there was a negative
association of SDC1 with TGF-β1, Smad-3 and SMA. The activation of
HSCs was reduced following treatment with SDC1 recombinant protein.
Concomitantly, the levels of the TGF-β1/Smad3 pathway protein
expression were reduced, in accordance with a recent study
demonstrating that the overexpression of SDC1 decreases TGF-β1
signaling (23). Treatment with
SDC1 inhibited the Smad3-siRNA-induced decrease in the levels of
α-SMA compared with that in the control group. These results
indicated that SDC1 is involved in the regulation of HSC
activation, which is mediated by the TGF-β1/Smad pathway.
In summary, the present study demonstrated that
reduced SDC1 expression induces activation of HSCs via the
TGF-β1/Smad3 signaling pathway. These findings provide evidence
supporting a regulatory role of SDC1 in the activation of HSCs via
TGF-β1/Smad3 signaling. The identification of this novel mechanism
may prove to be useful in the prevention and treatment of liver
fibrosis.
Acknowledgements
Not applicable.
Funding
The present study was supported, in part, by grants
from the Natural Science Foundation of Zhejiang Province (grant no.
LY17H090019), the Science and Technology Project of Jiaxing City
(grant no. 2017AY33014), the National Natural Science Foundation of
China (grant no. 81341035), the Construction Project of the
Anesthesiology Discipline and the Special Disease Center in the
Zhejiang North Region (grant no. 201524), the Construction Project
of Key Laboratory of Nerve and Pain Medicine in Jiaxing City, the
Construction Project of Key Discipline of Infectious Diseases in
Jiaxing City, and the Construction Project of Laboratory of
Hepatology in Jiaxing City.
Availability of materials and data
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MD, XS and MY performed the experiments and analyzed
the data; LX, XX and ZZ interpreted the experimental results; MD
and LX drafted the manuscript; MD and MY edited and revised the
manuscript; MY was responsible for the conception and design of the
research. All authors have read and approved the final version of
this manuscript.
Ethics approval and consent to
participate
Experiments were performed in accordance with the
Guide for the Care and Use of Laboratory Animals of China Jiaxing
University, and approved by the Ethical Committee of Animal
Experimentation. All the laboratory procedures were performed with
the permission and under the surveillance of the institutional
ethics committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HSC
|
hepatic stellate cell
|
|
SDC1
|
syndecan-1
|
|
α-SMA
|
α-smooth muscle actin
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
References
|
1
|
Han X, Hong Y and Zhang K: TUG1 is
involved in liver fibrosis and activation of HSCs by regulating
miR-29b. Biochem Biophys Res Commun. 503:1394–1400. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Friedman SL: Hepatic stellate cells:
Protean, multifunctional, and enigmatic cells of the liver. Physiol
Rev. 88:125–172. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ding H, Ma JJ, Wang WP, Zeng WJ, Jiang T,
Huang BJ and Chen SY: Assessment of liver fibrosis: The
relationship between point shear wave elastography and quantitative
histological analysis. J Gastroenterol Hepatol. 30:553–558. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Manon-Jensen T, Multhaupt HA and Couchman
JR: Mapping of matrix metalloproteinase cleavage sites on
syndecan-1 and syndecan-4 ectodomains. FEBS J. 280:2320–2331. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pasqualon T, Lue H, Groening S,
Pruessmeyer J, Jahr H, Denecke B, Bernhagen J and Ludwig A: Cell
surface syndecan-1 contributes to binding and function of
macrophage migration inhibitory factor (MIF) on epithelial tumor
cells. Biochim Biophys Acta. 1863:717–726. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Masola V, Gambaro G, Tibaldi E, Brunati
AM, Gastaldello A, D'Angelo A, Onisto M and Lupo A; Heparanase and
syndecan-1 interplay orchestrates fibroblast growth
factor-2-induced epithelial-mesenchymal transition in renal tubular
cells, : J Biol Chem. 287:1478–1488. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu Y, Duan J, Li Y, Li Y, Jing L, Yang M,
Wang J and Sun Z: Silica nanoparticles induce liver fibrosis via
TGF-beta1/Smad3 pathway in ICR mice. Int J Nanomedicine.
12:6045–6057. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee JH, Jang EJ, Seo HL, Ku SK, Lee JR,
Shin SS, Park SD, Kim SC and Kim YW: Sauchinone attenuates liver
fibrosis and hepatic stellate cell activation through TGF-beta/Smad
signaling pathway. Chem Biol Interact. 224:58–67. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hara T, Yoshida E, Shinkai Y, Yamamoto C,
Fujiwara Y, Kumagai Y and Kaji T: Biglycan intensifies ALK5-Smad2/3
signaling by TGF-beta1 and downregulates syndecan-4 in cultured
vascular endothelial cells. J Cell Biochem. 118:1087–1096. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao H, Zhang LE, Guo S, Yuan T, Xia B,
Zhang L and Zhang Y: Overexpression of DNA methyltransferase 1 as a
negative independent prognostic factor in primary gastrointestinal
diffuse large B-cell lymphoma treated with CHOP-like regimen and
rituximab. Oncology Letters. 9:2307–2312. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schuppan D and Afdhal NH: Liver cirrhosis.
Lancet. 371:838–851. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schafer DF and Jones EA: Potential neural
mechanisms in the pathogenesis of hepatic encephalopathy. Prog
Liver Dis. 7:615–627. 1982.PubMed/NCBI
|
|
13
|
Takegami Y, Ohkawara B, Ito M, Masuda A,
Nakashima H, Ishiguro N and Ohno K: R-spondin 2 facilitates
differentiation of proliferating chondrocytes into hypertrophic
chondrocytes by enhancing Wnt/beta-catenin signaling in
endochondral ossification. Biochem Biophys Res Commun. 473:255–264.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang JJ, Tao H, Huang C and Li J: Nuclear
erythroid 2-related factor 2: A novel potential therapeutic target
for liver fibrosis. Food Chem Toxicol. 59:421–427. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang CQ, Yang L, Yang WZ, Zhang Z, Zhang
H, Chang YZ, Yuan M and Chen XM: Mechanism of hepatic stellate cell
migration during liver fibrosis. Zhonghua Yi Xue Za Zhi.
88:119–122. 2008.(In Chinese). PubMed/NCBI
|
|
16
|
Park SM, Deering RP, Lu Y, Tivnan P,
Lianoglou S, Al-Shahrour F, Ebert BL, Hacohen N, Leslie C, Daley
GQ, et al: Musashi-2 controls cell fate, lineage bias, and TGF-β
signaling in HSC. J Exp Med. 211:71–87. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu F, Fan X, Chen B, Dong P and Zheng J:
Activation of hepatic stellate cells is inhibited by
microRNA-378a-3p via wnt10a. Cell Physiol Biochem. 39:2409–2420.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim KA, Kakitani M, Zhao J, Oshima T, Tang
T, Binnerts M, Liu Y, Boyle B, Park E, Emtage P, et al: Mitogenic
influence of human R-spondin1 on the intestinal epithelium.
Science. 309:1256–1259. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Beauvais DM, Jung O, Yang Y, Sanderson RD
and Rapraeger AC: Syndecan-1 (CD138) suppresses apoptosis in
multiple myeloma by activating IGF1 receptor: Prevention by
synstatinIGF1R inhibits tumor growth. Cancer Res. 76:4981–4993.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Goto K: CD138 expression is observed in
the urothelial epithelium and in various urothelial carcinomas, and
cannot be evidence for plasmacytoid urothelial carcinoma. Int J
Surg Pathol. 24:614–619. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu D, Hu J, Xu S, De Bruyne E, Menu E, Van
Camp B, Vanderkerken K and Van Valckenborgh E: Dll1/Notch
activation accelerates multiple myeloma disease development by
promoting CD138+ MM-cell proliferation. Leukemia. 26:1402–1405.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baghy K, Tátrai P, Regős E and Kovalszky
I: Proteoglycans in liver cancer. World J Gastroenterol.
22:379–393. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Regős E, Abdelfattah HH, Reszegi A, Szilák
L, Werling K, Szabó G, Kiss A, Schaff Z, Kovalszky I and Baghy K:
Syndecan-1 inhibits early stages of liver fibrogenesis by
interfering with TGFβ1 action and upregulating MMP14. Matrix Biol.
68-69:474–489. 2018. View Article : Google Scholar : PubMed/NCBI
|