Introduction
Liver cancer is currently the second leading cause
of cancer-associated mortality worldwide, with the highest
incidence being reported in Asia and sub-Saharan Africa (1). However, the pathophysiology of
mechanical pressure in liver cancer development has not been
definitively elucidated.
During the development of liver cancer, cancerous
cells may be subjected to various forms of mechanical pressures,
including pressure from the extracellular matrix (ECM) caused by
matrix stiffness (2); the
mechanical-pressure effect caused by rapid tumor growth (3); portal hypertension, caused when the
portal pressure gradient or the pressure difference between the
portal and inferior cava vein strongly increases (4); pressure generated by the blood and
lymphatic transport of shed cancer cells (5); or pressure from iatrogenic
intraoperative stimulation (4,5).
During recent decades, studies have provided evidence that pressure
serves a distinctive role in malignant cell proliferation and
invasion: Craig et al (5)
reported that activation of cancer cells by pressure promotes tumor
development and impaired tumor-free survival. Furthermore, Basson
et al (6) revealed that
increased extracellular pressure activates a mechanosensitive
calcium pathway to further enhance the proliferation of tumor
cells, and Fiering et al (7) demonstrated that the mechanical
pressure from cancer-associated fibroblasts (CAFs) leads to the
progression of metastasis. Fernández-Sánchez et al (8) explored the contribution of mechanical
pressure exerted by tumor growth onto non-tumorous adjacent
epithelium, and demonstrated that that the tumorigenic β-catenin
pathway could be mechanically activated in healthy epithelial cells
surrounding the tumor, suggesting an unexplored mode of tumor
propagation based on mechanical signaling pathways.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs
considered to be key post-transcriptional modulators of gene
expression, which target mRNA for translational repression or
destabilization (9). Mechanically
responsive miRNAs are sensitive or responsive to
mechanotransduction. To date, some mechanically induced miRNAs have
been associated with physiological or pathological processes
(10–13). The preliminary results of a
previous study confirmed that the mechanically responsive miR-9a-5p
regulates proliferation and migration of hepatic stellate cells
(HSCs) through inhibition of sirtuin 1 (Sirt1) (13). Clinical data has revealed that 90%
of patients with liver cancer have a background of liver cirrhosis
(14), and the median overall
survival rate of patients with liver cancer and a liver cirrhosis
background has significantly decreased (15). Currently, elevated portal pressure
has been exclusively considered a consequence of liver cirrhosis
(16), but whether
mechanosensitive miRNAs have a pivotal role in the subsequent
development of liver cancer remains unknown. In addition, it may be
hypothesized that the increased recurrence rate following
hepatectomy is associated with the increased biological activity of
liver cancer cells following intraoperative mechanical stimulation.
However, the role of miRNAs in this process should be further
evaluated.
To investigate alterations in the proliferation and
invasion of liver cancer cell lines following mechanical
stimulation, HepG2 and Huh-7 cell lines were subjected to gradually
increasing pressure (0, 5, 15, 30 and 60 mmHg) for different
periods of time (0, 12, 24 and 48 h) using 2-dimensional (2D) and
3-dimensional (3D) pressure-loading systems. Subsequently, the
differentially expressed miRNAs and mRNAs were screened under
optimal conditions (15 mmHg, 24 h) by microarray analysis. The
target genes of miRNAs and the differentially expressed mRNAs were
integrated, and 1,309 genes were bioinformatically predicted to
respond to mechanical pressure. Through Gene Ontology (GO) and
pathway analyses, it was revealed that the function of these target
genes was primarily associated with proliferation and invasion.
Materials and methods
Cell culture and reagents
The HepG2 cell line was purchased from American Type
Culture Collection (cat. no. HB-8065). The Huh-7 cell line was
purchased from the Japanese Collection of Research Bioresources
Cell Bank (cat. no. 0403). Mycoplasma testing was performed for all
cell lines and no infection was found. The cell lines were both
authenticated by short tandem repeat analysis. Cells were cultured
at 37°C in an atmosphere containing 5% CO2 in Dulbecco's
modified Eagle's medium (DMEM; HyClone; GE Healthcare Life
Sciences) containing 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.), 2 mM glutamine, 1 mM sodium pyruvate, 10
mM HEPES, 100 U/ml penicillin G and 100 mg/ml streptomycin
(Sigma-Aldrich; Merck KGaA). Cells were digested with EDTA-0.25%
trypsin (Thermo Fisher Scientific, Inc.) during cell passage.
Pressure loading
The 2D and 3D pressure-loading systems were used to
exert increasing pressure (0, 5, 15, 30 and 60 mmHg) for various
periods of time (0, 12, 24 and 48 h). The 2D pressure-loading
system used in the present study was designed by previous studies
(17–19). The container was made from
stainless steel, the cap of which contained an input port and an
output port that can be opened and tightly sealed with a rubber
ring. The inlet was connected to high-pressure helium by a silica
gel sorbent tube and a pressure regulator, and the outlet was
connected to a three-way control valve, of which one end was
connected to a buffer gas valve and the other end was connected to
a medical sphygmomanometer to read the pressure in the container.
When the cells were cultured under pressure, the system was placed
in a 5% CO2 incubator.
Pressure loading of 3D cultured cells was undertaken
using a Flexcell-5000 Compression system (Flexcell International
Corporation). Prior to pressure loading, the cells were cultured in
3D Life Biomimetic Hydrogels (Cellendes GmbH) for 4 h. The cell
gels were placed in a BioPres Compression Culture Plate (Flexcell
International Corporation), and the pressure parameters, including
compressive strength and intervention time, were adjusted digitally
using FlexSoft FX-5000™ (Flexcell International Corporation). The
cells in gel were detached by 1X Accutase enzymes (Innovative Cell
Technologies, Inc.), and centrifuged for 10 min at 111.8 × g at
room temperature. The cells were subsequently collected for
follow-up experiments.
Cell counting kit-8 (CCK-8) assay
Cell proliferation was analyzed using the CCK-8
assay (Dojindo Molecular Technologies, Inc.). Cells were plated in
96-well plates at 1×104 cells/well. At four different
time-points (0, 12, 24 and 48 h), 10 µl CCK-8 solution was added to
each well, and the cells were incubated for a further 2 h at room
temperature. The optical density value was obtained by determining
the differences in absorbance at a wavelength of 450 nm using a
microplate reader (KHB-ST-360; Shanghai Kehua Bio-engineering Co.,
Ltd.). The activity of cellular dehydrogenases generates formazan
dye, the amount of which is directly proportional to the number of
living cancer cells.
Cell cycle analysis
Cell cycle arrest was measured using a Cell Cycle
Assay kit (Hangzhou MultiSciences Biotech, Co., Ltd.). A total of
2×105−1×106 3D-cultured and pressure-treated
cells were collected and centrifuged for 10 min at 111.8 × g at
room temperature. After the supernatant was discarded, cells were
washed once with PBS (HyClone; GE Healthcare Life Sciences).
Afterwards, DNA staining solution (1 ml) was added to the cells.
The mixture was spun for 5–10 sec, and incubated at room
temperature for 30 min. Finally, the mixed solution was collected
for analysis via flow cytometry using FlowJo V10 software (FlowJo
LLC).
Cell invasion assay
The cell invasion assay was performed using
Transwell® Permeable Supports, which consisted of
Snapwell™ and Netwell™ inserts (Corning Inc.). The kit contained 24
inserts; each insert contained a polycarbonate membrane with 8-µm
pores coated with a thin layer of type I/III collagen-coated
polytetrafluoroethylene. Briefly, serum-free DMEM was used to
prepare cell suspension, and 200 µl cell suspension containing
0.5×104 cells was added to the upper chamber of each
pore; DMEM containing 10% FBS was added to each lower chamber as a
chemoattractant. The cells were cultured for 24 h. Subsequently,
non-migratory cells on the upper surface were scraped off. The
migrated cells were fixed with 4% paraformaldehyde for 30 min at
room temperature and stained with 0.1% crystal violet for 5 min at
room temperature. Acetic acid (10%) was used to elute the stained
inserts to detect the percentage of invaded cells. Images of the
cells were then captured using a light microscope (magnification,
×100).
Wound-healing assay
Cells (5×105/well) were seeded into
24-well plates (Corning, Inc.); after 24 h, the cells reached
70–80% confluence as a monolayer. A new pipette tip (100 µl) was
used to gently scratch the monolayer across the center of the well.
After scratching, the well was washed twice with DMEM to remove the
detached cells. The well was then replenished with fresh DMEM
containing 10% FBS, and incubated for an additional 24 h at 37°C.
The cells were subsequently washed twice with PBS, and fixed with
3.7% paraformaldehyde for 30 min at room temperature and stained
with 1% crystal violet in 2% ethanol for 30 min at room
temperature. Images of the cells were captured using a light
microscope (magnification, ×100). The gap distances were measured
using ImageJ V1.51 (National Institutes of Health).
Cell mortality assay
The HepG2 and Huh-7 cells in logarithmic growth
phase were inoculated in 6-cm culture dishes (Corning, Inc.) at a
density of 1×106 cells/ml; 5 ml cell suspension was
added to each dish. Cells were cultured in DMEM containing 10% FBS
and pressure-loaded (0, 5, 15, 30 and 60 mmHg) for a set period of
time (0, 12, 24 and 48 h). Cells were then digested with EDTA-0.25%
trypsin (Thermo Fisher Scientific, Inc.) to prepare a 0.2-ml cell
suspension. The cells were mixed evenly with 0.3 ml PBS and 0.5 ml
0.4% trypan blue staining solution (Sigma-Aldrich; Merck KGaA) for
5–15 min at room temperature. The total number of cells and the
number of cells stained with trypan blue were counted on a
hemocytometer. Finally, the cell survival rate was calculated
according to the following formula: Cell mortality=(no. of
blue-stained cells/total no. of cells)×100.
Microarray analysis of miRNA
expression
A total of three samples were allocated to the
control and pressure groups. Total RNA was extracted by
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol and purified using
miRNeasy Mini kit (Qiagen GmbH). After the assessment of RNA
quality and quantity, microarray analysis of miRNA expression
(Agilent Human miRNA V21.0 Array; Agilent Technologies, Inc.) was
performed, according to the manufacturer's protocol. Briefly, 1 µg
total RNA was labeled and hybridized with miRNA Complete Labeling
and Hyb kit (Agilent Technologies, Inc.). Slides were then washed
with Gene Expression Wash Buffer kit (Agilent Technologies, Inc.),
and scanned by Agilent Microarray Scanner (Agilent Technologies,
Inc.). The data were analyzed with Feature Extraction software
(version 10.7; Agilent Technologies, Inc.) using the default
settings.
Microarray analysis of mRNA
expression
For mRNA analysis, the array (OE BioTech) was
performed using an Agilent expression profile gene chip (Agilent
Technologies, Inc.). Two samples were allocated to the control and
pressure groups. Total RNA was extracted using TRIzol®
reagent (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol and quantified using NanoDrop 2000
(NanoDrop Technologies; Thermo Fisher Scientific, Inc.), and
detected with an Agilent Bioanalyzer 2100 (Agilent Technologies,
Inc.). The Quick Amp Labeling kit (One-Color; Agilent Technologies,
Inc.) was used to reverse transcribe total RNA to double-stranded
cDNA, prepare the label reaction and transcribe cRNAs from cDNAs,
all according to the manufacturer's protocol. Subsequently, an
RNeasy Mini kit (Qiagen GmbH) was used to purify the
labeled/amplified RNAs and quality control-labeled cRNAs. Each
slide was hybridized with cyanine 3-labeled RNA using the Agilent
Gene Expression Hybridization kit (Agilent Technologies, Inc.) in a
Hybridization Oven (Thermo Fisher Scientific, Inc.) at 65°C for 17
h. After hybridization, slides were washed in staining dishes with
Gene Expression Wash Buffer kit (Agilent Technologies, Inc.).
Slides were scanned using the Agilent Microarray Scanner (Agilent
Technologies, Inc.) and analyzed using the Feature Extraction
software (version 10.7; Agilent Technologies, Inc.) with default
settings.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
TRIzol® reagent (Thermo Fisher
Scientific, Inc.) was used to extract RNA from cancer cells,
according to the manufacturer's protocol. A NanoDrop 2000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.) was used to measure RNA quantity and quality. Standard
denaturing agarose gel electrophoresis was used to access RNA
integrity (20). An Mir-X™ miRNA
First Strand Synthesis kit (Clontech Laboratories, Inc.) was
applied to reverse transcribe miRNA, and a PrimeScript™ RT Reagant
kit (Takara Bio, Inc.) was applied to reverse transcribe mRNA. The
reverse transcription reactions for miRNA and mRNA were conducted
according to manufacturers' protocols. qPCR was performed on a
Rotor Gene 3000 real-time PCR system from Corbett Research (Qiagen
GmbH). Mir-X™ miRNA RT-qPCR SYBR® kit (Takara Bio, Inc.)
was used to amplify the cDNA of the detected miRNA. The reaction
system had a total volume of 10 µl, consisting of 5 µl SYBR
Advantage premix (2X), 3.6 µl double-distilled H2O, 0.2
µl ROX Dye (50X), 0.2 µl forward (F) primer, 0.2 µl reverse (R)
primer and 0.8 µl DNA sample. The thermocycling conditions were as
follows: 95°C, 10 sec, followed by 40 cycles at 95°C for 5 sec and
60°C for 20 sec. TB Green™ Premix Ex Taq™ II kit (Takara Bio, Inc.)
was used to amplify cDNA of the detected mRNA. The reaction system
also comprised a 10-µl total volume, consisting of 5 µl TB Green
Premix Ex Taq II (Tli RNaseH Plus; 2X), 0.4 µl F primer, 0.4 µl R
primer, 0.2 µl ROX Reference Dye (50X), 1 µl DNA sample and 3 µl
double-distilled H2O. The thermocycling conditions were
as follows: 95°C for 30 sec, followed by 40 cycles at 95°C for 5
sec and 60°C for 30 sec. The primers (Wcgene Biotech) used in this
study are listed in Tables I and
II. The reverse primers for miRNA
RT-qPCR were taken from the kit. GAPDH was used as an internal
reference for mRNA expression detection, whereas U6 was used as an
internal reference for miRNA expression detection. All tests were
independent and repeated three times. miRNA and mRNA expression
levels were calculated based on the 2−ΔΔCq method
(21). The results were
statistically analyzed and presented as the means ± standard
deviation (SD).
 | Table I.Primer sequences of miRNAs used for
reverse transcription-quantitative polymerase chain reaction. |
Table I.
Primer sequences of miRNAs used for
reverse transcription-quantitative polymerase chain reaction.
| Gene name | Forward primer
sequence |
|---|
| hsa-miR-7641 |
5′-TTGATCTCGGAAGCTAAGC-3′ |
|
hsa-miR-4485-3p |
5′-TAACGGCCGCGGTACCCTAA-3′ |
| hsa-miR-7-5p |
5′-TGGAAGACTAGTGATTTTGTTGTT-3′ |
| hsa-miR-5703 |
5′-AGGAGAAGTCGGGAAGGT-3′ |
| hsa-miR-630 |
5′-AGTATTCTGTACCAGGGAAG |
|
| GT-3′ |
 | Table II.Primer sequences of mRNAs used for
reverse transcription-quantitative PCR. |
Table II.
Primer sequences of mRNAs used for
reverse transcription-quantitative PCR.
| Gene name | Forward primer | Reverse primer |
|---|
| MMP1 |
5′-GTGTCTGGTCAATGGTTATCC-3′ |
5′-GCCAGATTATTTCCGTGG-3′ |
| MMP2 |
5′-CAGGACATTGTCTTTGATGGCATCGC-3′ |
5′-TACCGTCAAAGGGGTATCCAT-3′ |
| MMP7 |
5′-TCTCCTCCGAGACCTGTCC-3′ |
5′-GCTGACATCATGATTGGCTTT-3′ |
| MMP14 |
5′-GCCGGGGCATCCAGCAACTTTA-3′ |
5′-TCCTCACCCGCCAGAACCAG-3′ |
| TIMP1 |
5′-CAGAACCGCAGGGAGGAG-3′ |
5′-CCCAGGGAACCAGGAAGG-3′ |
| LAMA4 |
5′-GTGTAGGAATTGCTTACGCAACA-3′ |
5′-GCTAACCGCAGGTCATCAGT-3′ |
| SRC |
5′-GACAGGCTACATCCCCAGC-3′ |
5′-CGTCTGGTGATCTTGCCAAAA-3′ |
| ERBB3 |
5′-CGAGATTCTGTCAGGGGGTG-3′ |
5′-ATCTCAGCATCTCGGTCCCT-3′ |
| PIK3R1 |
5′-AGCATTGGGACCTCACATTACACA-3′ |
5′-ACTGGAAACACAGTCCATGCACATA-3′ |
| CTNNB1 |
5′-AGCTTCCAGACACGCTATCAT-3′ |
5′-CGGTACAACGAGCTGTTTCTAC-3′ |
| FAK |
5′-GCTTACCTTGACCCCAACTTG-3′ |
5′-ACGTTCCATACCAGTACCCAG-3′ |
| CLPTM1L |
5′-AGAAACAATGGGACGCTGTATG-3′ |
5′-GCTTGGGGACCATGTAGGTG-3′ |
| TP53I3 |
5′-AATGCTTTCACGGAGCAAATTC-3′ |
5′-TTCGGTCACTGGGTAGATTCT-3′ |
| FSCN1 |
5′-CTGCTACTTTGACATCGAGTGG-3′ |
5′-GGGCGGTTGATGAGCTTCA-3′ |
| GAPDH |
5′-CAATGACCCCTTCATTGACC-3′ |
5′-GACAAGCTTCCCGTTCTCAG-3 |
Bioinformatics analysis
Target genes of miRNAs were predicted from three
databases, including miRDB V6.0 (www.mirdb.org), MiRTarBase (mirtarbase.mbc.nctu.edu.tw/php/index.php) and
TargetScan V7.2 (www.targetscan.org/vert_72). All data were obtained
from the databases in June 2018. The annotations were collected
from the GO (geneontology.org) and the National
Center for Biotechnology Information (www.ncbi.nlm.nih.gov) databases. The Kyoto
Encyclopedia of Genes and Genomes (KEGG) database was used for
genome/metagenome annotation (www.genome.jp/kegg/annotation). On the basis of the
aforementioned analytical tools, specific biological processes and
pathways were found to be enriched. P<0.05 and false discovery
rate <0.25 were used to define the threshold of significance.
The miRNA-gene network was produced using Cytoscape 3.6 (22).
Statistical analysis
All data are expressed as the means ± SD. A
Student's t-test was used to analyze data from two groups. Data
from three or more groups were analyzed using one-way ANOVA
followed by least significant difference post hoc comparison test.
P≤0.05 was considered to indicate a statistically significant
difference.
Results
Mechanical pressure regulates the
proliferation of liver cancer cells
The proliferation of 2D-cultured liver cancer cell
lines was determined using the CCK-8 assay (Fig. 1A and B). The results demonstrated
that proliferation was increased under 15 mmHg pressure for 24 h in
the HepG2 (P<0.05) and Huh-7 (P<0.01) cell lines. Conversely,
60 mmHg pressure for 48 h decreased proliferation of HepG2
(P<0.05) and Huh-7 (P<0.01) cell lines.
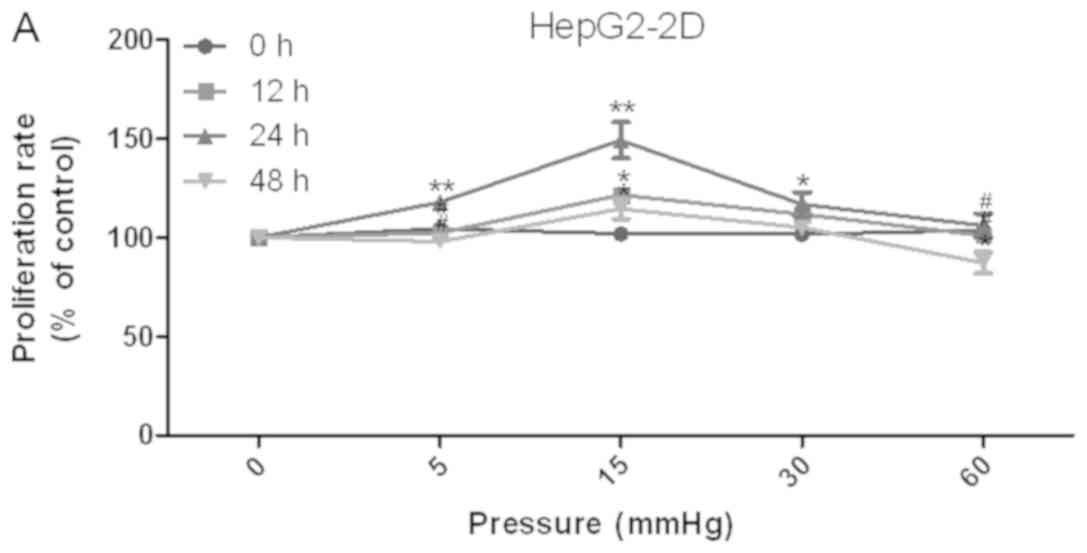 | Figure 1.Effects of elevated pressure on the
proliferation of HepG2 and Huh-7 liver cancer cells in
vitro. 2D-cultured (A) HepG2 and (B) Huh-7 liver cancer cells
were cultivated under pressure (0, 5, 15, 30 and 60 mmHg) and
observed at different time points (0, 12, 24 and 48 h); their
proliferation (relative to the control group) was assessed using
the Cell Counting kit-8 assay. (C) Morphology of 2D-cultured and
3D-cultured liver cancer cell lines (magnification, ×400). Uneven
illumination was corrected using a control image. Scale bar=50 µm.
(D) Flow cytometry results of 3D-cultured cells under pressure for
24 h; the ratio of (E) HepG2 and (F) Huh-7 cells in the S phase of
the cell cycle was evaluated using a cell cycle assay. Effects of
elevated pressure on the proliferation of HepG2 and Huh-7 liver
cancer cells in vitro. Mortality rate for HepG2 pressurized
at 15 mmHg for (G) 12, (H) 24 and (I) 48 h. All data are expressed
as the means ± standard deviation. The data were analyzed using
ANOVA, followed by the least significant difference post hoc test.
*P<0.05, **P<0.01 and ***P<0.001 vs. 0 mmHg.
#P<0.05, ##P<0.01 and
###P<0.001 vs. 15 mmHg. 2D, 2-dimensional; 3D,
3-dimensional; NS, not significant. |
The liver cancer cell lines were also cultured in 3D
Life Hydrogel (Fig. 1C) before
being pressurized and were treated with Accutase enzymes prior to
detection by flow cytometry (Fig.
1D). The results revealed that the ratio of cells in S phase
was significantly increased in response to 15 mmHg pressure for 24
h (P<0.001; Fig. 1E and F).
Mechanical pressure regulates the
invasion of liver cancer cells
The migration and invasion of 2D-cultured liver
cancer cell lines were examined using a wound-healing assay
(Fig. 2A-D) and Transwell assay
(Fig. 2E-G), respectively. The
results demonstrated that the percentage of invasive and migratory
cells was increased under 15 mmHg pressure for 24 h (P<0.001),
with no statistical difference between the 0 mmHg group and the 60
mmHg group.
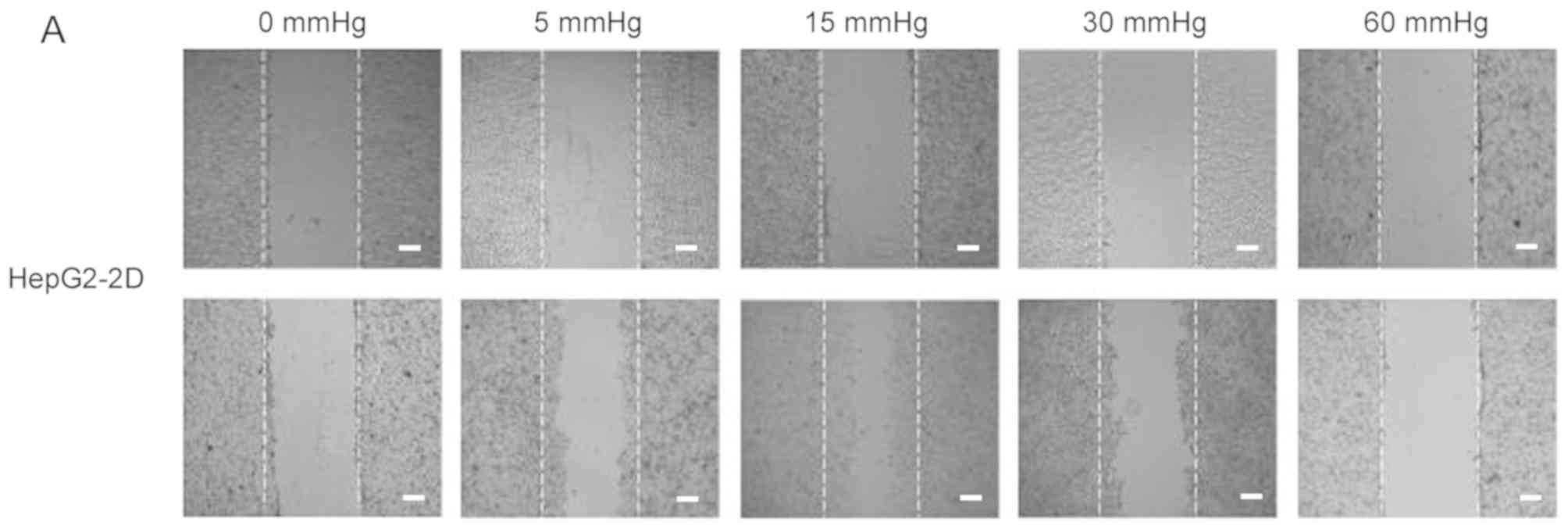 | Figure 2.Effect of pressure on HepG2 and Huh-7
liver cancer cell migration and invasion in vitro. (A) HepG2
and (B) Huh-7 confluent cell monolayers were wounded and cultured
under different levels of pressure (0, 5, 15, 30, 60 mmHg) for 24 h
(magnification, ×100); (C) HepG2 and (D) Huh-7 percentage of
migration was calculated. (E) Representative images of Transwell
assay results showing cells stained with 0.1% crystal violet
solution (magnification, ×100). Effect of pressure on HepG2 and
Huh-7 liver cancer cell migration and invasion in vitro. The
assay lasted 24 h, and after fixing and staining, the percentage of
(F) HepG2 and (G) Huh-7 cells on the lower chamber was calculated;
10% acetic acid was used to elute the stained inserts to detect the
percentage of invaded cells. mRNA expression levels of MMP1, MMP2,
MMP7, MMP14 and TIMP1 in 3D-cultured (H) HepG2 and (I) Huh-7 cells
exposed to different levels of pressure (0, 5, 15, 30, 60 mmHg) for
24 h. All data are expressed as the means ± standard deviation. The
data were analyzed using ANOVA, followed by the least significant
difference post hoc test. *P<0.05, **P<0.01 and ***P<0.001
vs. 0 mmHg. #P<0.05, ##P<0.01 and
###P<0.001 vs. 15 mmHg. 2D, 2-dimensional; 3D,
3-dimensional; MMP, matrix metalloproteinase; TIMP, tissue
inhibitor of metalloproteinases; NS, not significant. |
RT-qPCR was used to detect the expression of tumor
metastasis-related genes in 3D-cultured liver cancer cells. Matrix
metalloproteinases (MMPs) can degrade components of the ECM to
induce tumor metastasis; MMPs are in turn inhibited by tissue
inhibitors of metalloproteinases (TIMPs) (23). Therefore, the expression levels of
MMP1, MMP2, MMP7 and MMP14, which represent collagenase,
gelatinase, matrix degrading enzyme and membrane-type MMP,
respectively, and TIMP1, which can inhibit almost all subtypes of
MMPs, were evaluated. In HepG2 and Huh-7 cell lines, the expression
levels of MMP1, MMP7 and MMP14 in the cells pressurized at 15 mmHg
for 24 h were significantly higher compared with in the other
groups, whereas TIMP1 expression exhibited the opposite trend. In
addition, MMP2 was increased in cells pressurized at 15 mmHg for 24
h compared with in the control group (P<0.01; Fig. 2H and I).
Mechanical pressure regulates the
mortality of liver cancer cells
For 2D-cultured liver cancer cells, trypan blue
staining was used to determine the cell death rate under various
pressures. The results indicated that there was no significant
difference in the mortality rate between cells under different
pressures for 12 and 24 h (Fig. 1G and
H). Conversely, the mortality rate was increased after 48 h of
30 and 60 mmHg compression (P<0.05; Fig. 1I).
Identification and integrative
analysis of miRNAs and mRNAs
The HepG2 cell line was pressurized at 15 mmHg for
24 h, and mRNA and miRNA expression was evaluated via microarray
analysis. Five miRNAs were identified using the miRNA chip (fold
change ≥1.2, P≤0.05; Fig. 3A).
miR-7-5p, miR-7641 and miR-4485-3p were upregulated, whereas
miR-5703 and miR-630 were downregulated under pressure (Table III). In addition, 10,150 mRNAs
were differentially expressed according to mRNA chip analysis (fold
change ≥2, P≤0.05; Fig. 3B). A
total of 5,102 genes were upregulated, including oncogenes such as
sarcoma gene (SRC), focal adhesion kinase (FAK), phosphoinositide
3-kinase (PI3K) regulatory subunit 1 (PIK3R1), integrin subunit α V
(ITGAV), son of sevenless 1 (SOS1), insulin receptor substrate 1
(IRS1) and serum/glucocorticoid regulated kinase 3 (SGK3). 5,048
genes were downregulated, including tumor suppressor genes such as
SMAD family member 2 (SMAD2), casein kinase 1 ε (CSNK1E) and
forkhead box O 1 (FOXO1).
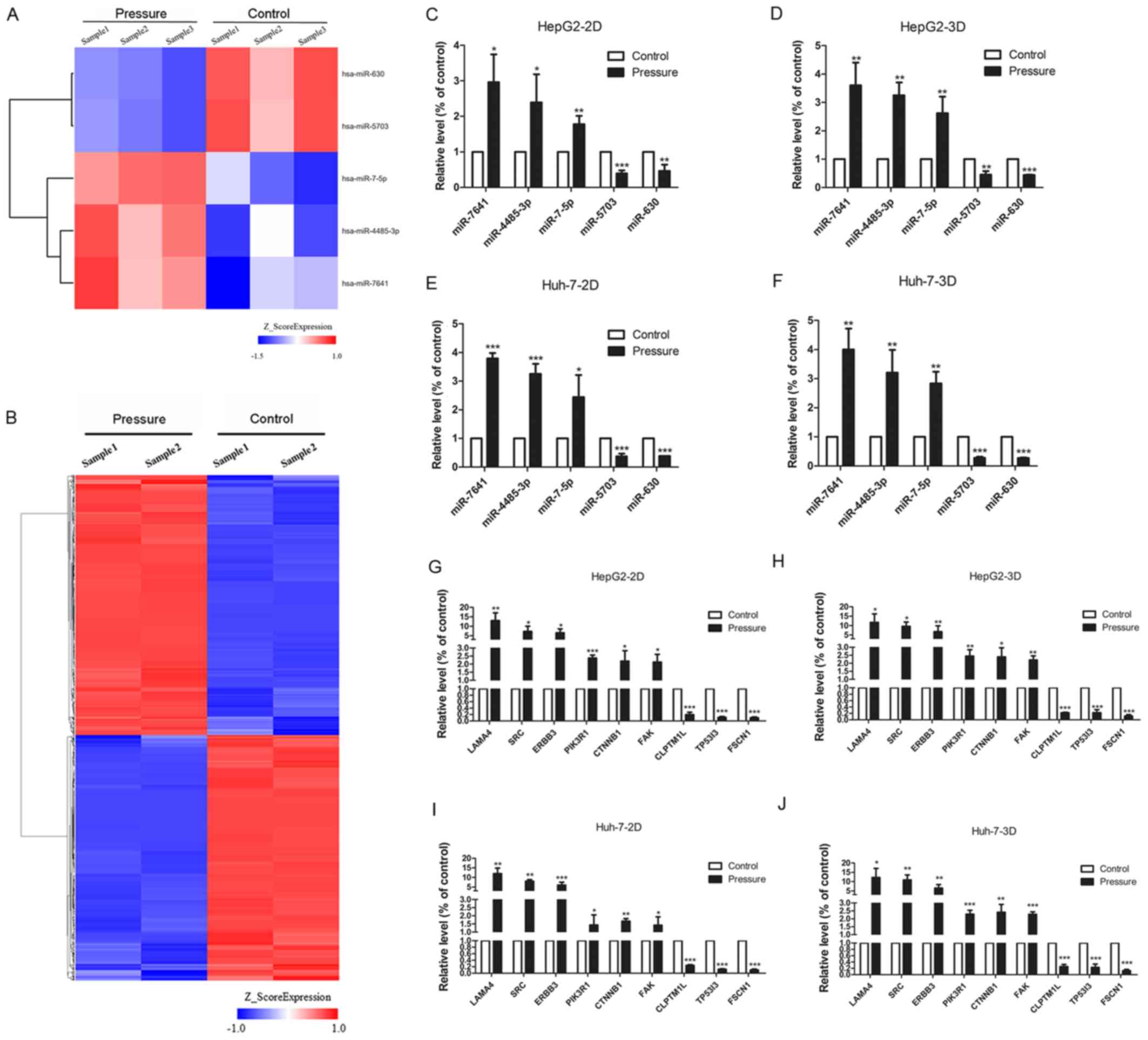 | Figure 3.Differently expressed miRNAs and
mRNAs in cells treated with 15 mmHg for 24 h, as determined by
microarray analysis and RT-qPCR. (A) In the cluster heat map,
changes in miRNA expression (fold change≥1.2, P≤0.05) in HepG2
cells were illustrated, and three pressure-upregulated miRNAs and
two pressure-downregulated miRNAs were identified. (B) In the
cluster heat map, changes in mRNA expression (fold change≥2,
P≤0.05) in HepG2 cells are illustrated. A total of 5,102
pressure-activated mRNAs and 5,048 pressure-repressed mRNAs were
identified. Quantitative analysis of the transcript levels
(relative to the control) of five miRNAs identified by miRNA chip
of HepG2 cells cultured in (C) 2D and (D) 3D and Huh-7 cells
cultured in (E) 2D and (F) 3D. The expression levels of nine genes
identified by mRNA chip in HepG2 cells, cultured in (G) 2D and (H)
3D, and Huh-7 cells, also cultured in (I) 2D and (J) 3D, were
evaluated via RT-qPCR. All data are expressed as the means ±
standard deviation, and analyzed using Student's t-test.
*P<0.05, **P<0.01 and ***P<0.001. 2D, 2-dimensional; 3D,
3-dimensional; miRNA, microRNA; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction. |
 | Table III.Differentially expressed miRNAs in
pressure-stimulated cells compared with control cells. |
Table III.
Differentially expressed miRNAs in
pressure-stimulated cells compared with control cells.
| A, Downregulated
microRNAs |
|---|
|
|---|
| Systematic
name | P-value | Fold-change |
|---|
| hsa-miR-630 | 0.004232634 | 0.679781 |
| hsa-miR-5703 | 0.006772327 | 0.727266 |
|
| B, Upregulated
microRNAs |
|
| Systematic
name | P-value |
Fold-change |
|
| hsa-miR-7641 | 0.047364561 | 1.517495 |
| hsa-miR-7-5p | 0.025880795 | 1.291842 |
|
hsa-miR-4485-3p | 0.041582116 | 1.234361 |
The expression of laminin subunit α 4, SRC, Erb-B2
receptor tyrosine kinase 3, PIK3R1, catenin β 1, FAK, cleft lip and
palate transmembrane protein 1-like protein, tumor protein P53
inducible protein 3, fascin actin-bundling protein 1 and all five
miRNAs was evaluated by RT-qPCR in the HepG2 and Huh-7 cell lines,
which were treated under 2D and 3D pressure-loading conditions. The
results revealed a similar trend to the microarray analyses,
confirming the changes detected in the expression levels of mRNAs
and miRNAs (Fig. 3C-J).
Three online databases were used to predict the
target genes of the five pressure-responsive miRNAs, and the sum
aggregate of the predictive target genes was integrated with the
differentially expressed mRNAs from the mRNA chip. With this
approach, a total of 1,309 genes (642 downregulated, 667
upregulated) were obtained and used to construct the miRNA-gene
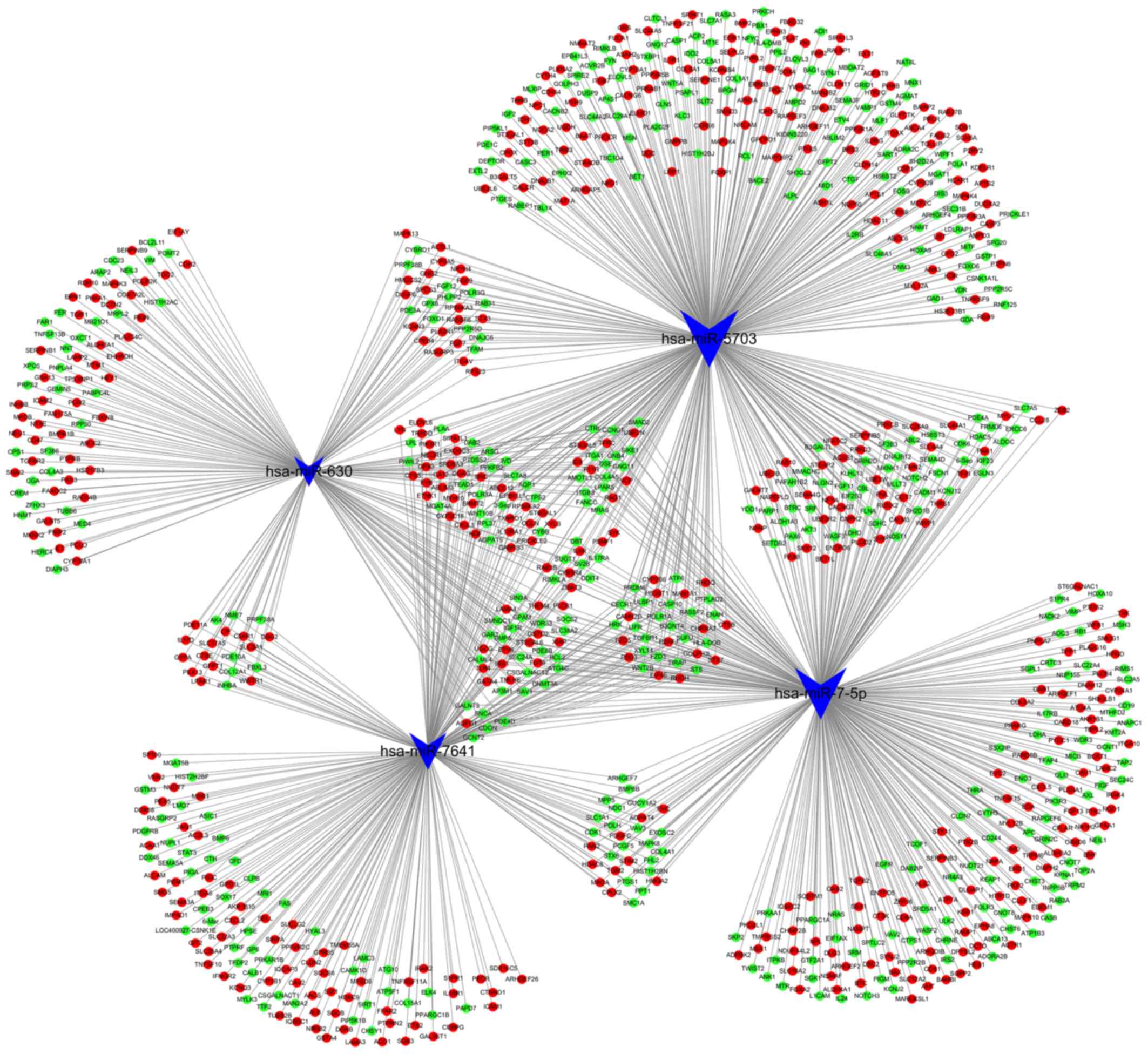
network (Fig. 4). Notably, there
was no predictive target gene of miR-4485-3p in the integrated
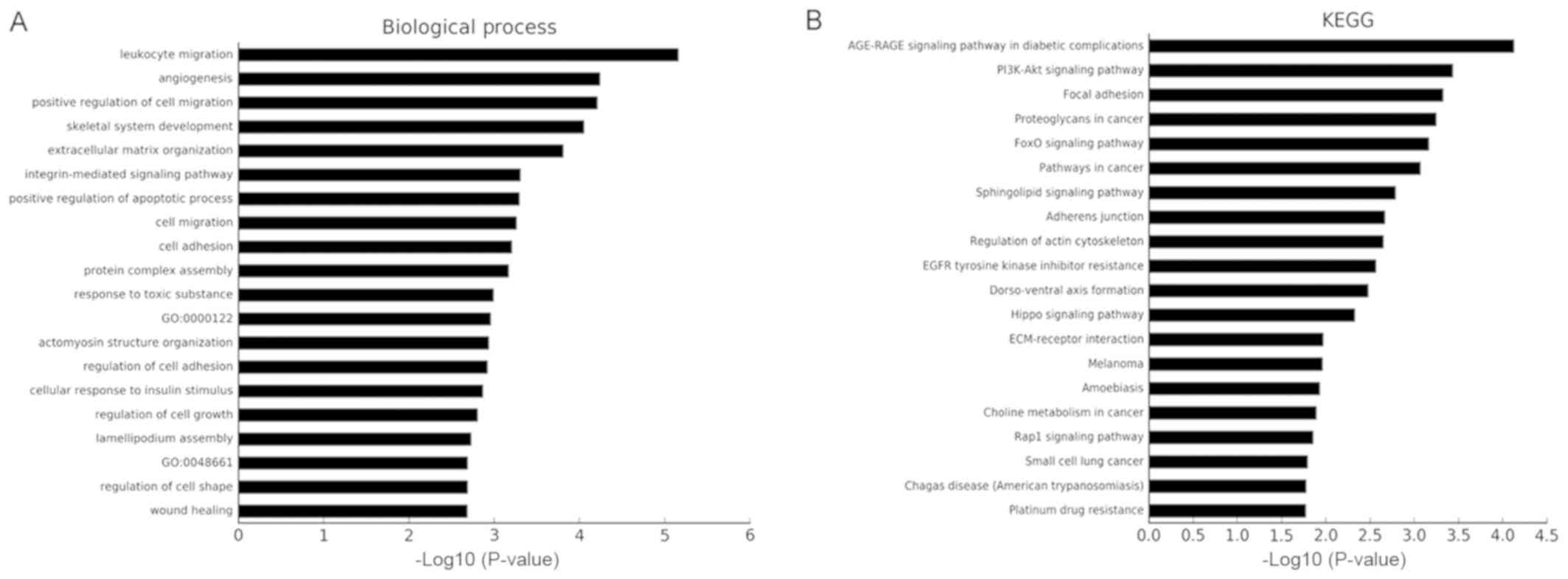
results. The results of the GO and KEGG pathway analyses of the
1,309 genes are shown in Fig. 5.
The top three cancer development-associated ‘biological process’
terms identified by GO analysis were ‘positive regulation of cell
migration’, ‘integrin-mediated signaling pathway’ and ‘cell
migration’. The top three cancer development-associated terms
identified by pathway analysis were ‘PI3K-Akt signaling pathway’,
‘Focal adhesion’ and ‘FoxO signaling pathway’.
Discussion
In the present study, the effects of elevated
pressure on HepG2 and Huh-7 cells cultured in 2D and 3D conditions
were evaluated, and it was observed that a pressure of 15 mmHg for
24 h increased the proliferation, migration and invasion of liver
cancer cells. Notably, the proliferation of 2D-cultured liver
cancer cells under 30 and 60 mmHg compression for 48 h was
decreased compared with that at 0 mmHg, suggesting that there is an
optimal pressure range that favors liver cancer cell proliferation.
Excessive pressure or its prolonged application may constitute
unfavorable conditions for liver cancer cell survival, and only
under moderate pressure and exposure time (15 mmHg, 24 h) can these
cells thrive. For 3D-cultured liver cancer cells, the number of
cells in S phase following 24 h of 60 mmHg compression was higher
than that of cells grown with no compression. It is therefore
possible that the encapsulation of liver cancer cells in a 3D
matrix may protect them from damage caused by higher pressures,
and/or attenuate the pressure-induced inhibition of DNA
synthesis.
The wound-healing assay showed that, compared with
in the control group, the percentage of migratory cells was
significantly increased under 15 or 30 mmHg pressure for 24 h,
whereas 60 mmHg pressure did not affect cell migration. This
indicated that pressure may promote the migratory ability of liver
cancer cells at appropriate conditions (15 mmHg for 24 h). Notably,
the levels of cell mortality were unchanged under elevated pressure
for 12 and 24 h, but were increased following compression for 48 h.
As trypan blue can only stain cells with membrane defects, and the
membranes of cells undergoing apoptosis may be undamaged (24), it is therefore possible that high
pressure (30 and 60 mmHg) may also induce liver cancer cell
apoptosis, which may reduce the number of living cells able to
migrate during the wound-healing assay. Furthermore, it is possible
that excessive pressure may activate the expression of
migration-suppressing genes. The mechanisms underlying the
mechanically induced changes in cell migration require further
study.
Under the optimal conditions identified using the
aforementioned tests (15 mmHg, 24 h), the microarray analysis
identified five miRNAs that exhibited differences in expression;
these differences were also detected by RT-qPCR. Alterations in the
mRNA expression profile were more complex, which suggests that the
effect of pressure on liver cancer cells may affect various
physiological processes, including endocytosis, apoptosis and
metabolism. Following integrative analysis of miRNAs and mRNAs, the
1,309 target genes were functionally evaluated using GO analysis,
and these were mostly associated with cell proliferation and
migration.
In the KEGG pathway analysis of the 1,309
differentially expressed mRNAs, ‘PI3K/Akt signaling pathway’ and
‘focal adhesion’ were second and third of the top 20 most enriched
annotations, while the ‘integrin-mediated signaling pathway’ ranked
sixth in GO analysis annotations. There are several interactions
between these pathways that may explain the migration and invasion
of malignant cells (25,26). The expression levels of the key
genes of these three pathways, including SRC, FAK, PI3K and ITGAV,
were all significantly upregulated after 15 mmHg compression for 24
h. Src and ITGAV were the predicted target genes of downregulated
miR-5703 and miR-630, respectively. Previous biomechanical studies
have confirmed that Src, FAK, PI3K and ITGAV are vital proteins
involved in cell mechanical stress transduction. Thamilselvan and
Basson (4) reported that
extracellular pressure may increase integrin affinity and promote
colon cancer adhesion via actin-dependent inside-out FAK and Src
signals, which may regulate metastatic tumor cell adhesion
(27). In a separate study, it was
reported that the Src inhibitor PP2 could inhibit the
phosphorylation of PI3K and protein kinase B (Akt) in pressure
models of colon cancer, and that the application of LY294002 (a
PI3K inhibitor) inhibited the pressure-mediated adhesion process of
colon cancer cells. Accordingly, the Src-PI3K-FAK-Akt signaling
axis may respond to pressure in colon cancer cells (28). In addition, extracellular pressures
reaching 29 mmHg have been reported in rapidly growing breast
cancers (29). Downey et al
(30) observed breast cancer cells
under 15 mmHg and demonstrated that cancer cells display increased
adhesion, stimulated by phosphorylated (p)-FAK, which is the same
as for colon cancer. Genes that were responsive to pressure in the
pressure models of colon cancer and breast cancer may also function
in the pressure model of liver cancer. They may act as target genes
of pressure-responsive microRNAs, regulating the proliferation,
migration and invasion of pressure-treated liver cancer cells. The
phosphorylation of FAK, Src and PI3K may serve an important role in
regulating other downstream pathways, including the FOXO signaling
pathway, which ranked fifth in pathway analysis annotations. PI3K
can phosphorylate SGKs by activating pyruvate dehydrogenase kinase
1, and SGKs then deactivate FOXOs. Furthermore, p-FOXOs have been
reported to inhibit the expression of p27kip and p21, which prevent
cell cycle arrest and apoptosis (31). SOS1 (32) and IRS1 (33), which are upstream of FOXOs, may be
targeted by miRNA-630 and upregulated in pressure-exposed liver
cancer cells according to the microarray. SOS1 can activate Ras
(34), thereby phosphorylating
downstream genes and inhibiting the expression of FOXO. IRS1 may
act as an oncogene, as it has been reported to be involved in tumor
initiation and progression by regulating PI3K (35). Therefore, the FOXO signaling
pathway may be downregulated by a reduction in SRC expression
induced by mechanically responsive miR-5703, or by a reduction in
ITGAV, SOS1 and IRS1 expression via pressure-induced upregulation
of miR-630.
In addition to the downregulated miR-630 and
miR-5703, upregulated miR-7641 and miR-7-5p may also be associated
with the development of liver cancer in a high-pressure tumor
microenvironment. Downregulation of CSNK1E and SMAD2 was observed
in liver cancer cells exposed to 15 mmHg pressure for 24 h
according to the microarray analysis. CSNK1E is a predictive target
gene of miR-7-5p, and higher CSNK1E levels are associated with a
better prognosis in subsets of patients with breast cancer
(36). SMAD2 is a putative target
gene of miRNA-7641, and as an intracellular mediator of the
transforming growth factor β signal transduction pathway, it may
have a relevant role in hepatic fibro-carcinogenesis (37).
In summary, it was hypothesized that pressure
signals may act on cell membranes to deform them, which may lead to
activation of proteins on the surface of cell membranes, which then
transfer pressure stimulation to the cytoskeleton, organelles and
distal cell membranes. The activation of pressure-sensitive cell
membrane surface proteins may induce the tyrosine-mediated
phosphorylation of cytoskeletal proteins, including paxillin and
vinculin, and may trigger the upregulation or downregulation of
other signaling factors (such as PI3K, FAK, IRS1, SOS1, FOXOs,
CSNK1E and SMAD2). This process may therefore occur in liver cancer
cells under pressure, and mechanically responsive miRNAs may be
recruited to regulate the proliferation, migration and invasion of
liver cancer cells. Other miRNAs and mRNAs screened using
microarray analyses were identified in the present study, and these
will be evaluated in future studies regarding the function of
pressure in liver cancer.
HSCs are the primary producer of liver ECM
components. It has been reported that metastatic colorectal cancer
stem cells in the liver are capable of initiating the formation of
a metastatic niche by reprogramming HSCs into CAFs (38). A previous study also revealed that
pressure interventions at 10 mmHg for 1 h may significantly enhance
the proliferation, activation and migration of HSCs via miR-9a-5p,
which targets Sirt1 (13).
Regarding the effects of pressure and mechanically responsive
miRNAs, further experiments consisting of liver cancer cells, HSCs
and CAFs, in combination or separately, should be designed to
broaden our knowledge of liver cancer evolution and malignancy.
In conclusion, to the best of our knowledge, no
previous studies have demonstrated the effects of mechanical
pressure on liver cancer. The present study reported that pressure
induced an aggressive cancer phenotype, promoting proliferation and
migration of liver cancer cells. Potential pressure-responsive
miRNAs and mRNAs were identified by gene chips, which may provide a
large number of potential therapeutic targets for clinical
treatments, and revealed how intrahepatic pressure may promote
cancer development. However, the effect of these specific
pressure-responsive miRNAs and their targets should be verified in
further in vitro and in vivo experiments, and the
molecular mechanism underlying pressure-induced proliferation and
migration of liver cancer cells requires further study. Answering
these questions may be of significance for the treatment of liver
cancer with portal hypertension and may help reduce the levels of
intraoperative mechanical stimulation-induced metastasis of liver
cancer. Overall, the present study aimed to understand the fate of
liver cancer cells following mechanically induced miRNA expression,
and may provide a strong theoretical basis for the prevention and
treatment of liver cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 11472300).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus
repository, www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE119881
and www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120194.
The datasets used and/or analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
SS proposed the study. SS and XL performed the
research and wrote the first draft. SS, KWG and YCS collected and
analyzed the data. All authors contributed to the design and
interpretation of the study and to further drafts. LZ and DKY were
the guarantors, and made substantial contributions to conception
and design of the study, and the interpretation of data. LZ and DKY
revised the contents of the manuscript prior to submission, and
were responsible for all aspects of the research work.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Author information
LZ is a Professor in the Second Affiliated Hospital
of the Second Military Medical University, a member of the Shanghai
Biomechanics Professional Committee and a peer-reviewed expert of
the National Natural Science Foundation of China (NSFC). His main
research direction is the biomechanical study of digestive system
diseases and tumors, with emphasis on liver hemodynamics and
gastrointestinal dynamics. For many years, his team has studied the
hemodynamics of portal hypertension before and after TIPS
(Transjugular intrahepatic portosystemic shunt), and completed the
NSFC project entitled: ‘The role and mechanism of microRNAs in the
biomechanical response of hepatic stellate cells’. The research
reported by this manuscript is part of another NSFC project
entitled: ‘The role of microRNAs regulating the differentiation and
invasion of hepatoma in response to mechanical force’.
Glossary
Abbreviations
Abbreviations:
|
2D
|
2-dimensional
|
|
3D
|
3-dimensional
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
PI3K
|
phosphoinositide 3-kinase
|
|
FOXO
|
forkhead box O
|
|
ECM
|
extracellular matrix
|
|
CAFs
|
cancer-associated fibroblasts
|
|
HSCs
|
hepatic stellate cells
|
|
miRNA
|
microRNA
|
|
GO
|
Gene Ontology
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
CCK-8
|
Cell Counting kit-8
|
|
MMPs
|
matrix metalloproteinase
|
|
TIMPs
|
tissue inhibitor of
metalloproteinase
|
|
Src
|
SRC proto-oncogene, non-receptor
tyrosine kinase
|
|
Akt
|
protein kinase B
|
|
ITGAV
|
integrin subunit α V
|
|
p
|
phosphorylated
|
|
SGKs
|
serum/glucocorticoid regulated
kinases
|
|
SOS1
|
son of sevenless 1
|
|
IRS1
|
insulin receptor substrate 1
|
|
EMT
|
epithelial-mesenchymal transition
|
|
CSNK1E
|
casein kinase 1 ε
|
|
SMAD2
|
SMAD family member 2
|
|
CAMs
|
cell adhesion molecules
|
References
|
1
|
Omata M, Cheng AL, Kokudo N, Kudo M, Lee
JM, Jia J, Tateishi R, Han KH, Chawla YK, Shiina S, et al:
Asia-Pacific clinical practice guidelines on the management of
hepatocellular carcinoma: A 2017 update. Hepatol Int. 11:317–370.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kalli M and Stylianopoulos T: Defining the
role of solid stress and matrix stiffness in cancer cell
proliferation and metastasis. Front Oncol. 8:552018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jain RK, Martin JD and Stylianopoulos T:
The role of mechanical forces in tumor growth and therapy. Annu Rev
Biomed Eng. 16:321–346. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thamilselvan V and Basson MD: Pressure
activates colon cancer cell adhesion by inside-out focal adhesion
complex and actin cytoskeletal signaling. Gastroenterology.
126:8–18. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Craig DH, Owen CR, Conway WC, Walsh MF,
Downey C and Basson MD: Colchicine inhibits pressure-induced tumor
cell implantation within surgical wounds and enhances tumor-free
survival in mice. J Clin Invest. 118:3170–3180. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Basson MD, Zeng B, Downey C, Sirivelu MP
and Tepe JJ: Increased extracellular pressure stimulates tumor
proliferation by a mechanosensitive calcium channel and PKC-β. Mol
Oncol. 9:513–526. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fiering S, Ang LH, Lacoste J, Smith TD and
Griner E; Reproducibility Project, : Cancer Biology: Registered
report: Biomechanical remodeling of the microenvironment by stromal
caveolin-1 favors tumor invasion and metastasis. Elife.
4:e047962015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fernández-Sánchez ME, Barbier S, Whitehead
J, Béalle G, Michel A, Latorre-Ossa H, Rey C, Fouassier L, Claperon
A, Brullé L, et al: Mechanical induction of the tumorigenic
β-catenin pathway by tumour growth pressure. Nature. 523:92–95.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou J, Li YS, Nguyen P, Wang KC, Weiss A,
Kuo YC, Chiu JJ, Shyy JY and Chien S: Regulation of vascular smooth
muscle cell turnover by endothelial cell-secreted microRNA-126:
Role of shear stress. Circ Res. 113:40–51. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou J, Wang KC, Wu W, Subramaniam S, Shyy
JY, Chiu JJ, Li JY and Chien S: MicroRNA-21 targets peroxisome
proliferators-activated receptor-alpha in an autoregulatory loop to
modulate flow-induced endothelial inflammation. Proc Natl Acad Sci
USA. 108:10355–10360. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang KC, Garmire LX, Young A, Nguyen P,
Trinh A, Subramaniam S, Wang N, Shyy JY, Li YS and Chien S: Role of
microRNA-23b in flow-regulation of Rb phosphorylation and
endothelial cell growth. Proc Natl Acad Sci USA. 107:3234–3239.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qi F, Hu JF, Liu BH, Wu CQ, Yu HY, Yao DK
and Zhu L: MiR-9a-5p regulates proliferation and migration of
hepatic stellate cells under pressure through inhibition of Sirt1.
World J Gastroenterol. 21:9900–9915. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Seitz HK and Stickel F: Risk factors and
mechanisms of hepatocarcinogenesis with special emphasis on alcohol
and oxidative stress. Biol Chem. 387:349–360. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fattovich G, Stroffolini T, Zagni I and
Donato F: Hepatocellular carcinoma in cirrhosis: Incidence and risk
factors. Gastroenterology 127 (5 Suppl 1). S35–S50. 2004.
|
|
16
|
Iwakiri Y: Pathophysiology of portal
hypertension. Clin Liver Dis. 18:281–291. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Perelló A, Escorsell A, Bru C, Gilabert R,
Moitinho E, García-Pagán JC and Bosch J: Wedged hepatic venous
pressure adequately reflects portal pressure in hepatitis C
virus-related cirrhosis. Hepatology. 30:1393–1397. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Okada Y, Tsuzuki Y, Hokari R, Miyazaki J,
Matsuzaki K, Mataki N, Komoto S, Watanabe C, Kawaguchi A, Nagao S,
et al: Pressure loading and ethanol exposure differentially
modulate rat hepatic stellate cell activation. J Cell Physiol.
215:472–480. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Asaumi H, Watanabe S, Taguchi M, Tashiro M
and Otsuki M: Externally applied pressure activates pancreatic
stellate cells through the generation of intracellular reactive
oxygen species. Am J Physiol Gastrointest Liver Physiol.
293:G972–G978. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fleige S and Pfaffl MW: RNA integrity and
the effect on the real-time qRT-PCR performance. Mol Aspects Med.
27:126–139. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nagase H, Visse R and Murphy G: Structure
and function of matrix metalloproteinases and TIMPs. Cardiovasc
Res. 69:562–573. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Martini M, De Santis MC, Braccini L,
Gulluni F and Hirsch E: PI3K/AKT signaling pathway and cancer: An
updated review. Ann Med. 46:372–383. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kerr JF, Wyllie AH and Currie AR:
Apoptosis: A basic biological phenomenon with wide-ranging
implications in tissue kinetics. Br J Cancer. 26:239–257. 1972.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guan X: Cancer metastases: Challenges and
opportunities. Acta Pharm Sin B. 5:402–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kavic SM and Basson MD: Environmental
factors of temperature, humidity, serum accumulation and cell
seeding increase colon cancer cell adhesion in vitro, with partial
characterization of the serum component responsible for
pressure-stimulated. J Surg Res. 98:89–96. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thamilselvan V, Craig DH and Basson MD:
FAK association with multiple signal proteins mediates
pressure-induced colon cancer cell adhesion via a Src-dependent
PI3KAkt pathway. FASEB J. 21:1730–1741. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nathanson SD and Nelson L: Interstitial
fluid pressure in breast cancer, benign breast conditions and
breast parenchyma. Ann Surg Oncol. 1:333–338. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Downey C, Alwan K, Thamilselvan V, Zhang
L, Jiang Y, Rishi AK and Basson MD: Pressure stimulates breast
cancer cell adhesion independently of cell cycle and apoptosis
regulatory protein (CARP)-1 regulation of focal adhesion kinase. Am
J Surg. 192:631–635. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nakamura Y, Hibino K, Yanagida T and Sako
Y: Switching of the positive feedback for RAS activation by a
concerted function of SOS membrane association domains. Biophys
Physicobiol. 13:1–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li P, Tang T, Liu T, Zhou J, Cui H, He Z,
Zhong Y, Hu E, Yang A, Wei G, et al: Systematic Analysis of
tRNA-Derived Small RNAs reveals novel potential therapeutic targets
of traditional Chinese medicine (Buyang-Huanwu-Decoction) on
intracerebral hemorrhage. Int J Biol Sci. 15:895–908. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Papatheodorou I, Petrovs R and Thornton
JM: Comparison of the mammalian insulin signalling pathway to
invertebrates in the context of FOXO-mediated ageing.
Bioinformatics. 30:2999–3003. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu X, Cui CL, Chen WL, Fu ZY, Cui XY and
Gong X: miR-144 suppresses the growth and metastasis of laryngeal
squamous cell carcinoma by targeting IRS1. Am J Transl Res. 8:1–11.
2016.PubMed/NCBI
|
|
35
|
Coomans de Brachène A and Demoulin JB:
FOXO transcription factors in cancer development and therapy. Cell
Mol Life Sci. 73:1159–1172. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lopez-Guerra JL, Verdugo-Sivianes EM,
Otero-Albiol D, Vieites B, Ortiz-Gordillo MJ, De León JM,
Praena-Fernandez JM, Marin JJ and Carnero A: High CSNK1E levels are
correlated with better prognosis in subsets of patients with breast
cancer. Oncotarget. 6:30343–30356. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yoshida K, Murata M, Yamaguchi T and
Matsuzaki K: TGF-β/Smad signaling during hepatic
fibro-carcinogenesis (review). Int J Oncol. 45:1363–1371. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tauriello DV, Calon A, Lonardo E and
Batlle E: Determinants of metastatic competency in colorectal
cancer. Mol Oncol. 11:97–119. 2017. View Article : Google Scholar : PubMed/NCBI
|