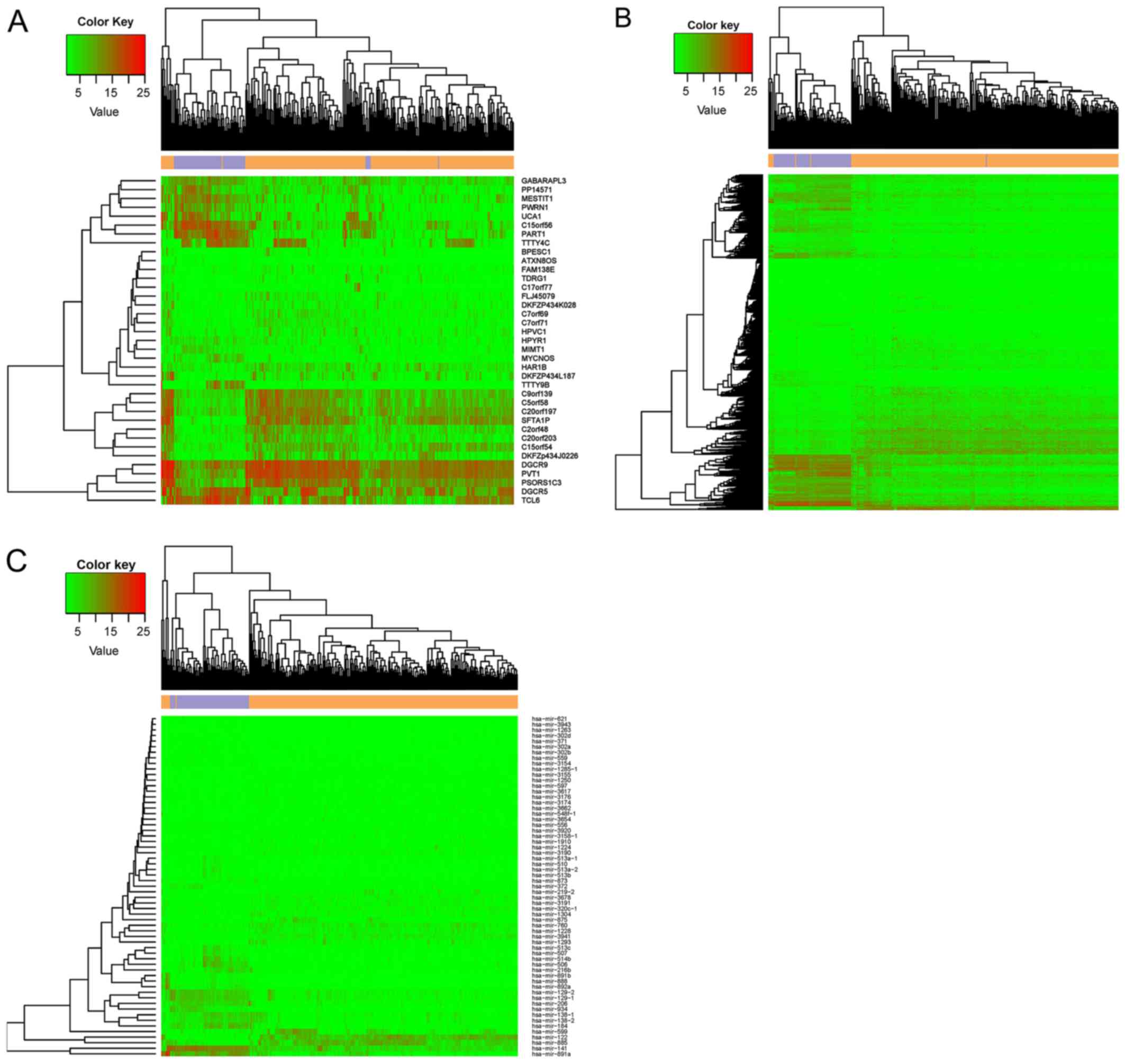
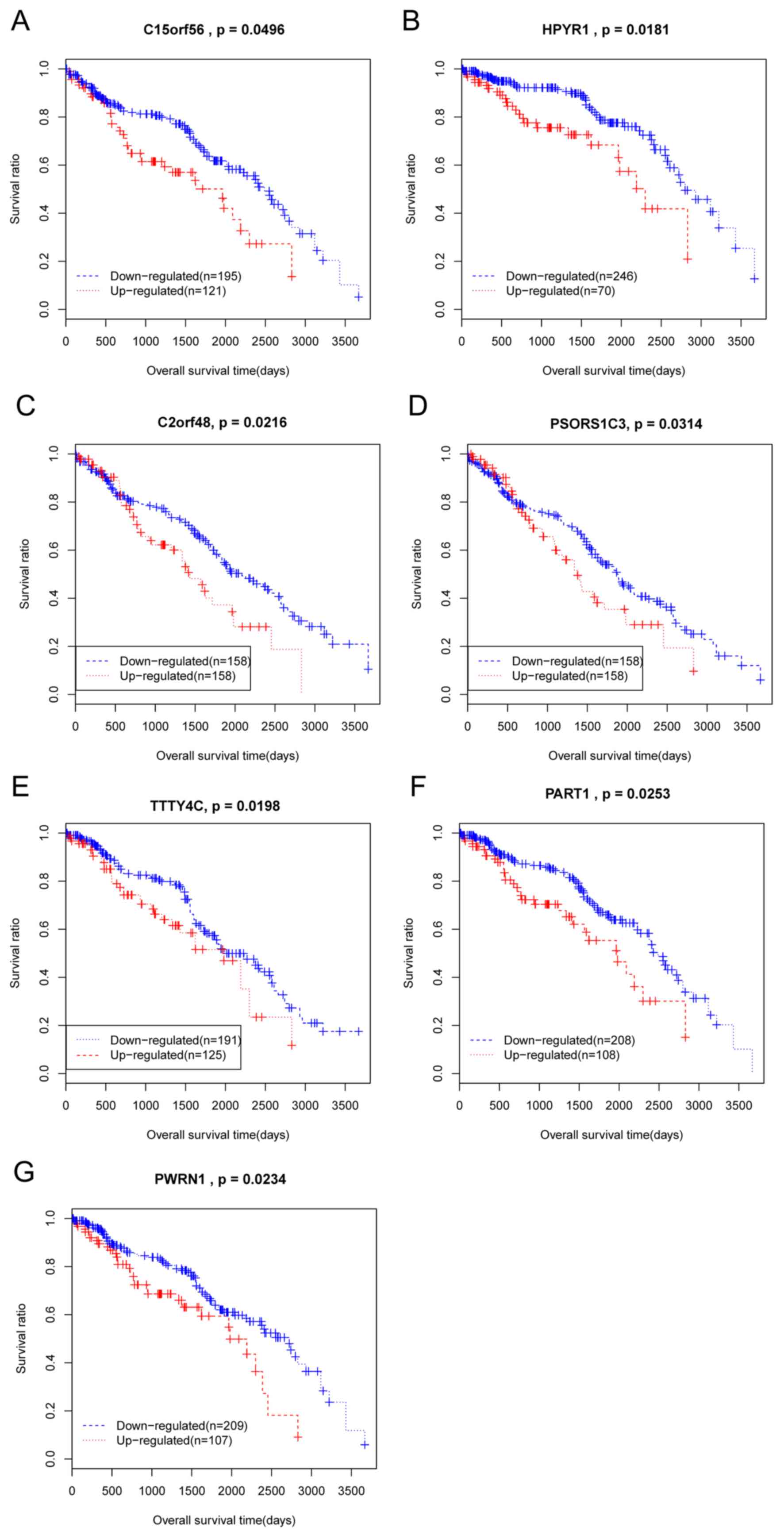
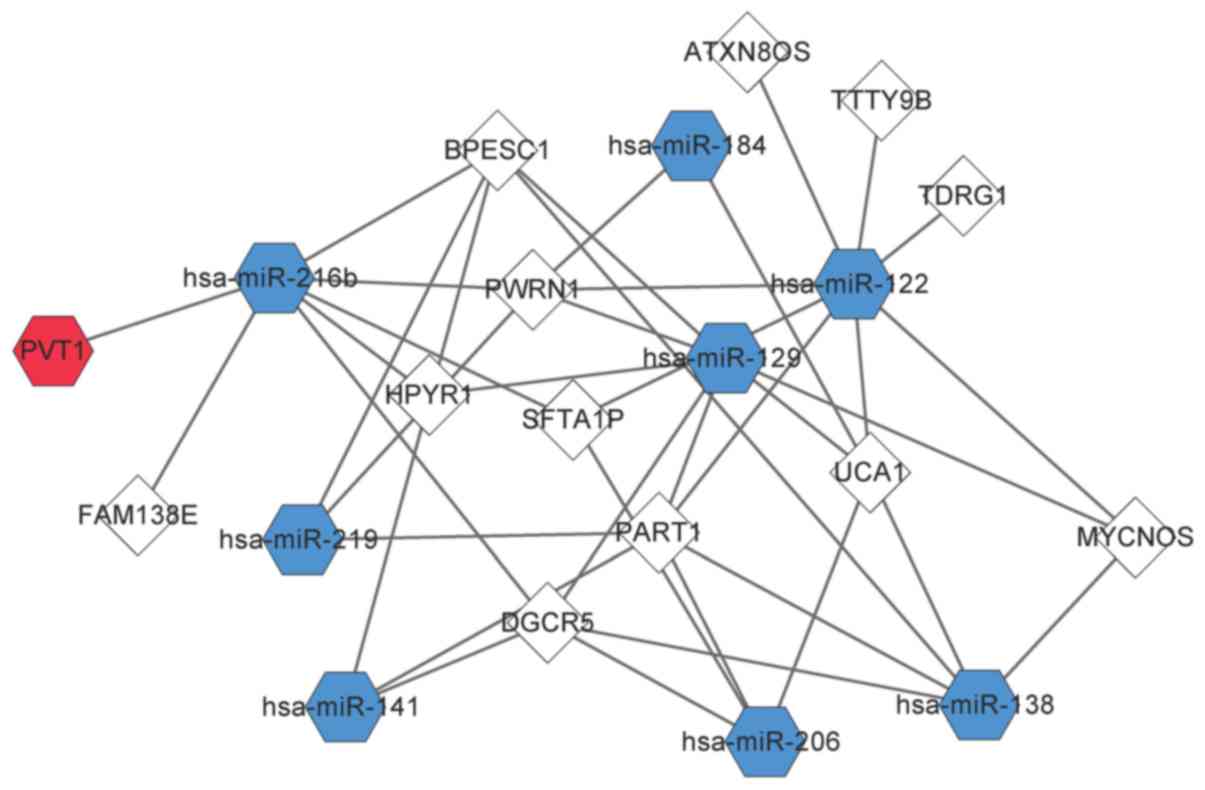
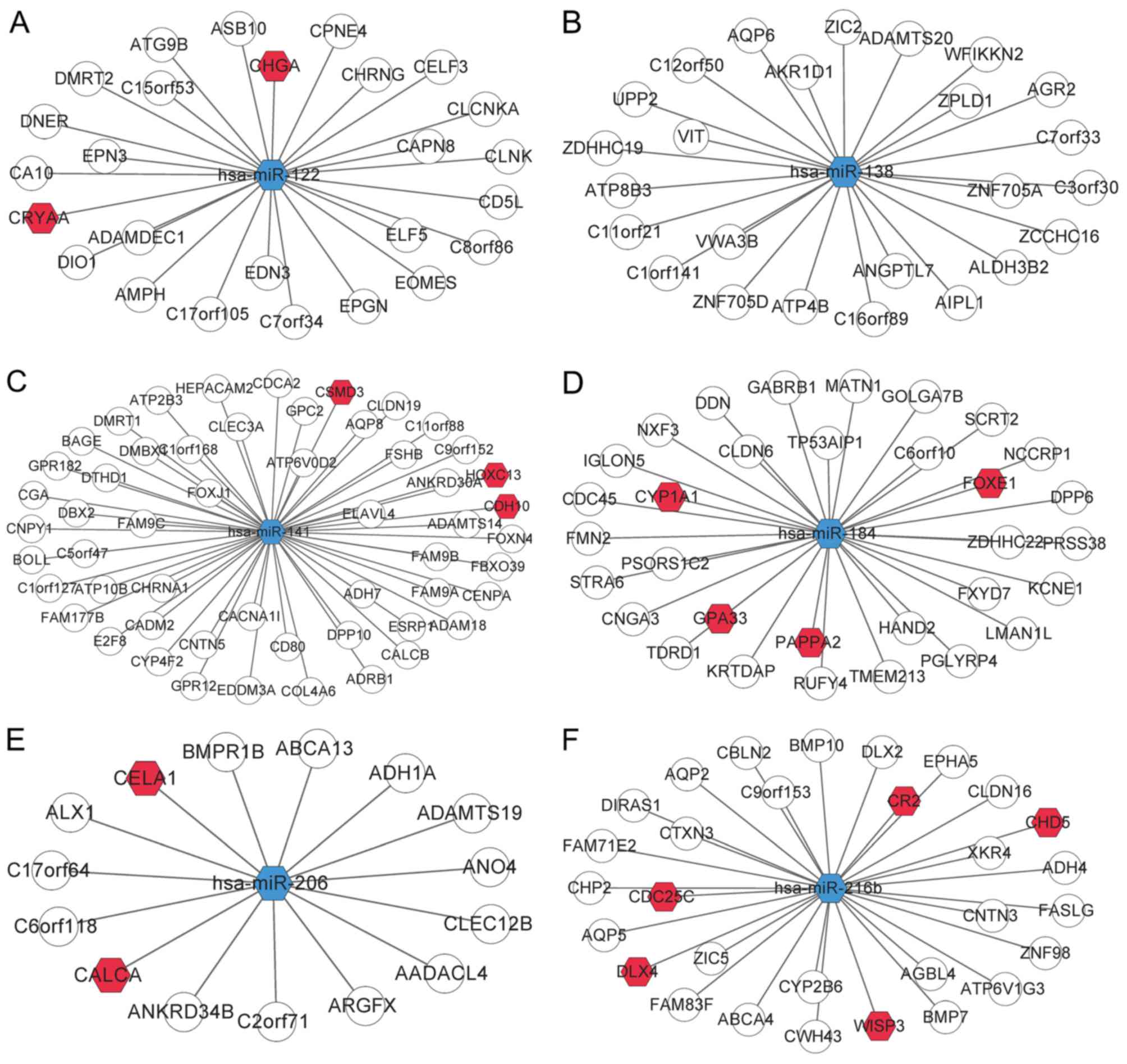
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gago-Dominguez M, Castelao JE, Yuan JM,
Ross RK and Yu MC: Increased risk of renal cell carcinoma
subsequent to hysterectomy. Cancer Epidemiol Biomarkers Prev.
8:999–1003. 1999.PubMed/NCBI
|
|
3
|
Linehan WM: Genetic basis of kidney
cancer: Role of genomics for the development of disease-based
therapeutics. Genome Res. 22:2089–2100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kwok ZH and Tay Y: Long noncoding RNAs:
Lincs between human health and disease. Biochem Soc Trans.
45:805–812. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Crea F, Watahiki A, Quagliata L, Xue H,
Pikor L, Parolia A, Wang Y, Lin D, Lam WL, Farrar WL, et al:
Identification of a long non-coding RNA as a novel biomarker and
potential therapeutic target for metastatic prostate cancer.
Oncotarget. 5:764–774. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ebert MS and Sharp PA: Emerging roles for
natural microRNA sponges. Curr Biol. 20:R858–R861. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Arvey A, Larsson E, Sander C, Leslie CS
and Marks DS: Target mRNA abundance dilutes microRNA and siRNA
activity. Mol Syst Biol. 6:3632010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M and Zatloukal K: Characterization of HULC, a novel
gene with striking up-regulation in hepatocellular carcinoma, as
noncoding RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bertozzi D, Iurlaro R, Sordet O, Marinello
J, Zaffaroni N and Capranico G: Characterization of novel antisense
HIF-1α transcripts in human cancers. Cell Cycle. 10:3189–3197.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Frevel MA, Sowerby SJ, Petersen GB and
Reeve AE: Methylation sequencing analysis refines the region of H19
epimutation in Wilms tumor. J Biol Chem. 274:29331–29340. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chiesa N, De Crescenzo A, Mishra K, Perone
L, Carella M, Palumbo O, Mussa A, Sparago A, Cerrato F, Russo S, et
al: The KCNQ1OT1 imprinting control region and non-coding RNA: New
properties derived from the study of Beckwith-Wiedemann syndrome
and Silver-Russell syndrome cases. Hum Mol Genet. 21:10–25. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cameron AC and Trivedi PK:
Regression-based tests for overdispersion in the Poisson model. J
Econ. 46:347–364. 1990. View Article : Google Scholar
|
|
21
|
Maritz JS and Lwin T: Empirical bayes
methods with applications (Chapman & Hall/CRC Monographs on
Statistics & Applied Probability). (2nd). 2018.
|
|
22
|
Solari A and Goeman JJ: Minimally adaptive
BH: A tiny but uniform improvement of the procedure of Benjamini
and Hochberg. Biom J. 59:776–780. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Koletsi D and Pandis N: Survival analysis,
part 2: Kaplan-Meier method and the log-rank test. Am J Orthod
Dentofacial Orthop. 152:569–571. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jeggari A, Marks DS and Larsson E:
miRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang W, Zhao E, Yu Y, Geng B, Zhang W and
Li X: MiR-216a exerts tumor-suppressing functions in renal cell
carcinoma by targeting TLR4. Am J Cancer Res. 8:476–488.
2018.PubMed/NCBI
|
|
27
|
Ricketts CJ and Linehan WM: Gender
specific mutation incidence and survival associations in clear cell
renal cell carcinoma (CCRCC). PLoS One. 10:e01402572015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen EG, Zhang JS, Xu S, Zhu XJ and Hu HH:
Long non-coding RNA DGCR5 is involved in the regulation of
proliferation, migration and invasion of lung cancer by targeting
miR-1180. Am J Cancer Res. 7:1463–1475. 2017.PubMed/NCBI
|
|
29
|
Dong HX, Wang R, Zeng J and Pan J: LncRNA
DGCR5 promotes lung adenocarcinoma (LUAD) progression via
inhibiting hsa-mir-22-3p. J Cell Physiol. 233:4126–4136. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao X, Li D, Pu J, Mei H, Yang D, Xiang
X, Qu H, Huang K, Zheng L and Tong Q: CTCF cooperates with
noncoding RNA MYCNOS to promote neuroblastoma progression through
facilitating MYCN expression. Oncogene. 35:3565–3576. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Malakootian M, Mirzadeh-Azad F, Naeli P,
Fouani Y and Mowla SJ: A long non-coding RNA, PSORS1C3, located
upstream of the human Oct4 gene is expressed in pluripotent and
tumor cell lines. Modares J Med Sci Pathobiol. 17:105–117.
2014.
|
|
32
|
Huang Y, Murakami T, Sano F, Kondo K,
Nakaigawa N, Kishida T, Kubota Y, Nagashima Y and Yao M: Expression
of aquaporin 1 in primary renal tumors: A prognostic indicator for
clear-cell renal cell carcinoma. Eur Urol. 56:690–698. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma J, Zhou C, Yang J, Ding X, Zhu Y and
Chen X: Expression of AQP6 and AQP8 in epithelial ovarian tumor. J
Mol Histol. 47:129–134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu F, Wu L, Wang A, Xu Y, Luo X, Liu X,
Hua Y, Zhang D, Wu S, Lin T, et al: MicroRNA-138 attenuates
epithelial-to-mesenchymal transition by targeting SOX4 in clear
cell renal cell carcinoma. Am J Transl Res. 9:3611–3622.
2017.PubMed/NCBI
|
|
35
|
Xiao H, Xiao W, Cao J, Li H, Guan W, Guo
X, Chen K, Zheng T, Ye Z, Wang J and Xu H: miR-206 functions as a
novel cell cycle regulator and tumor suppressor in clear-cell renal
cell carcinoma. Cancer Lett. 374:107–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fan Y, Ma X, Li H, Gao Y, Huang Q, Zhang
Y, Bao X, Du Q, Luo G, Liu K, et al: miR-122 promotes metastasis of
clear-cell renal cell carcinoma by downregulating Dicer. Int J
Cancer. 142:547–560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang J, Kong W, Zhang J, Chen Y, Xue W,
Liu D and Huang Y: c-Myc modulates glucose metabolism via
regulation of miR-184/PKM2 pathway in clear-cell renal cell
carcinoma. Int J Oncol. 49:1569–1575. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mitsui Y, Chang I, Fukuhara S, Hiraki M,
Arichi N, Yasumoto H, Hirata H, Yamamura S, Shahryari V, Deng G, et
al: CYP1B1 promotes tumorigenesis via altered expression of CDC20
and DAPK1 genes in renal cell carcinoma. BMC Cancer. 15:9422015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zaravinos A, Pieri M, Mourmouras N,
Anastasiadou N, Zouvani I, Delakas D and Deltas C: Altered
metabolic pathways in clear cell renal cell carcinoma: A
meta-analysis and validation study focused on the deregulated genes
and their associated networks. Oncoscience. 1:117–131. 2014.
View Article : Google Scholar : PubMed/NCBI
|