Introduction
The incidence and mortality of lung cancer are the
highest among all malignant tumours (1). Non-small cell lung cancer (NSCLC) is
the primary pathological type of lung cancer, and the majority of
tumours are lung adenocarcinomas (LAD). Although significant
progress has been made in the diagnosis and treatment of lung
cancer in recent years (2), the
prognosis of lung cancer remains poor, and the 5-year survival rate
is <16% (3). Therefore, it is
urgent that early diagnostic indicators and novel therapeutic
targets for lung cancer be actively sought.
With the rapid development of whole-genome
sequencing technology and bioinformatics methods, a large number of
long non-coding RNAs (lncRNAs) have been identified. Previous
studies have demonstrated that lncRNAs regulate gene expression at
multiple levels and participate in the progression of various
diseases (4–6). lncRNAs serve an important role in
tumour progression and participate in the regulation of tumour cell
proliferation, migration, invasion and differentiation (7,8).
These macromolecules may also be used as potential biomarkers and
therapeutic targets for a number of types of tumours (9), and their potential value in lung
cancer is gradually being revealed.
linc00467, which has 4 transcripts and a full length
of 3,508 bp, belongs to the group of long intergenic non-coding
RNAs and is located on lq32.3. At present, little is known about
linc00467. Previous studies on neuroblastoma have identified that
linc00467 may promote Wnt signalling pathway activation through a
transacting mechanism, thereby promoting the proliferation of
neuroblastoma (10). To the best
of our knowledge, there have been no studies on the association
between linc00467 and lung cancer at present.
Through RNA sequencing, HtrA serine peptidase 3
(HTRA3) was identified to be the downstream target gene of LAD in
the present study. HTRA3 belongs to the conserved HTRA family of
oligomeric serine proteases and has been demonstrated to inhibit
tumour cell proliferation, migration and invasion. Previous studies
have revealed that HTRA3 is downregulated in different types of
tumours (11), and its
upregulation may inhibit the growth of pancreatic cancer (12). In addition, a previous study
demonstrated that downregulation of HTRA3 promoted lymph node
metastasis in breast cancer (13).
However, little is known about HTRA3 and linc00467 in LAD.
The present study investigated the role of linc00467
in LAD. It was identified that linc00467 was markedly upregulated
in LAD tissues. Knockdown of linc00467 resulted in the decreased
proliferation, migration and invasion of LAD cells, while linc00467
overexpression increased LAD cell proliferation, migration and
invasion. The results of the present study elucidated the crucial
role of linc00467 in LAD progression and suggests that linc00467
may serve as a novel candidate target for LAD diagnosis and a
potential therapeutic target in LAD.
Materials and methods
Gene expression data sets
The GSE19804, GSE19188, GSE30219, GSE27262 data sets
from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) and The Cancer Genome
Atlas (TCGA) TANRIC database (http://ibl.mdanderson.org/tanric/_design/basic/query.html)
were analysed using bioinformatic methods. The gene expression
analyses were assessed using a linear model with Limma package in R
software (version 3.3.2) (14).
Differentially expressed genes were identified using the threshold
absolute fold change (FC) >1.5 and false discovery rate <0.05
as cut-offs.
RNA transcriptome sequencing
For RNA-Seq, transcript alignments were conducted by
HISAT2 (15) (version 2.0.5),
assembly and quantification were conducted by StringTie (16,17)
(version 1.3.3b), and differential expression analyses were
conducted based on the negative binomial distributions with edgeR
package in R software (version 3.18.1). Differential expression
genes were identified as the threshold |log2(FC)| >1 and FDR
<0.05. RNA transcriptome sequencing was performed by Novogene
Bioinformatics Technology Co. Ltd.
Tissue samples and clinical data
collection
A total of 35 paired LAD and normal lung tissues
were obtained from patients (10 females and 25 males) who underwent
primary surgical resection at the Jinling Clinical Medical College
of Nanjing Medical University (Nanjing, China) from January 2015 to
December 2016. The median age was 64 years and the range from 38 to
83 years. No treatments were conducted in these patients prior to
surgery, and all tissues were immersed in RNALater stabilization
solution (Qiagen GmbH) and stored at −80°C. All patients with LAD
were diagnosed according to histopathological evaluation, which was
based on the 2011 International Association for the Study of Lung
Cancer/American Thoracic Society/European Respiratory Society:
international multidisciplinary classification of lung
adenocarcinoma (18). The study
protocol and methods were approved by the Research Ethics Committee
of Jinling Clinical Medical College of Nanjing Medical University.
All of the participants provided written informed consent form and
agreed to the use of their samples in scientific research.
Cell lines
A total of 3 LAD H1299, A549, PC9 and 16HBE cell
lines were purchased from the Shanghai Institute of Biochemistry
and Cell Biology, Chinese Academy of Sciences. H1299 and A549 cells
were maintained in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.). PC9 and 16HBE cells were cultured in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.).
Both media were supplemented with 10% foetal bovine serum (FBS,
HyClone; GE Healthcare Life Sciences). All of the cells were grown
in a humidified atmosphere at 37°C with 5% CO2.
RNA isolation and reverse
transcription quantitative polymerase chain reaction (RT-qPCR)
analyses
Total RNA was isolated from cultured LAD cells or
frozen tissue samples using TRIzol® reagent (Thermo
Fisher Scientific, Inc.). cDNA was synthesized using a Reverse
Transcription kit (Takara Biotechnology Co., Ltd.). RT-qPCR assays
were performed using a SYBR Premix Ex Taq II kit (Takara
Biotechnology Co., Ltd.), and the target gene expression values
were normalized to the expression of GAPDH. The results were
analysed based on the comparative cycle threshold (Cq)
(2−ΔCq or 2−ΔΔCq) methods (19). The primer sequences are listed in
Table SI.
Transfection of LAD cells
Small interfering RNAs (siRNAs; si-linc00467 1#,
si-linc00467 2# and si-linc00467 3#) and plasmid vectors (pcDNA3.1
and pcDNA3.1-linc00467) were designed and synthesized by Shanghai
GenePharma Co., Ltd., and the empty pcDNA3.1 vector was used as the
control. The concentrations of siRNA and plasmids were 50 µM and 2
µg/ml, respectively. siRNAs were transfected into cells using
HiPerFect Transfection Reagent (Qiagen GmbH). The plasmid vectors
were transfected into cells using X-tremeGENE HP DNA transfection
reagent (Roche Diagnostics). After 48 h of incubation in a
humidified atmosphere at 37°C with 5% CO2, the cells
were harvested for subsequent experiments. Short hairpin RNA
(shRNA) lentiviruses (linc00467 shRNA, control shRNA) were also
designed and synthesized by Shanghai GenePharma Co., Ltd. The LV3
lentiviral vector (Shanghai GenePharma Co., Ltd.) to express shRNA
via the H1 promoter. Synthetic oligonucleotide primers [forward
(5′-GATCCGCTGGCAAATATGAAGGTATTCAAGAGATACCTTCATATTTGCCAGCTTTTTTG-3′)
and reverse
(5′AATTCAAAAAAGCTGGCAAATATGAAGGTATCTCTTGAATACCTTCATATTTGCCAGCG-3′)],
and the shRNA sequence expressed by the vector were sheared to form
a target sequence 5′-GCTGGCAAATATGAAGGTA-3′. A fragment targeting
linc00467 (5′-GCTGGCAAATATGAAGGTA-3′) inserted into a lentiviral
vector was used as linc00467-shRNA, and a non-targeting fragment
(5′-TTCTCCGAACGTGTCACGT-3′) inserted into a lentiviral vector was
used as control-shRNA. The shRNA lentiviruses were transfected into
cells in the presence of 2 µg/ml Polybrene, and
linc00467-shRNA-transfected cells were screened with 1, 2, 4 or 8
µg/ml puromycin for 8 days post-transfection. siRNA was used in the
preliminary functional experiments, whereas the experiments
investigating the mechanisms of action required longer culture
times. Therefore, shRNA was used in the later mechanism studies.
The primer sequences are listed in Table SII.
Nuclear and cytoplasmic RNA
isolation
Cytoplasmic and nuclear RNA were separated and
purified using a PARIS kit (Thermo Fisher Scientific, Inc.),
following the manufacturers' protocol. The buffer was pre-cooled
using ice and added to the cultured cells; lysate was then
collected in an enzyme-free Eppendorf (EP) tube, and cytoplasmic
lysate was collected by centrifugation at 10,000 × g for 1 min at
room temperature into an enzyme-free EP tube. Cell disruption
buffer was then added, and the nuclear lysate was collected by
centrifugation at 10,000 × g for 1 min at room temperature into an
enzyme-free EP tube. An equal volume of 2X lysis/binding solution
(Invitrogen; Thermo Fisher Scientific, Inc.) and absolute ethanol
were added into each tube, following which the mixture was
transferred into an enzyme-free EP tube and centrifuged at 10,000 ×
g for 1 min at room temperature. The liquid flow-through was then
discarded, 3 wash steps were performed and 95–100°C Elution
Solution (Qiagen GmbH). was added to obtain RNA. GAPDH and U6 were
used as internal controls, and RT-qPCR was used to measure the
expression of linc00467 in the nucleus and cytoplasm.
Cell proliferation assays
An MTT assay was used to measure cell proliferation
using a Cell Proliferation Reagent kit I (Roche Diagnostics). After
48 h of transfection, the cells were plated on 96-well plates at a
density of 3,000 cells per well. The 96-well culture plates were
taken at 0, 24, 48, 72 and 96 h, respectively, and proliferation
was detected at the corresponding time points. MTT (20 µl) was
added into each well, and the cells were incubated for 4 h in a
humidified atmosphere containing 5% CO2 at 37°C. The
reaction was then terminated by removal of the supernatant, and 150
µl dimethyl sulfoxide (DMSO) solution was added to dissolve the
purple formazan crystals. The plate was shaken for 20 min to
promote the dissolution of the crystals. The absorbance was
measured at a wavelength of 490 nm using a microplate reader, and
cell proliferation was analysed based on the absorbance value. The
clonogenic ability of the LAD cells was examined using the colony
formation method. The transfected cells were digested with 0.25%
trypsin and thoroughly pipetted into a single-cell suspension.
Following cell counting, the cells were densely seeded (1,000
cells/well) in a 6-well culture plate, evenly dispersed and plated
at 37°C. After 4 days, the supernatant was discarded and replaced
with new complete medium. When the colonies were visible to the
naked eye, the culture was terminated. The supernatant was
aspirated, the plates were carefully rinsed with PBS, and cells
were fixed with 4% paraformaldehyde for 15 min at room temperature.
Subsequent to removal of the fixative, 1 ml 0.1% crystal violet
stain solution was added into each well for 15 min at room
temperature, following which the stain was discarded. The remaining
stain was washed off with PBS, and the plates were dried at room
temperature. The number of colonies >10 cells was counted using
an optical light microscope at ×100 magnification (Olympus, Tokyo,
Japan), and statistical analyses were performed.
Flow cytometry (FCM) apoptosis
assay
The cells were densely seeded (2×105
cells/well) in a 6-well culture plate. H1299, A549 and PC9 cells
transfected with si-NC/si-linc00467 or empty
vector/pcDNA3.1-linc00467 were collected 48 h after transfection.
The cells were diluted with the cell binding buffer (BD Pharmingen;
BD Biosciences) and stained with Annexin V-allophycocyanin (APC)
and 7 amino-actinomycin (7-AAD) in the dark at room temperature for
15 min, prior to being analysed with a flow cytometer
(FACScan®; BD Biosciences). A tube containing only
Annexin V-APC and 7-AAD was used as the negative control. The data
were analysed using CellQuest software (version 6.0 (BD
Biosciences).
Transwell assay
Transwell chambers (EMD Millipore) were used for
cell migration and invasion assays. For the migration assay,
3×104 cells in 300 µl serum-free medium were seeded into
the top chamber of each insert, and 700 µl medium supplemented with
10% FBS was added into the lower chamber. For the invasion assay,
the upper chamber membrane was first uniformly coated with Matrigel
(BD Biosciences) at 37°C for 24 h. Then, 1×105 cells in
300 µl serum-free medium were placed into the top chamber of each
insert, and 700 µl medium supplemented with 10% FBS was added into
the lower chamber. Following incubation of the A549 and H1299 cells
at 37°C for 48 h, and the PC9 cells were incubated at 37°C for 24
h, the cells in the top chamber were removed, and cells that had
migrated or invaded through the membrane were stained for 15 min
with 0.1% crystal violet at 37°C. The stained cells were counted
under a light microscope (magnification, ×100; Olympus
Corporation).
RNA-protein interaction
prediction
The probability of an interaction between linc00467
and RBPs was determined using an lncRNA prediction website
(http://annolnc.cbi.pku.edu.cn/index.jsp). The sequence
of linc00467 was entered into the AnnoLnc database and into the
protein interaction page; from this, the interaction score between
lncRNA and protein could be predicted.
Western blot analysis
Cells were collected, washed twice with ice-cold PBS
and lysed in 100 µl RIPA buffer (Servicebio) supplemented with
protease inhibitor (Sigma-Aldrich; Merck KGaA) for 30 min, and a
bicinchoninic protein assay kit (CWBIO Corporation) was used to
detect the protein concentration. Total protein samples (20 µg)
were electrophoresed on 10% SDS-PAGE and transferred onto
polyvinylidene fluoride membranes (EMD Millipore). Following
blocking with 5% milk for 1 h at room temperature, the membranes
were incubated overnight at 4°C with primary antibody EZH2
(1:1,000; ab191250; Abcam) and HTRA3 (1:1,000; ab227463; Abcam);
GAPDH was used as the loading control (1:3,000; ab181602; Abcam).
The blots were extensively washed five times with TBST (0.5%
Tween-20) and incubated with horseradish peroxidase-conjugated goat
anti-rabbit secondary antibody (1:3,000; ab150081; Abcam) for 1 h
at room temperature. Following washing, an ECL chromogenic
substrate and densitometry using Quantity One software (version
4.6.9, Bio-Rad Laboratories, Inc.) were used to measure the
blots.
RNA immunoprecipitation (RIP)
assay
The RNA immunoprecipitation (RIP) assay was
performed using a Magna RIP RNA-Binding Protein Immunoprecipitation
kit (17–701; EMD Millipore) in accordance with the manufacturer's
protocol. Whole-cell extract from A549 and H1299 cells was used for
immunoprecipitations by using RIP lysis buffer (CS203176; EMD
Millipore). For each immunoprecipitation reaction, 50 µl protein A
Sepharose beads and 5 µg antibody against histone-lysine
N-methyltransferase EZH2 (ab191250; Abcam) were used.
Immunoprecipitation was performed using a relevant antibody to the
DNase-treated extract and incubating at 4°C overnight. Then samples
were sequentially treated with Proteinase K and RQ1 RNase-free
DNase to digest protein and genomic DNA, respectively. Purified RNA
samples were re-suspended in water and stored at 80°C. Finally, the
purified RNA was used for RT-qPCR.
Chromatin immunoprecipitation (ChIP)
assay
A549 and H1299 cells were cross-linked with 1%
formaldehyde for 10 min at room temperature. Crosslinking was
halted by addition of 125 mM glycine. Pellets were washed twice
with complete proteinase inhibitor cocktail (Roche Diagnostics) in
PBS, and then resuspended in 400 µl SDS lysis buffer
(Sigma-Aldrich; Merck KGaA) for 30 min. Then, 2% of supernatant was
saved as input and 100 µl of supernatant was incubated with 5 µg
EZH2 (ab191250; Abcam) or IgG (ab172730; Abcam) antibodies for 16
hours at 4°C. Then 30 µl of protein G magnetic beads (Thermo Fisher
Scientific, Inc.) was added to each IP reaction and incubated for 2
h at 4°C with rotation. Immunoprecipitates were then processed with
a series of washes and the cross-linking reversed. The DNA was
purified by phenol/chloroform/isoamyl extraction and RT-qPCR was
performed as aforementioned. The sequences of the ChIP primers for
the HTRA3 promoter region are listed in Table SI.
Statistical analysis
All of the statistical analyses were performed using
SPSS 18.0 software (SPSS, Inc.). P<0.05 was considered to
indicate a statistically significant difference, and P<0.01 was
considered to indicate a highly statistically significant
difference. Data are presented as mean ± standard deviation. A
two-tailed Student's t-test was applied to compare differences
between two groups. The comparison of multiple groups were
performed using one-way analysis of variance, and the post-hoc
multiple comparisons were performed using the Least Significant
Difference test.
Results
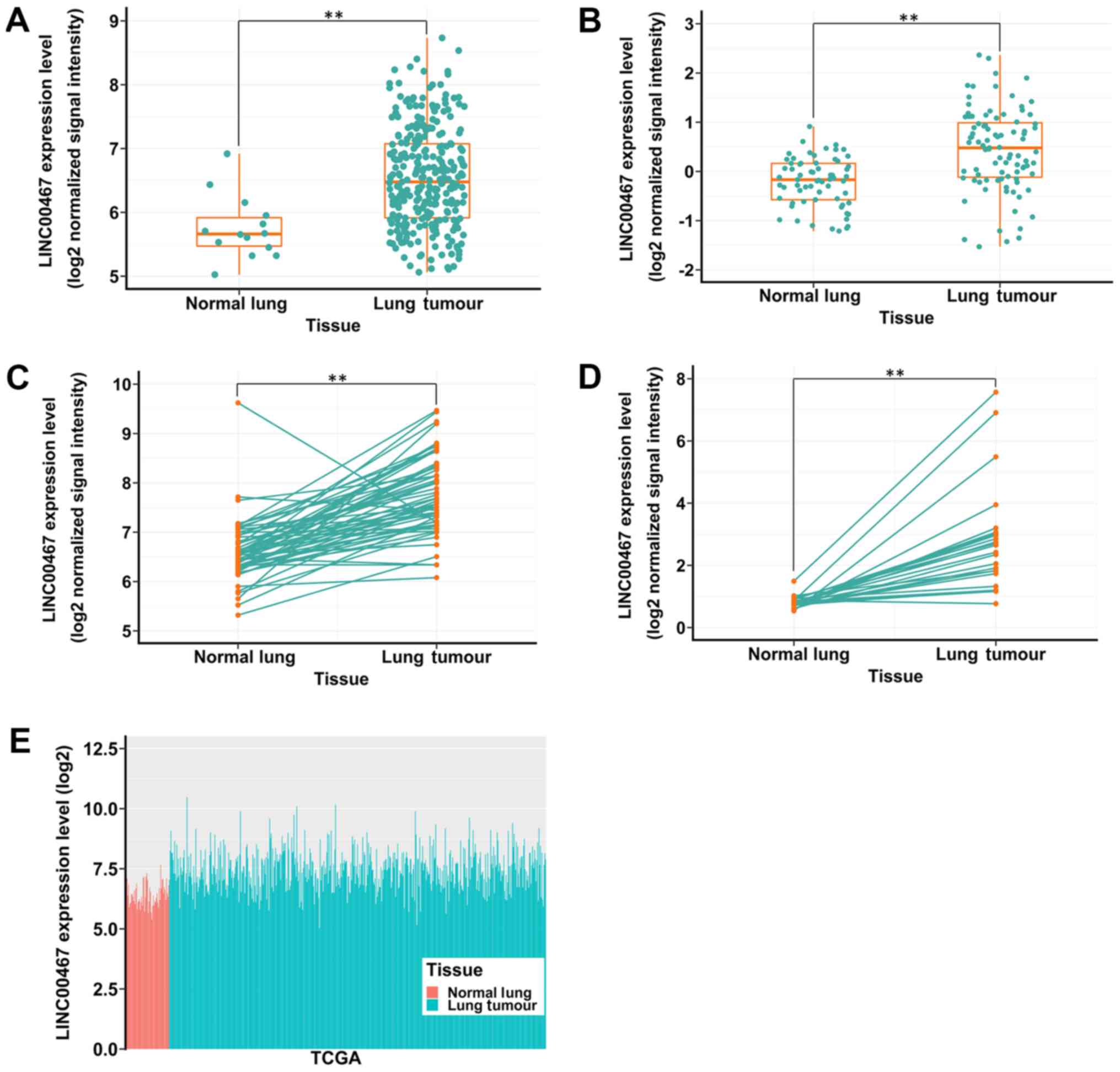
Expression profile of linc00467 in LAD
tissues from GEO and TCGA datasets
A total of 4 microarray datasets (GSE19804,
GSE19188, GSE30219 and GSE27262) obtained from the GEO were used to
analyse linc00467 expression in lung cancer tissues and normal
tissues. It was identified that linc00467 was upregulated in the 4
data sets (Fig. 1A-D). In
addition, TCGA data also indicated that linc00467 expression was
upregulated in lung cancer tissues compared with normal tissues
(Fig. 1E).
linc00467 is upregulated in LAD
tissues and is associated with the clinical characteristics of
LAD
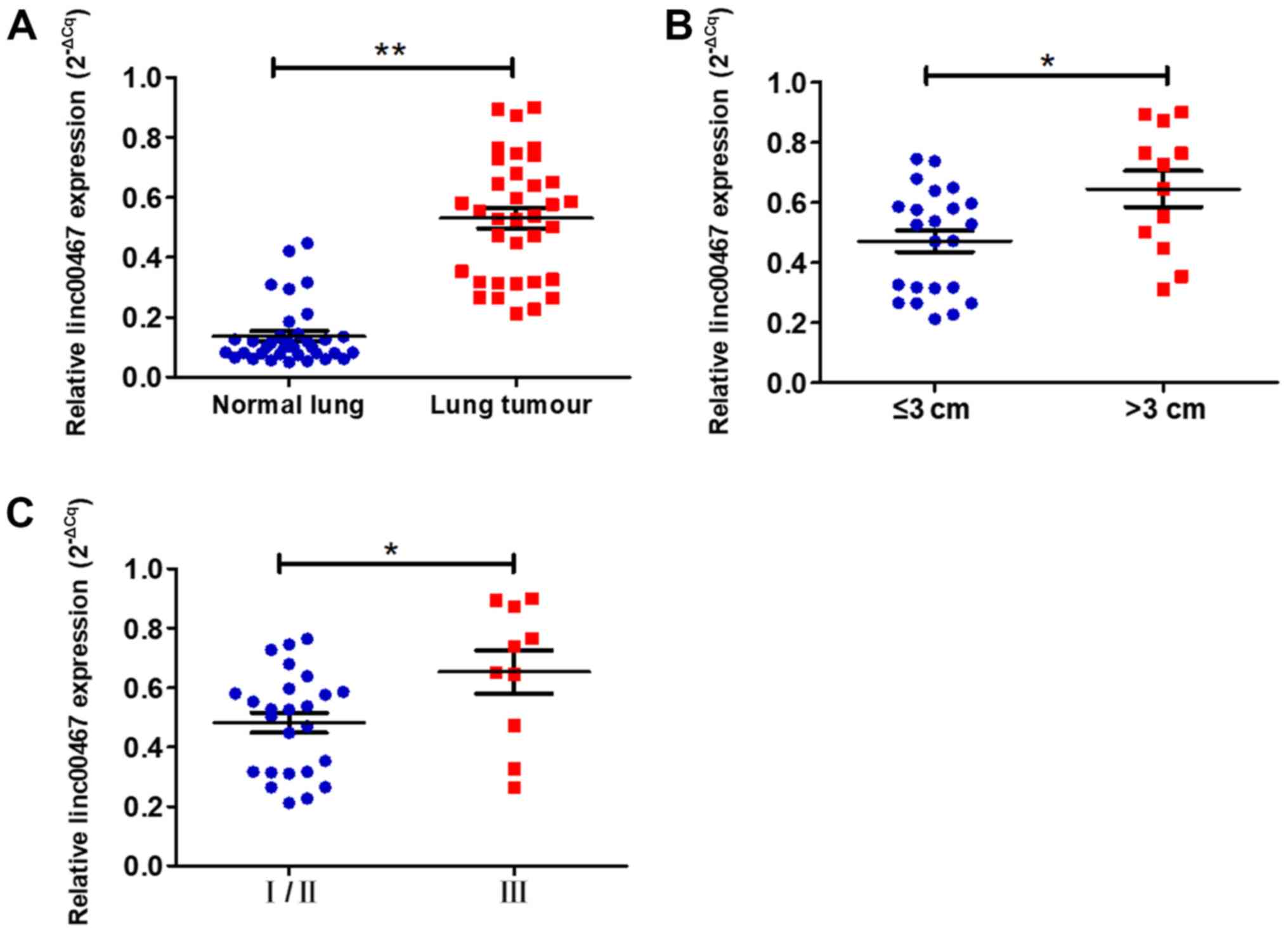
To validate the GEO and TCGA results, RT-qPCR assays
were used to measure linc00467 expression in 35 paired LAD and
adjacent normal lung tissues. The relative expression of lncRNA was
calculated using the Cq value (2−ΔCq) method. The
expression level of linc00467 in LAD tissues was increased compared
with that in adjacent normal lung tissues (P<0.001; Fig. 2A). Then, the association between
linc00467 expression and patients' clinical characteristics were
evaluated (Table I). The linc00467
expression levels were significantly increased in patients with
larger tumours compared with smaller tumours (P=0.013; Fig. 2B), and in those with more advanced
TNM stages (P=0.021; Fig. 2C).
However, there were no significant associations between linc00467
expression and other clinical parameters, including age, sex,
differentiation, lymph node metastasis and smoking history.
 | Table I.Associations between linc00467
expression and clinicopathological parameters of lung
adenocarcinoma. |
Table I.
Associations between linc00467
expression and clinicopathological parameters of lung
adenocarcinoma.
|
|
| linc00467
expression (2−ΔCq) |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
parameters | Number of
patients | Median ± standard
deviation | P-value |
|---|
| Sex |
|
| 0.963 |
|
Male | 25 | 0.530±0.205 |
|
|
Female | 10 | 0.533±0.202 |
|
| Age, years |
|
| 0.601 |
|
≤65 | 23 | 0.544±0.120 |
|
|
>65 | 12 | 0.506±0.210 |
|
| Size of tumour,
cm |
|
| 0.013 |
| ≤3 | 23 | 0.471±0.173 |
|
|
>3 | 12 | 0.645±0.209 |
|
|
Differentiation |
|
| 0.637 |
|
Well/moderate | 24 | 0.520±0.199 |
|
|
Poor | 11 | 0.555±0.213 |
|
| Lymph node
metastasis (pN) |
|
| 0.253 |
| N0 | 16 | 0.490±0.117 |
|
|
N1-N3 | 19 | 0.565±0.250 |
|
| TNM stage |
|
| 0.021 |
|
I/II | 25 | 0.482±0.170 |
|
|
III | 10 | 0.653±0.230 |
|
| Smoking |
|
| 0.467 |
|
Yes | 22 | 0.550±0.218 |
|
| No | 13 | 0.498±0.173 |
|
linc00467 promotes LAD cell
proliferation by inhibiting apoptosis in vitro
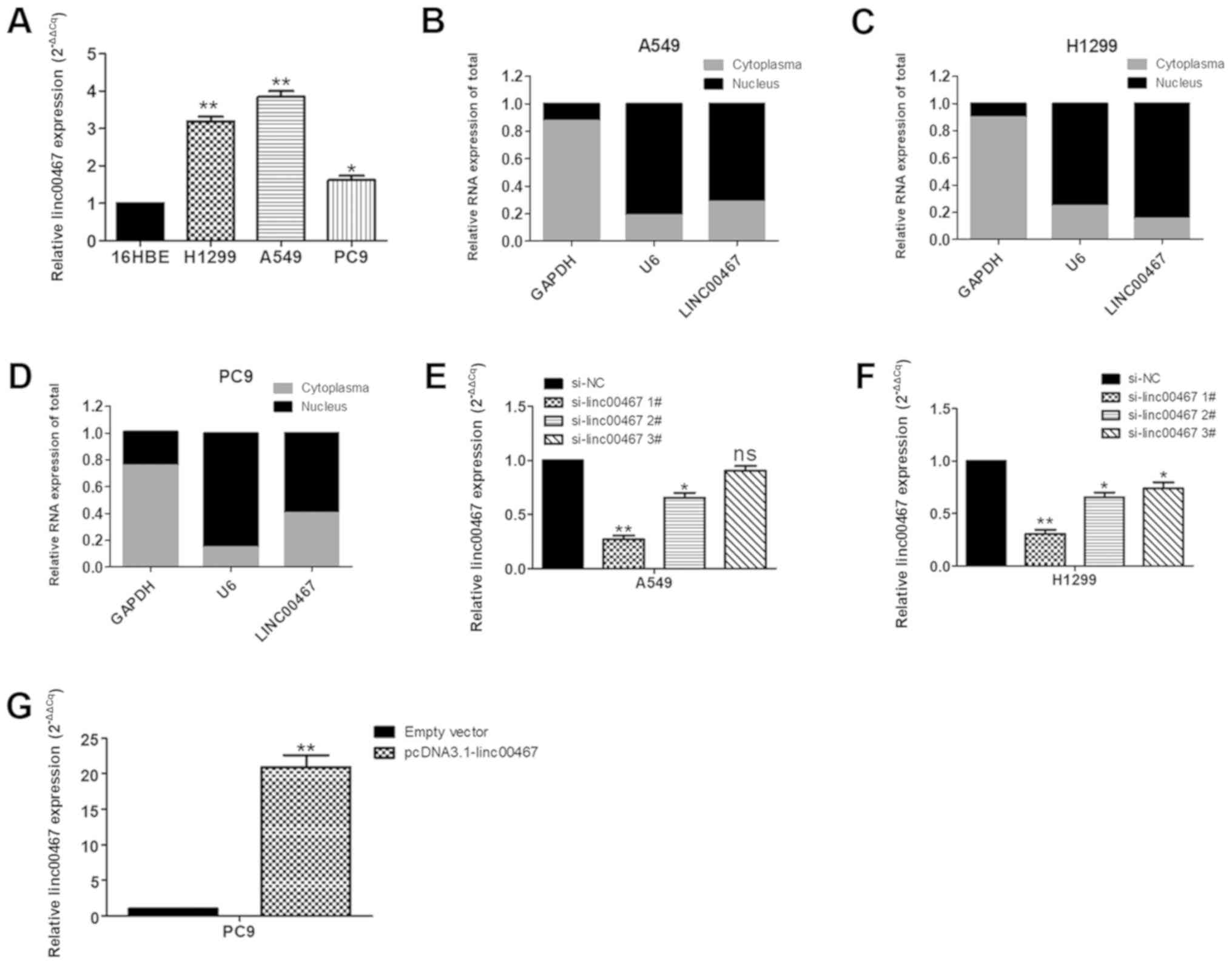
To investigate the function of linc00467 in LAD
cells, its expression levels in the human bronchial epithelial
16HBE cell line and in 3 human LAD A549, PC9 and H1299 cell lines
were assessed using RT-qPCR. A549 and H1299 cells expressed
increased levels of linc00467 compared with 16HBE cells, while the
PC9 cells expressed relatively decreased linc00467 levels compared
with A549 and H1299 cells (Fig.
3A). Therefore, linc00467 was knocked down with siRNA in the
A549 and H1299 cell lines and overexpressed in PC9 cell lines. A
localization assay for linc00467 in cells indicated that linc00467
was localized in the nucleus and cytosol but was localized to a
greater extent in the nucleus (Fig.
3B-D). The results of the linc00467 knockdown assay in the A549
and H1299 cells by transfection with siRNAs are presented in
Fig. 3E and F; ‘si-linc00467 1#’
exhibited the best knockdown efficiency, so it was used for
subsequent functional experiments. linc00467 expression was
upregulated in PC9 cells by transfection with pcDNA3.1-linc00467
(Fig. 3G).
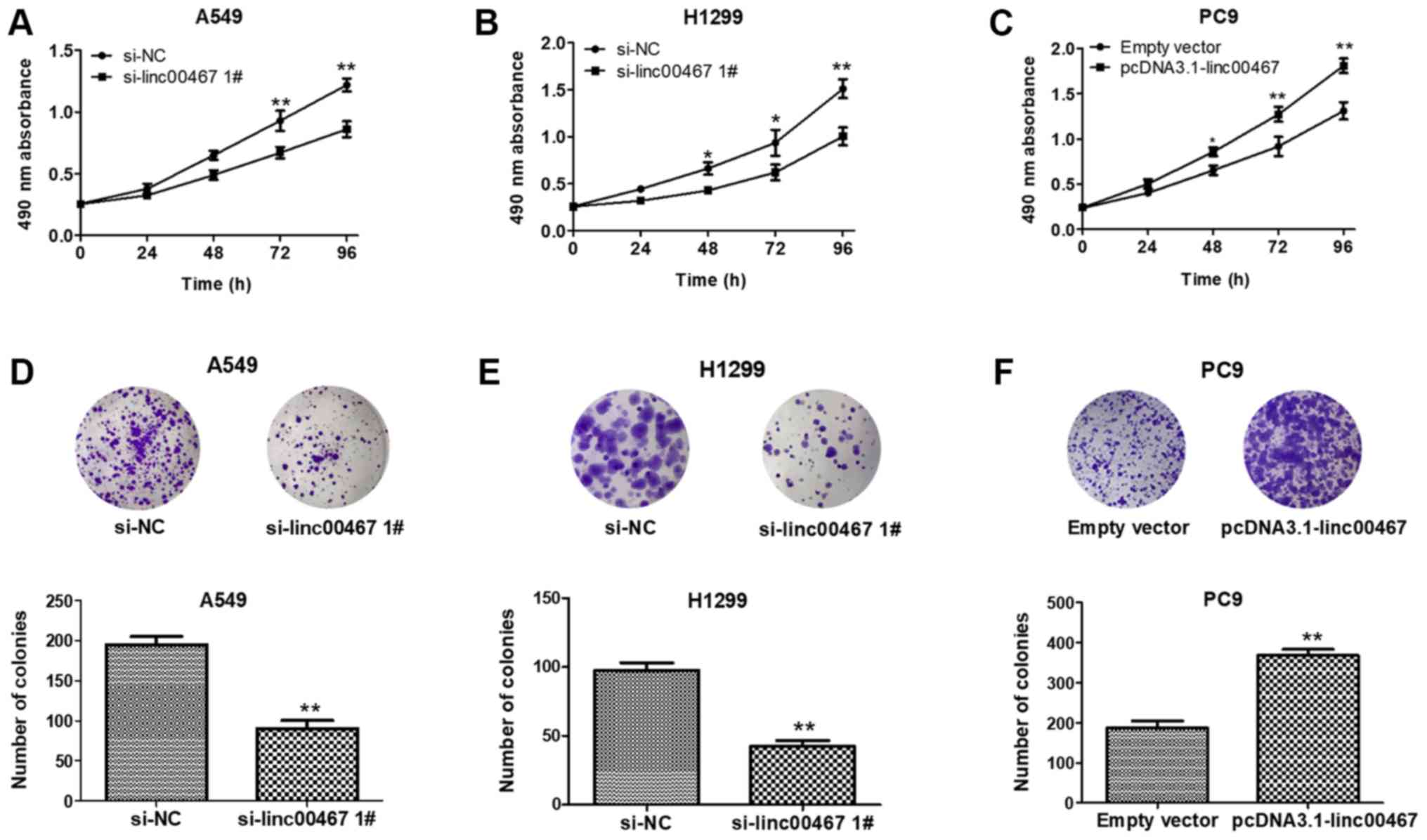
The role of linc00467 in LAD development was next
examined. linc00467 was knocked down in A549 and H1299 by
transfection with siRNAs and overexpressed in PC9 with
pcDNA3.1-linc00467. The MTT assays indicated that A549 and H1299
cell viability was decreased following knockdown of linc00467
(Fig. 4A and B), while cell
viability was increased in pcDNA3.1-linc00467-treated PC9 cells
(Fig. 4C). The colony formation
ability of A549 and H1299 cells was consistently and significantly
impaired following linc00467knockdown, while linc00467
overexpression increased PC9 cell colony formation ability
(Fig. 4D-F). Overall, linc00467
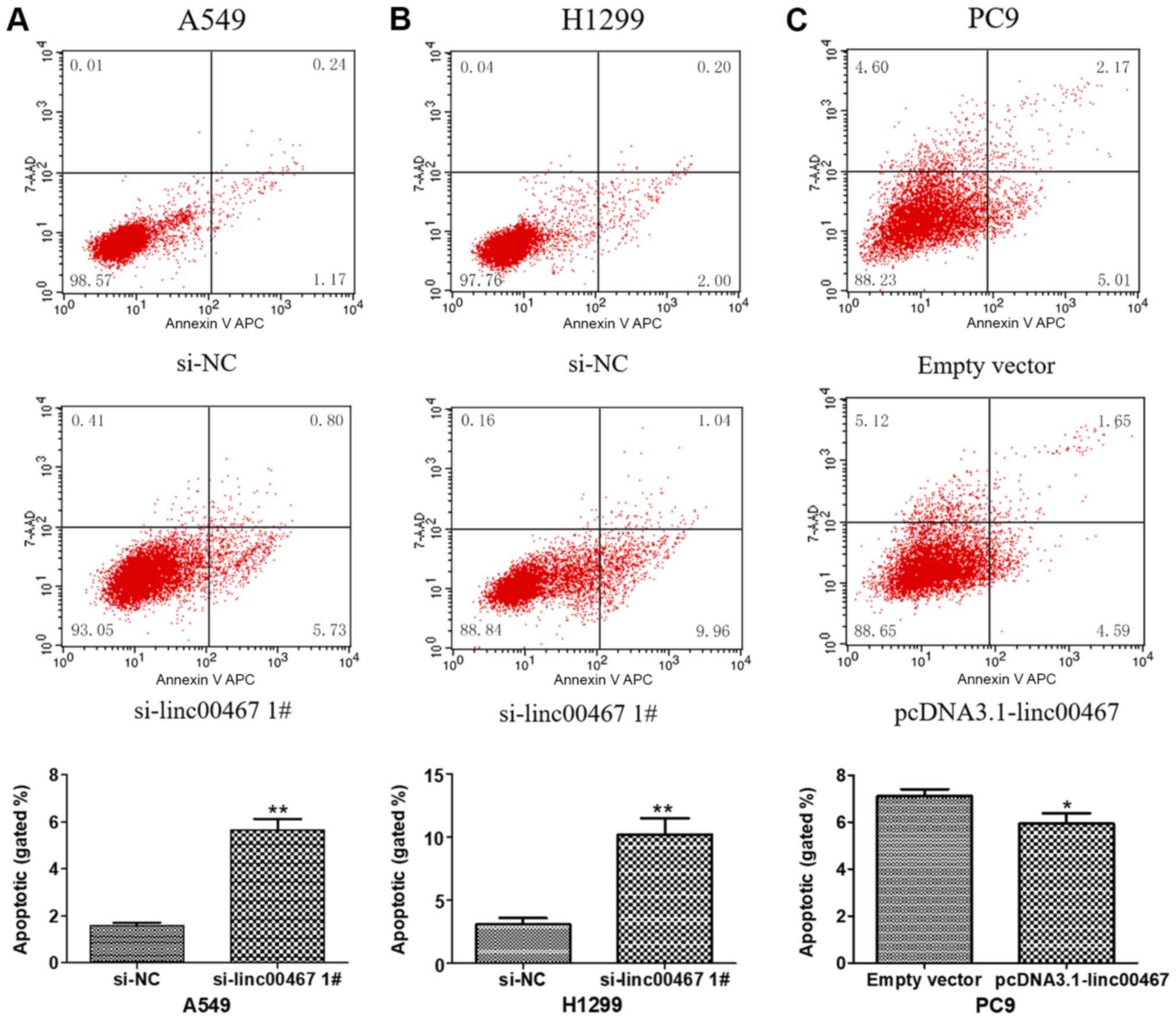
served as an oncogene to promote LAD cell proliferation. Flow
cytometric analyses indicated that linc00467 knockdown
significantly increased the levels of apoptosis in A549 and H1299
cells (Fig. 5A and B). By
contrast, linc00467 overexpression decreased the levels of
apoptosis in PC9 cells (Fig. 5C).
The results confirmed that linc00467 promoted LAD cell
proliferation by affecting LAD cell apoptosis.
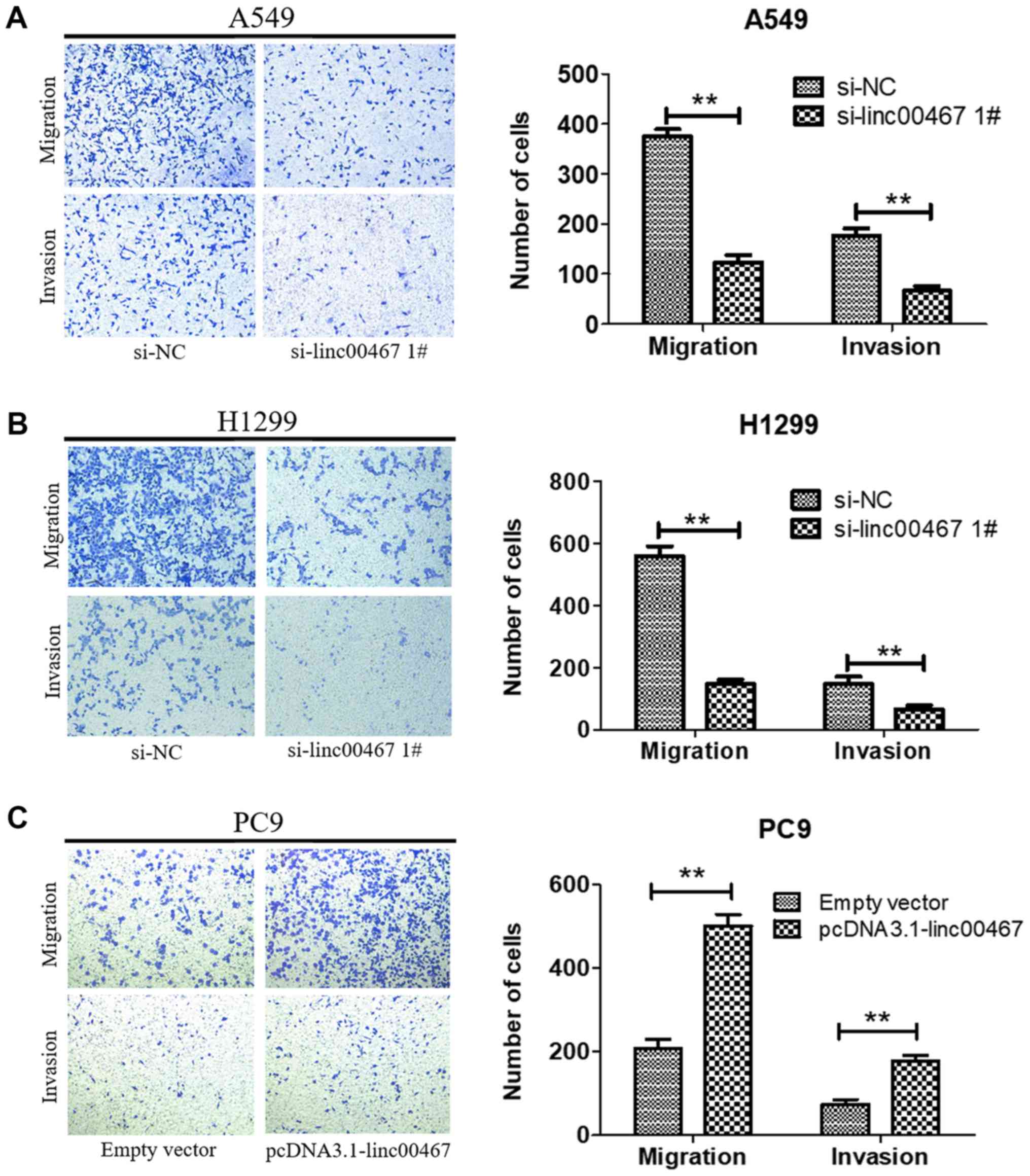
linc00467 promotes LAD cell migration
and invasion in vitro
As linc00467 was confirmed to promote LAD cell
proliferation in vitro, its effects on LAD cell migration
and invasion was additionally investigated. A Transwell assay was
used to evaluate the biological effect of linc00467 on the LAD cell
metastasis. linc00467 knockdown significantly suppressed the
migratory and invasive abilities of A549 and H1299 cells (Fig. 6A and B), while linc00467
overexpression increased the migratory and invasive abilities of
PC9 cells (Fig. 6C). These results
suggested that linc00467 promoted the migration and invasion of LAD
cells.
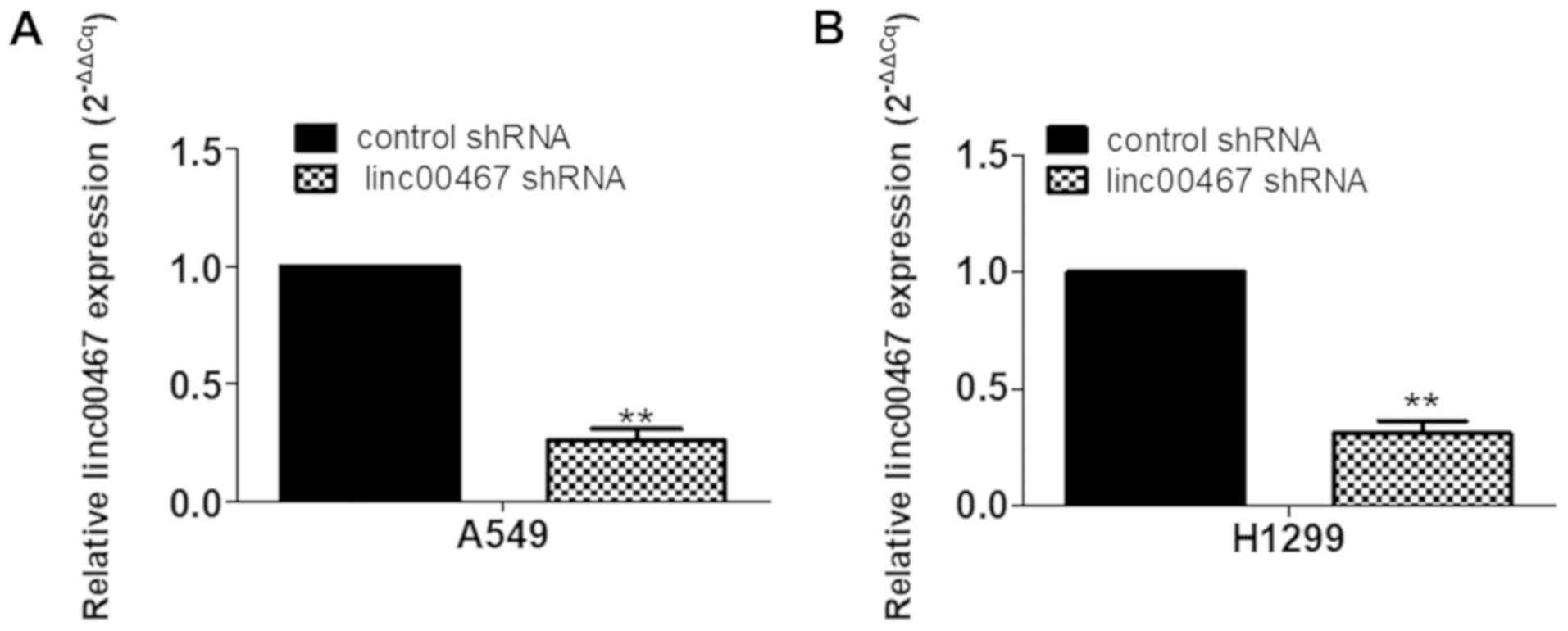
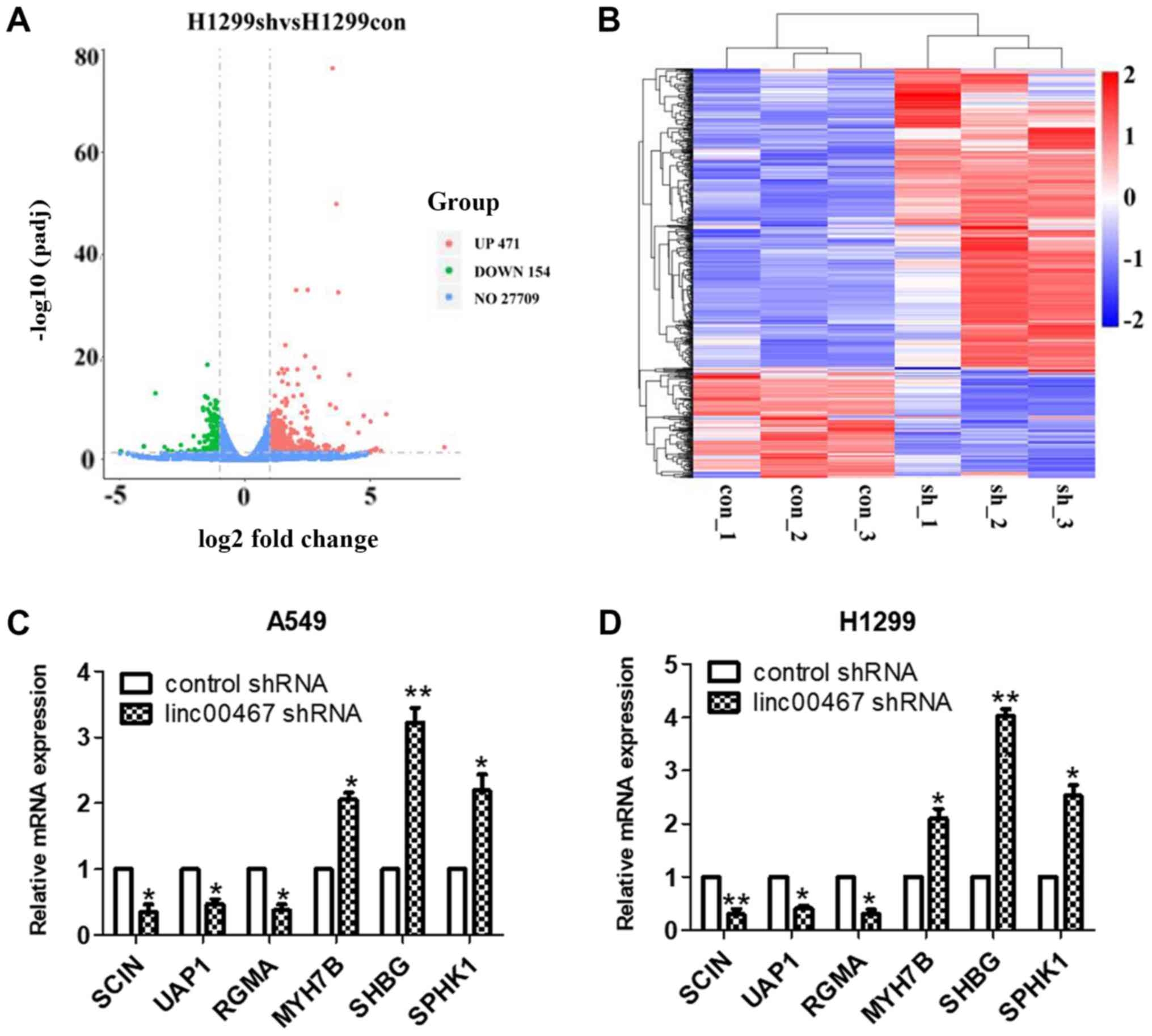
Gene expression profiling
linc00467 was knocked down in H1299 cells by
transfection with shRNA lentiviruses. The knockdown efficiency of
shRNA was verified by RT-qPCR (Fig. 7A
and B). RNA transcriptome sequencing was used to identify genes
that were differentially expressed between linc00467-knockdown
H1299 and control cells. A total of 625 differentially expressed
transcripts were identified using database analysis (471
upregulated and 154 downregulated transcripts, |log2 (FC)| >1
and FDR<0.05) (Fig. 8A and B).
RT-qPCR was used to verify the expression of 6 randomly selected
genes expression in A549 and H1299 cells, and the results were
consistent with the RNA transcriptome sequencing results (Fig. 8C and D).
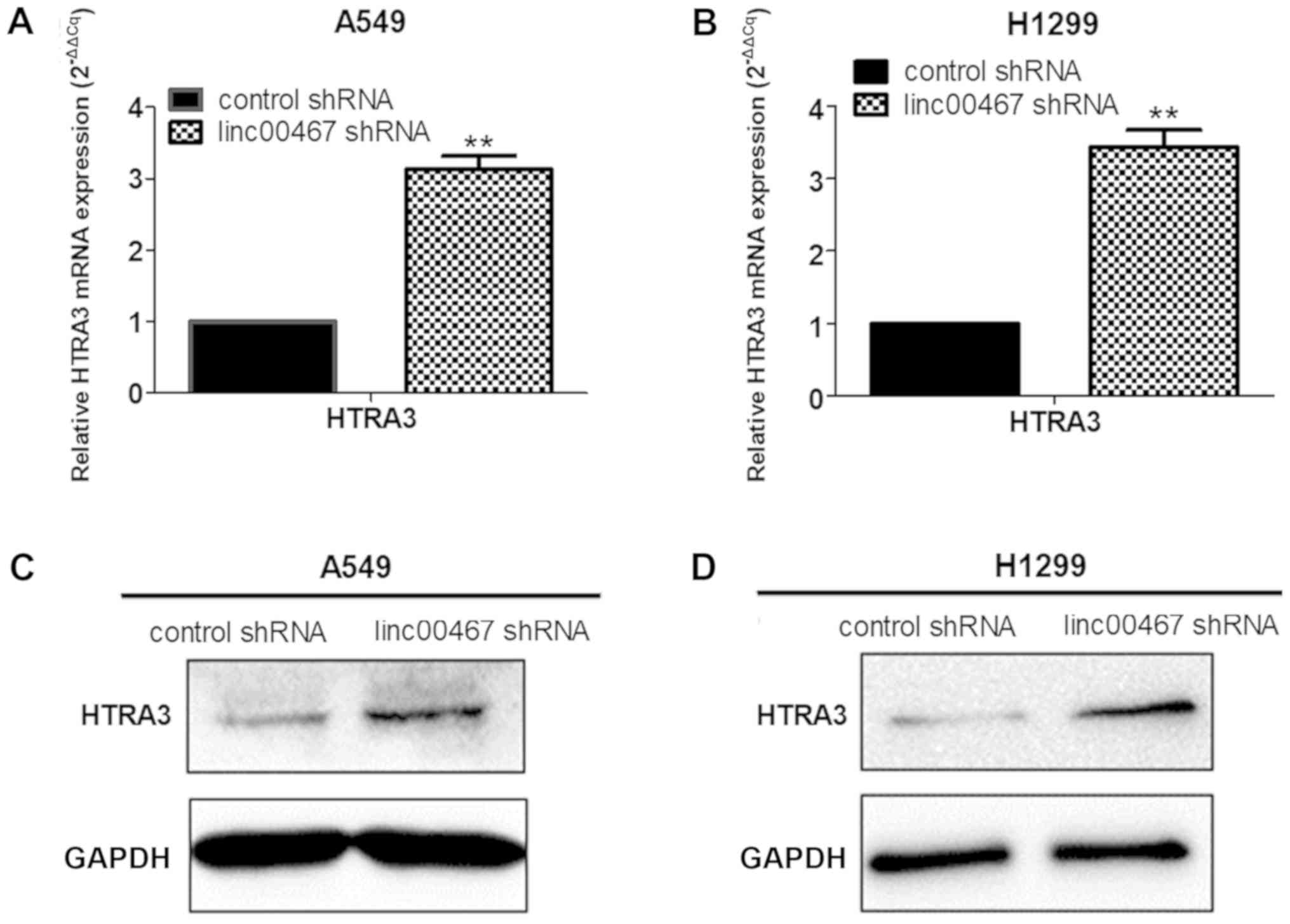
RNA transcriptome sequencing indicated that HTRA3
was upregulated to a greater extent in linc00467-depleted H1299
cells compared with the control cells. It was then additionally
verified that HTRA3 exhibited higher mRNA expression in
linc00467-knockdown A549 cells and H1299 cells compared with the
control cells using RT-qPCR, and protein levels were
correspondingly elevated (Fig.
9A-D).
linc00467 represses HTRA3 expression
by binding with EZH2
lncRNAs are able to bind RNA-binding proteins (RBPs)
to regulate the expression of downstream genes. Therefore, the
present study investigated whether linc00467 regulated HTRA3
expression by binding to RBPs. The probability of an interaction
between linc00467 and RBPs was first determined using an lncRNA
prediction website (http://annolnc.cbi.pku.edu.cn/index.jsp). The sequence
of linc00467 was input into the AnnoLnc database, and entered into
the protein interaction page; subsequently, the interaction score
between lncRNA and protein could be predicted. The interaction
score for linc00467 and EZH2 was 96.0705, close to the maximum
value of 100. Whether linc00467 repressed HTRA3 transcription was
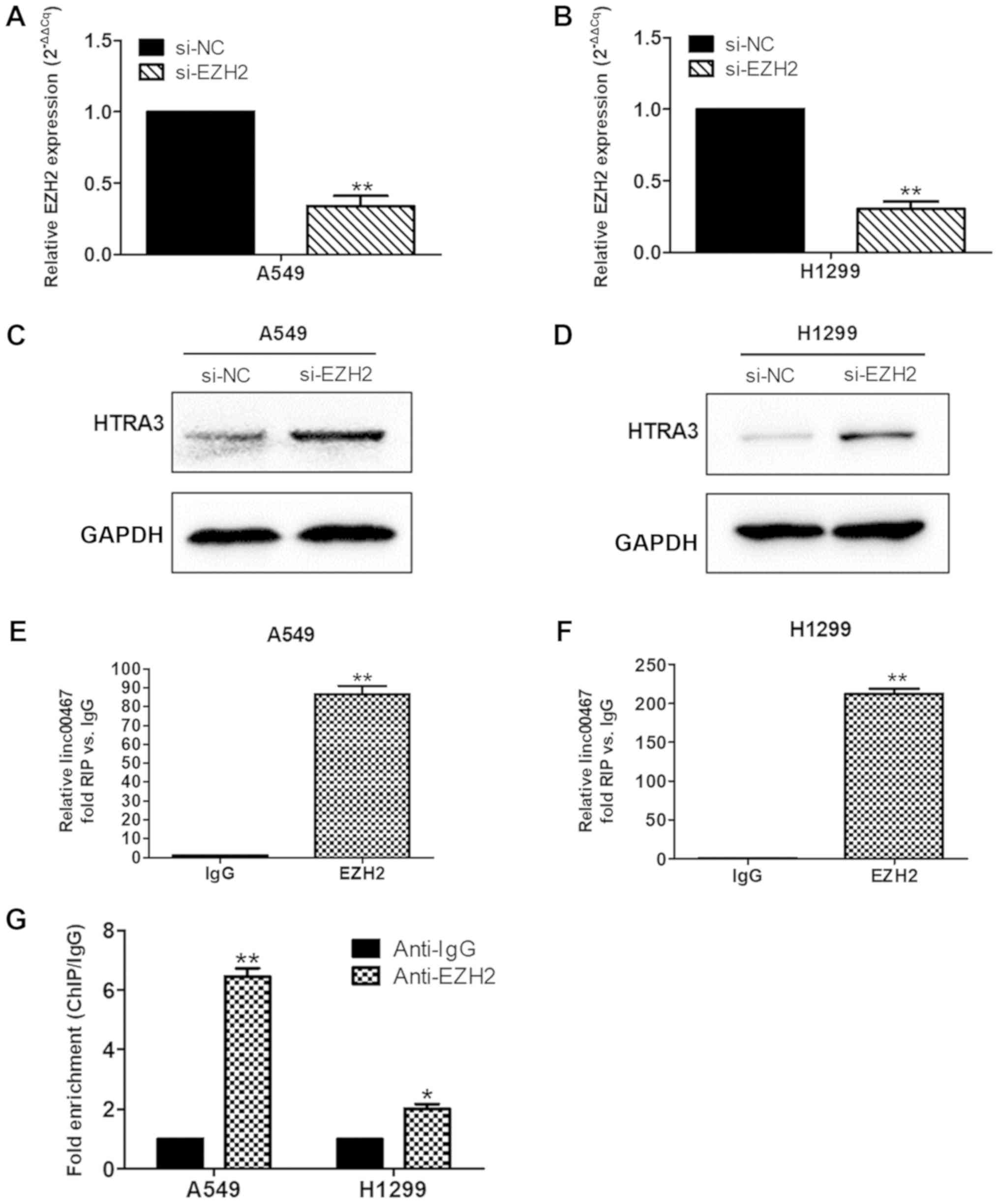
next investigated by recruiting EZH2 to the HTRA3 promoter. EZH2
expression was first knocked down in A549 and H1299 cells using
transfection with siRNA (Fig. 10A and
B). In addition, western blot analysis was performed to measure
HTRA3 protein expression levels when EZH2 was downregulated. The
results indicated that HTRA3 protein expression levels were
increased in the EZH2-downregulated group compared with in the
control group (Fig. 10C and D).
Then, an RIP assay was performed to directly address whether
linc00467 and EZH2 are binding partners. The RIP assay suggested
that linc00467 was able to directly bind EZH2 in A549 and H1299
cells (Fig. 10E and 10F). In addition, a ChIP assay was used
to verify that EZH2 bound to the promoter of HTRA3 (Fig. 10G). The results demonstrated that
linc00467 repressed HTRA3 expression by binding with EZH2.
HTRA3 knockdown partially reverses
linc00467-depletion induced inhibition of LAD cell proliferation,
migration and invasion
Rescue experiments were used to examine the effect
of inhibiting HTRA3 expression in linc00467-knockdown A549 and
H1299 cells. The rescue experiments demonstrated that cell
proliferation, migration and invasion were inhibited in
linc00467-knockdown A549 and H1299 cells, whereas knockdown of
HTRA3 partially reversed all of these effects (Fig. 11A-D).
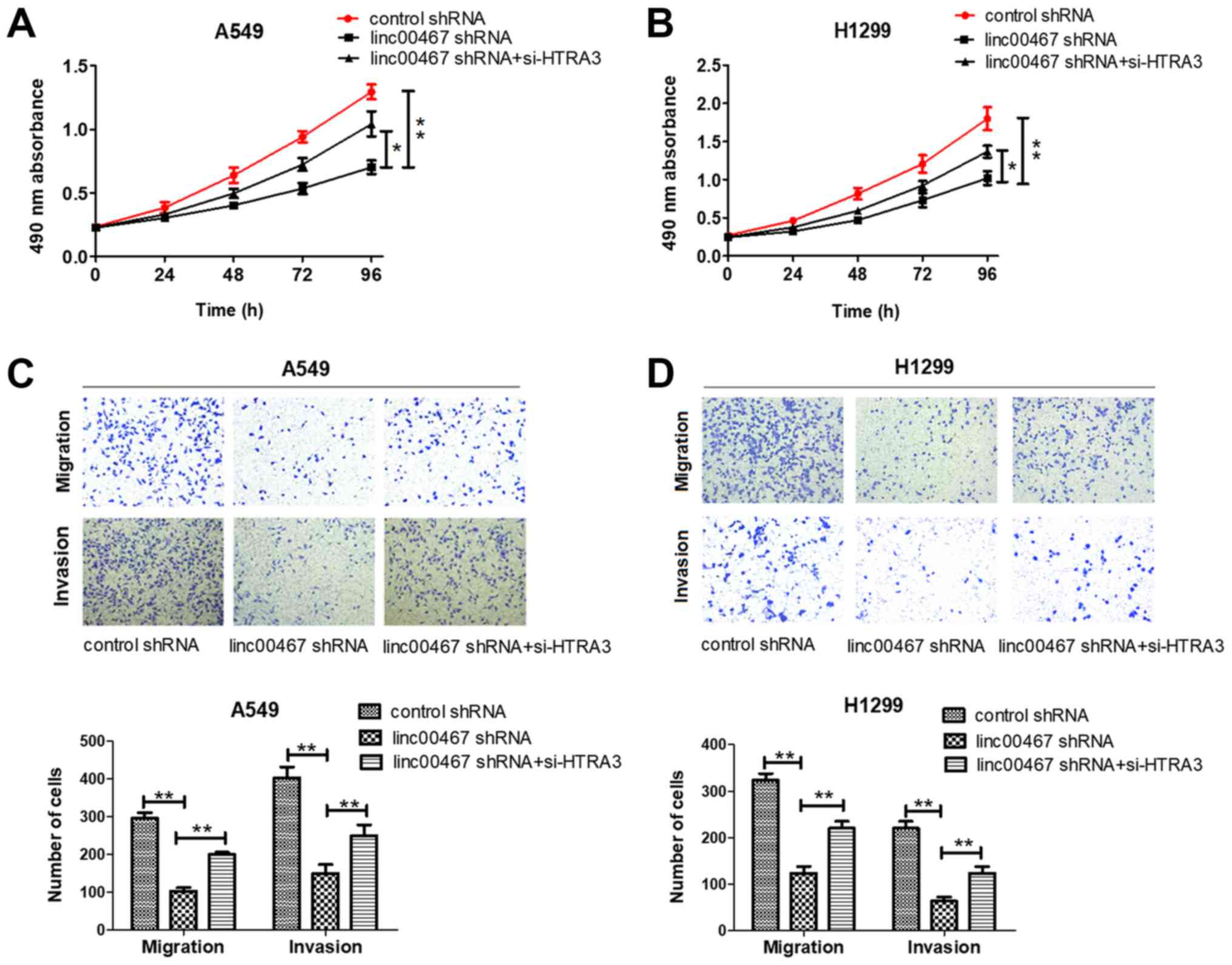 | Figure 11.HTRA3 inhibition attenuates the
inhibitory effect on proliferation, migration and invasion of LAD
cells induced by linc00467 depletion. (A and B) Cell proliferation,
migration and invasion were measured in (A) A549 and (B) H1299
cells transfected with control shRNA, linc00467 shRNA and linc00467
shRNA+si-HTRA3. (C and D) Cell proliferation, migration and
invasion were inhibited in linc00467-knockdown (C) A549 and (D)
H1299 cells, whereas knockdown of HTRA3 partially reversed all of
these inhibitory effects. The data were obtained from at least 3
independent experiments and are presented as the mean ± standard
deviation. Magnification, ×100. *P<0.05 and **P<0.01. HTRA3,
HtrA serine peptidase 3; linc, long intergenic non-coding RNA;
shRNA, short hairpin RNA; siRNA, small interfering RNA; NC,
negative control. |
HTRA3 is downregulated in LAD tissues
and is associated with the clinical characteristics of LAD
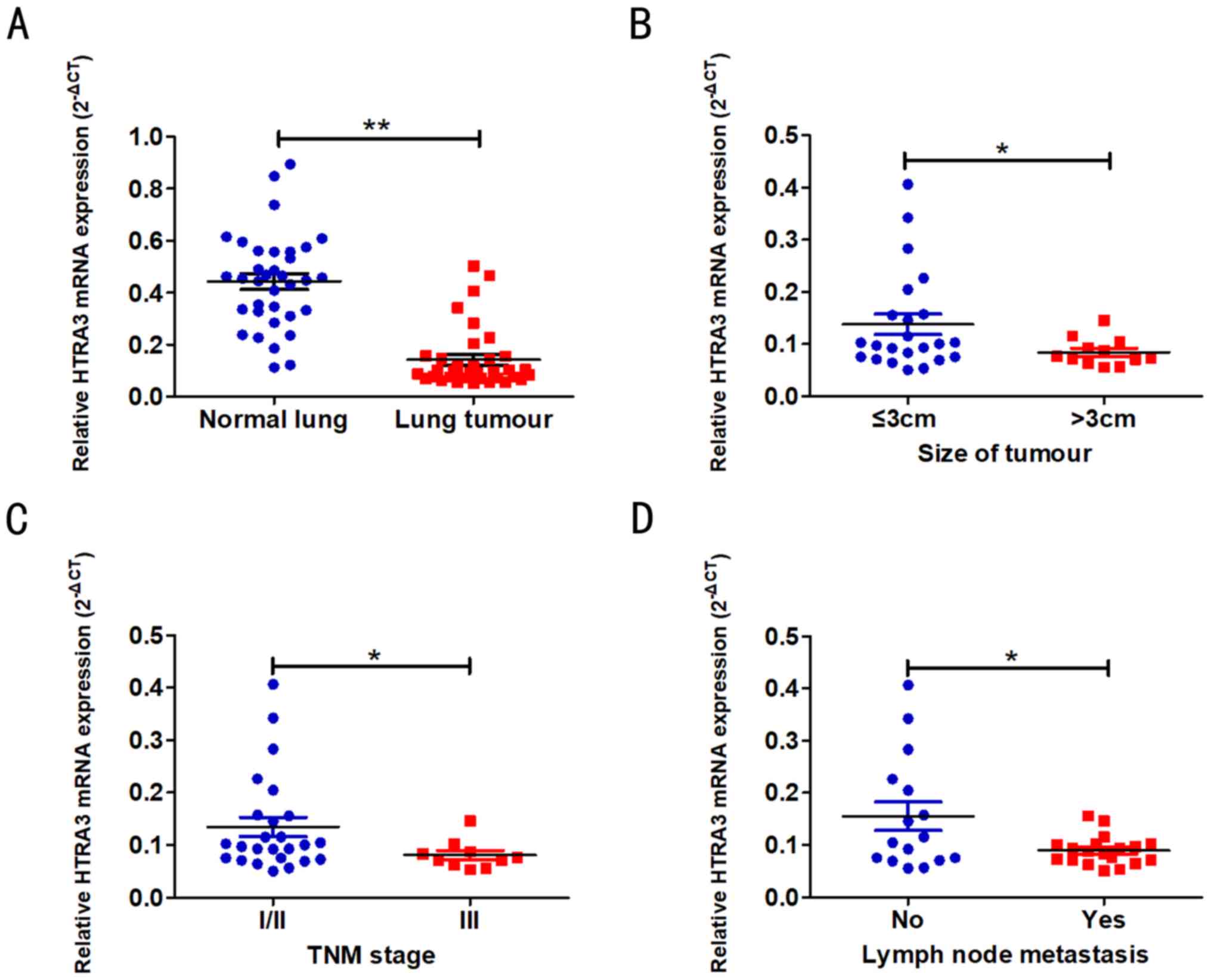
The expression level of HTRA3 in LAD tissues was
decreased compared with the adjacent normal lung tissues
(P<0.001; Fig. 12A). The
association between HTRA3 expression and LAD clinical
characteristics was additionally evaluated (Table II). Decreased HTRA3 expression was
associated with larger tumour sizes (P=0.018; Fig. 12B), increased lymph node
metastasis (P=0.031; Fig. 12C)
and advanced TNM stages (P=0.012; Fig. 12D).
 | Table II.Associations between HTRA3 expression
and clinicopathological parameters of lung adenocarcinoma. |
Table II.
Associations between HTRA3 expression
and clinicopathological parameters of lung adenocarcinoma.
|
|
| HTRA3 mRNA
expression (2−ΔCq) |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
parameters | Number of
patients | Median ± standard
deviation | P-value |
|---|
| Sex |
|
|
|
|
Male | 25 | 0.121±0.092 | 0.856 |
|
Female | 10 | 0.115±0.055 |
|
| Age, years |
|
|
|
|
≤65 | 23 | 0.122±0.091 | 0.736 |
|
>65 | 12 | 0.113±0.065 |
|
| Size of tumor,
cm |
|
|
|
| ≤3 | 23 | 0.138±0.095 | 0.018 |
|
>3 | 12 | 0.084±0.027 |
|
|
Differentiation |
|
|
|
|
Well/moderate | 24 | 0.117±0.091 | 0.799 |
|
Poor | 11 | 0.125±0.061 |
|
| Lymph node
metastasis |
|
|
|
| N0 | 16 | 0.155±0.109 | 0.031 |
|
N1-N3 | 19 | 0.090±0.028 |
|
| TNM stage |
|
|
|
|
I/II | 25 | 0.135±0.092 | 0.012 |
|
III | 10 | 0.081±0.027 |
|
| Smoking |
|
|
|
|
Yes | 22 | 0.116±0.080 | 0.738 |
| No | 13 | 0.126±0.088 |
|
Discussion
Previous studies have suggested that a large number
of lncRNAs affect tumour cell proliferation, migration, invasion
and apoptosis, and are closely associated with tumour development,
progression and metastasis (20,21).
In the present study, bioinformatics analysis of GEO and TCGA
datasets demonstrated linc00467 to be highly expressed in human
lung cancer tissues. Subsequently, RT-qPCR analyses confirmed that
linc00467 was overexpressed in LAD tissues. In particular, the
expression of linc00467 was significantly increased in larger and
later-stage tumours. Additionally, it was identified that knockdown
of linc00467 inhibited LAD cell proliferation, migration and
invasion, consistent with the results of Xia et al (22). These data suggested that linc00467
may exert an oncogenic function and serve an important role in LAD
development. Consequently, it is necessary to explore the molecular
mechanisms of linc00467 in LAD carcinogenesis.
lncRNAs do not encode proteins but are able to
regulate the expression levels of genes at the epigenetic and
transcriptional levels (23,24).
lncRNAs may be used as important regulatory factors to bind
proteins and affect transcription. Previous studies have identified
that lncRNAs recruit EZH2 (25)
and WDR5 (26) to downstream
target gene promoters to inhibit or activate target gene
expression. EZH2 is encoded by the Drosophila EZH2 gene on human
chromosome 7q35. This protein is the key subunit of the polycomb
repressor complex 2 and mediates its catalytic action. The
methylation of histone H3 mediated by EZH2 is associated with the
occurrence and development of multiple tumours (27,28).
Initial studies first identified the role of EZH2 in hematologic
malignancies (29), with
subsequent studies identifying important roles in the development
of prostate (30) and breast
cancer (31), digestive system
malignancies (32), head and neck
(33) and lung cancer (34).
RNA sequencing of linc00467-knockdown LAD cells
revealed that the anti-oncogene HTRA3 is a novel linc00467 target
in LAD cells. HTRA3 is involved in important physiological
processes, including apoptosis, cell signalling and mitochondrial
homeostasis (35). Zhao et
al (36) identified that HTRA3
may be used as a biomarker for the postoperative recurrence and
prognosis of NSCLC. Wenta et al (37) additionally confirmed that HTRA3
should be considered a target of novel anticancer therapies. In the
present study, it was identified that HTRA3 was downregulated in
LAD compared to normal lung tissues, consistent with the results of
the study by Zhao et al (36). Specifically, the expression of
HTRA3 was significantly decreased in larger tumours, those with
lymph node metastases and in later-stage tumours.
In the present study, RIP experiments confirmed that
linc00467 bound EZH2 in LAD cells, suggesting that linc00467
regulated underlying targets at the transcriptional level. In
addition, EZH2 knockdown in LAD cells led to the upregulation of
HTRA3, and ChIP assays revealed that EZH2 may be recruited to the
HTRA3 promoter to repress HTRA3 transcription. Together, these data
demonstrated that linc00467 served an important role in the
EZH2-mediated repression of HTRA3 in LAD cells.
In summary, to the best of our knowledge, the
present study is the first to demonstrate that linc00467 expression
is upregulated in LAD tissues and that this lncRNA promoted LAD
cell proliferation, migration and invasion. In addition, linc00467
was demonstrated to mediate oncogenic effects by binding with EZH2
and regulating HTRA3. The results of the present study strengthen
the understanding of LAD pathogenesis, and support the hypothesis
that linc00467 acts as an oncogene and serves an oncogenic role in
LAD. However, there are certain limitations of the present study.
Firstly, the biological roles of linc00467 were only examined in
cell lines. Secondly, linc00467 may regulate multiple genes or
miRNAs; for this reason, additional studies should be conducted to
investigate the comprehensive linc00467 regulatory network.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the financial
support from the National Natural Science Foundation of China
(grant no. 81572273) and the Jiangsu Key Disease Special Research
Projects (grant no. BL2013026).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW and YS conceived and designed the experiment. XW
wrote the manuscript. XW, HL, KS, XP and YW performed the
experiments. XW and TL analyzed the data. XW and YS revised the
manuscript. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of the Jinling Clinical Medical College of Nanjing
Medical University (Nanjing, China). All of the participants
provided written informed consent form and agreed to the use of
their samples in scientific research.
Patient consent for publication
All of the participants provided written informed
consent form and agreed to the use of their samples in scientific
research.
Competing interests
All authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
D'Antonio C, Passaro A, Gori B, Del
Signore E, Migliorino MR, Ricciardi S, Fulvi A and de Marinis F:
Bone and brain metastasis in lung cancer: Recent advances in
therapeutic strategies. Ther Adv Med Oncol. 6:101–114. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang PQ, Wu YX, Zhong XD, Liu B and Qiao
G: Prognostic significance of overexpressed long non-coding RNA
TUG1 in patients with clear cell renal cell carcinoma. Eur Rev Med
Pharmacol Sci. 21:82–86. 2017.PubMed/NCBI
|
|
5
|
Wang Y, Zhang W, Wang Y and Wang S:
HOXD-AS1 promotes cell proliferation, migration and invasion
through miR-608/FZD4 axis in ovarian cancer. Am J Cancer Res.
8:170–182. 2018.PubMed/NCBI
|
|
6
|
Ricciuti B, Mencaroni C, Paglialunga L,
Paciullo F, Crino L, Chiari R and Metro G: Long noncoding RNAs: New
insights into non-small cell lung cancer biology, diagnosis and
therapy. Med Oncol. 33:182016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y and Tang L: The application of
lncRNAs in cancer treatment and diagnosis. Recent Pat Anticancer
Drug Discov. 13:292–301. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen L, Dzakah EE and Shan G: Targetable
long non-coding RNAs in cancer treatments. Cancer Lett.
418:119–124. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Atmadibrata B, Liu PY, Sokolowski N, Zhang
L, Wong M, Tee AE, Marshall GM and Liu T: The novel long noncoding
RNA linc00467 promotes cell survival but is down·regulated by
N-Myc. Plos One. 9:e881122014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Singh H, Li Y, Fuller PJ, Harrison C, Rao
J, Stephens AN and Nie G: HtrA3 is downregulated in cancer cell
lines and significantly reduced in primary serous and granulosa
cell ovarian tumors. J Cancer. 4:152–164. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Y, Gong L, Qi R, Sun Q, Xia X, He H,
Ren J, Zhu O and Zhou D: Paeoniflorin suppresses pancreatic cancer
cell growth by upregulating HTRA3 expression. Drug Des Devel Ther.
11:2481–2491. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yin Y, Wu M, Nie G, Wang K, Wei J, Zhao M
and Chen Q: HtrA3 is negatively correlated with lymph node
metastasis in invasive ductal breast cancer. Tumour Biol.
34:3611–3617. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pertea M, Pertea GM, Antonescu CM, Chang
TC, Mendell JT and Salzberg SL: StringTie enables improved
reconstruction of a transcriptome from RNA-seq reads. Nat
Biotechnol. 33:290–295. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pertea M, Kim D, Pertea GM, Leek JT and
Salzberg SL: Transcript-level expression analysis of RNA-seq
experiments with HISAT, StringTie and Ballgown. Nat Protoc.
11:1650–1667. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Travis WD, Brambilla E, Noguchi M,
Nicholson AG, Geisinger K, Yatabe Y, Powell CA, Beer D, Riely G,
Garg K, et al: International Association for the Study of Lung
Cancer/American Thoracic Society/European Respiratory Society:
International multidisciplinary classification of lung
adenocarcinoma: Executive summary. Proc Am Thorac Soc. 8:381–385.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Malek E, Jagannathan S and Driscoll JJ:
Correlation of long non-coding RNA expression with metastasis, drug
resistance and clinical outcome in cancer. Oncotarget. 5:8027–8038.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen J, Wang R, Zhang K and Chen LB: Long
non-coding RNAs in non-small cell lung cancer as biomarkers and
therapeutic targets. J Cell Mol Med. 18:2425–2436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xia H, Jing H, Li Y and Lv X: Long
noncoding RNA HOXD-AS1 promotes non-small cell lung cancer
migration and invasion through regulating miR-133b/MMP9 axis.
Biomed Pharmacother. 106:156–162. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Di Gesualdo F, Capaccioli S and Lulli M: A
pathophysiological view of the long non-coding RNA world.
Oncotarget. 5:10976–10996. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang L, Ge D, Chen X, Qiu J, Yin Z, Zheng
S and Jiang C: FOXP4-AS1 participates in the development and
progression of osteosarcoma by downregulating LATS1 via binding to
LSD1 and EZH2. Biochem Biophys Res Commun. 502:493–500. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu
TC, Tang JY, Bao YJ, Hu Y, Lin Y, et al: A novel lncRNA GClnc1
promotes gastric carcinogenesis and may act as a modular scaffold
of WDR5 and KAT2A complexes to specify the histone modification
pattern. Cancer Discov. 6:784–801. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Takashina T, Kinoshita I, Kikuchi J,
Shimizu Y, Sakakibara-Konishi J, Oizumi S, Nishimura M and
Dosaka-Akita H: Combined inhibition of EZH2 and histone
deacetylases as a potential epigenetic therapy for non-small-cell
lung cancer cells. Cancer Sci. 107:955–962. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu S, Chen D, Shen W, Chen L, Yu A, Fu H,
Sun K and Sun X: EZH2 mediates the regulation of S100A4 on
E-cadherin expression and the proliferation, migration of gastric
cancer cells. Hepatogastroenterology. 62:737–741. 2015.PubMed/NCBI
|
|
29
|
McDevitt MA: Clinical applications of
epigenetic markers and epigenetic profiling in myeloid
malignancies. Semin Oncol. 39:109–122. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
van Leenders GJ, Dukers D, Hessels D, van
den Kieboom SW, Hulsbergen CA, Witjes JA, Otte AP, Meijer CJ and
Raaphorst FM: Polycomb-group oncogenes EZH2,BMIl and RINGl are
overexpressed in prostate cancer with adverse pathologic and
clinical features. Eur Urol. 52:455–463. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yoo KH and Hennighausen L: EZH2
methyltransferase and H3K27 methylation in breast cancer. Int J
Biol Sci. 8:59–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Benard A, Goossens-Beumer IJ, van Hoesel
AQ, Horati H, Putter H, Zeestraten EC, van de Velde CJ and Kuppen
PJ: Prognostic value of polycomb proteins EZH2, BMI1 and SUZ12 and
histone modification H3K27me3 in colorectal cancer. PLoS One.
9:e1082652014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gannon OM, Merida de Long L, Endo-Munoz L,
Hazar-Rethinam M and Saunders NA: Dysregulation of the repressive
H3K27 trimethylation mark in head and neck squamous cell carcinoma
contributes to dysregulated squamous differentiation. Clin Cancer
Res. 19:428–441. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Saito M, Saito K, Shiraishi K, Maeda D,
Suzuki H, Minamiya Y, Kono K, Kohno T and Goto A: Identification of
candidate responders for anti-PD-L1/PD-1 immunotherapy, Rova-T
therapy, or EZH2 inhibitory therapy in small-cell lung cancer. Mol
Clin Oncol. 8:310–314. 2018.PubMed/NCBI
|
|
35
|
Clausen T, Southan C and Ehrmann M: The
HtrA family of proteases: Implications for protein composition and
cell fate. Mol Cell. 10:443–455. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao J, Zhang J, Zhang X, Feng M and Qu J:
High temperature requirement A3 (HTRA3) expression predicts
postoperative recurrence and survival in patients with
non-small-cell lung cancer. Oncotarget. 7:40725–40734.
2016.PubMed/NCBI
|
|
37
|
Wenta T, Zurawa-Janicka D, Rychlowski M,
Jarzab M, Glaza P, Lipinska A, Bienkowska-Szewczyk K,
Herman-Antosiewicz A, Skorko-Glonek J and Lipinska B: HtrA3 is a
cellular partner of cytoskeleton proteins and TCP1α chaperonin. J
Proteomics. 177:88–111. 2018. View Article : Google Scholar : PubMed/NCBI
|