Introduction
Preeclampsia (PE) is a pregnancy-related disorder,
with symptoms including new-onset hypertension, proteinuria
commonly occurring after 20 weeks of gestation. PE affects 3–8% of
pregnancies worldwide (1) and is a
major contributor to maternal and fetal morbidity and mortality
(2). As methods to mitigate,
prevent and treat PE are lacking, terminating the pregnancy and
treating the symptoms have become effective approaches. Therefore,
PE has become a primary cause of fetal mortality. Invasive villous
trophoblastic cells invade the maternal endometrium and myometrium
reshaping the uterine spiral artery. These cells ultimately provide
effective and stable placental blood flow and play a vital role
during the process of forming the placenta. Similar to tumor cells,
invasiveness is a feature of trophoblasts; however, unlike tumor
invasion, trophoblastic invasion is a tightly controlled
physiological event (2,3). Abnormalities in the differentiation
of trophoblasts are associated with a variety of diseases during
pregnancy. Insufficiencies in the invasion and proliferation of
trophoblasts are known to be correlated with the development of PE
(4,5). Human chorionic gonadotropin (hCG) is
a glycoprotein hormone composed of α and β subunits that is
secreted by trophoblasts. hCG is necessary for the implantation of
embryos and the maintenance of pregnancy. Retinol-binding protein 4
(RBP4) is secreted by the liver and adipose tissue, and was
originally identified as a specific transporter of vitamin A
(6). A number of studies have
suggested that RBP4 is involved in the development of obesity and
insulin resistance, as such RBP4 is considered to be a new
adipose-derived factor (7,8). A fusion protein involves merging two
or more different protein domains into a single protein molecule
using recombinant DNA technology. The goal of fusion protein
technology is to achieve an increase in performance by combining
the functions of the different proteins (9). In this present study, an anti-hCG
antibody was used to target RBP4 to placental tissue by
constructing an anti-hCG antibody: RBP4 [single-chain antibody
variable fragment (scFv)-RBP4] fusion protein. This fusion protein
could thus improve the local RBP4 concentration in the placenta and
may lead to new strategies for the treatment of PE.
Materials and methods
Construction and purification of the
scFv-RBP4 fusion protein
The RBP4 and anti-hCG scFv sequences were obtained
from the NCBI database (RBP4 chain pro0000017961). The amino acid
sequence of the scFv-RB4 protein was optimized and a full-length
splicing primer was designed by Detai Biotechnology. The template
was optimized using MaxCodon Optimization Program (version 13)
(Detai Biotechnology). The scFv-RBP4 gene was inserted into the
expression vector proEM (Detai Biotechnology) by double enzyme (T4
DNA ligase, TaqDNA polymerase) digestion. The accuracy of the final
expression vector was confirmed by restriction enzyme (EcoR
I and BamHI; New England BioLabs, Inc.) digestion and
sequencing. The final expression vector was transformed into DH5α
competent cells (Detai Biotechnology) and transfection grade
plasmid was extracted using a plasmid purification kit (Qiagen
GmbH). Following extraction, the expression plasmids were analyzed
by agarose gel electrophoresis using a 1% gel. The plasmid was
transfected into the mammalian 293T cell line (Xi Bei Hong Cheng
Biological Technology Co., Beijing, China) using Qiagen Plasmid
Maxi kit (Qiagen GmbH) for transient expression and the scFv-RBP4
fusion protein was purified by nickel-iminodiacetic acid (Ni-IDA)
affinity chromatography.
Cell culture
The immortalized human trophoblast cell line
HTR8/SVneo was obtained from the American Type Culture Collection.
Cells were cultured in RPMI-1640 medium (HyClone; GE Healthcare
Life Sciences) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin
in a 37°C humidified incubator with 5% CO2. The cells
were sub-cultured at a ratio of 1:3 when the cultures reached
80–90% confluence.
The 293T cells were cultured in suspension 2 days
before transfection. The inoculation density was 4–5×105
cells/ml. The cells were incubated in suspension after 4–6 days at
37°C with 5% CO2 at 110 × g.
Transfection of the scFv-RBP4 plasmid
into 293T cells
The extracted scFv-RBP4 plasmid was transfected into
1 liter of 293T cells using a transfection-grade plasmid extraction
kit (Qiagen GmbH) and the cells were incubated in suspension at 110
rpm at 37°C in a 5% CO2 incubator. The cell density was
maintained at 1.5–2×106 cells/ml. For the transfection,
the DNA (1,500 bp)-polyethylenimine (PEI) mixture was at a mass
ratio of 1:5; the DNA was first added to the transfection buffer
and was then added to the incubation buffer. Between 4 and 6 days
after transfection at 37°C in a 5% CO2 incubator the
cell suspension was centrifuged at 800 × g at 37°C for 6 min to
collect the supernatant and cells.
Protein purification
The supernatant was filtered through a 0.22 µm
membrane and dialyzed in buffer 1 (25 mM Tris, 150 mM NaCl, pH 8.0)
at 4°C for 30 min. After dialysis, purification was performed using
a Ni-IDA affinity chromatography column. The fusion protein was
collected and dialysis repeated in buffer 2 (1X PBS, 10% glycerol,
pH 7.4). After dialysis, the fusion protein was filtered through a
0.22 µm membrane and stored at −80°C.
Freeze-thaw stability test
The scFv-RBP4 protein that had been frozen at −80°C
was placed in an ice-water mixture until the sample slowly thawed;
no abnormality occurred after thawing. The scFv-RBP4 protein was
subjected to western blot analysis verification after repeated
freeze-thaw cycles and demonstrated no significant change in
protein content, indicating that the scFv-RBP4 protein had normal
characteristics.
Protein extraction
Total cellular proteins were extracted using RIPA
200 µl, Pi 22 µl and PMSF 5 µl, (Abcam). Following centrifugation
at 12,000 × g at 4°C for 15 min, the protein concentrations of the
supernatants were determined using the Bradford assay. A set of BSA
(Abcam) standards at 1.0, 0.8, 0.6, 0.4 and 0.2 mg/ml in PBS was
used. Another 1 ml of the PBS solution (BSA solution concentration
of 0 mg/ml) was used as a control. A volume of 50 µl was removed
from each BSA solution and was added to a 96-well plate. The same
volume of the protein solutions to be tested was placed in the
other wells of the plate and 200 µM Coomassie Brilliant Blue was
then added. After incubating the plate for 10 min at 37°C, the
optical density values of the BSA standards were measured at 595 nm
using a microplate reader. A standard curve was generated and the
protein concentrations were calculated.
SDS-PAGE and western blot
analysis
Total cellular proteins were extracted (as detailed
above). Following centrifugation at 12,000 × g and 4°C for 15 min,
the protein concentrations of the supernatants were determined
using a Bradford assay. Equal amounts of the total proteins (3 µg)
were loaded per lane and separated via 10% SDS-PAGE. After
electrophoresis, the gel was placed in a clean glass container, and
5 times the gel volume of Coomassie Brilliant Blue G-250 dye was
added. The gel was incubated for 1 h at room temperature with
agitation to disperse the large colloid particles of the added
Coomassie Brilliant Blue G-250 dye. The liquid was then drained.
After boiling and incubating overnight at room temperature, the gel
was rinsed with water to observe the decolonization effect. After
electrophoresis, the samples were transferred to nitrocellulose
membranes using standard procedures. The primary antibody used was
rabbit polyclonal anti-RBP4 (1:1,000; Abcam). Equal amounts of
total protein (3 µg) were probed with primary antibody in 5% SBS
(Abcam)/PBS-Tween-20 solution overnight at 4°C. The membranes were
rinsed several times with PBS-Tween-20 solution and then incubated
with Goat anti-rabbit, HRT conjugated secondary antibody (1:3,000;
ab6721; Abcam) for 2 h at room temperature. Excess secondary
antibody was removed by washing four times in PBS-Tween 20
solution. Bands were visualized and imaged using an ECL Western
Blot kit (CW Biotech) and densitometry was performed using ImageJ
software (version 1.49e, National Institutes of Health).
Identification of fusion proteins by
matrix assisted laser desorption/ionization time-of-flight
(MALDI-TOF) mass spectrometry (MS)
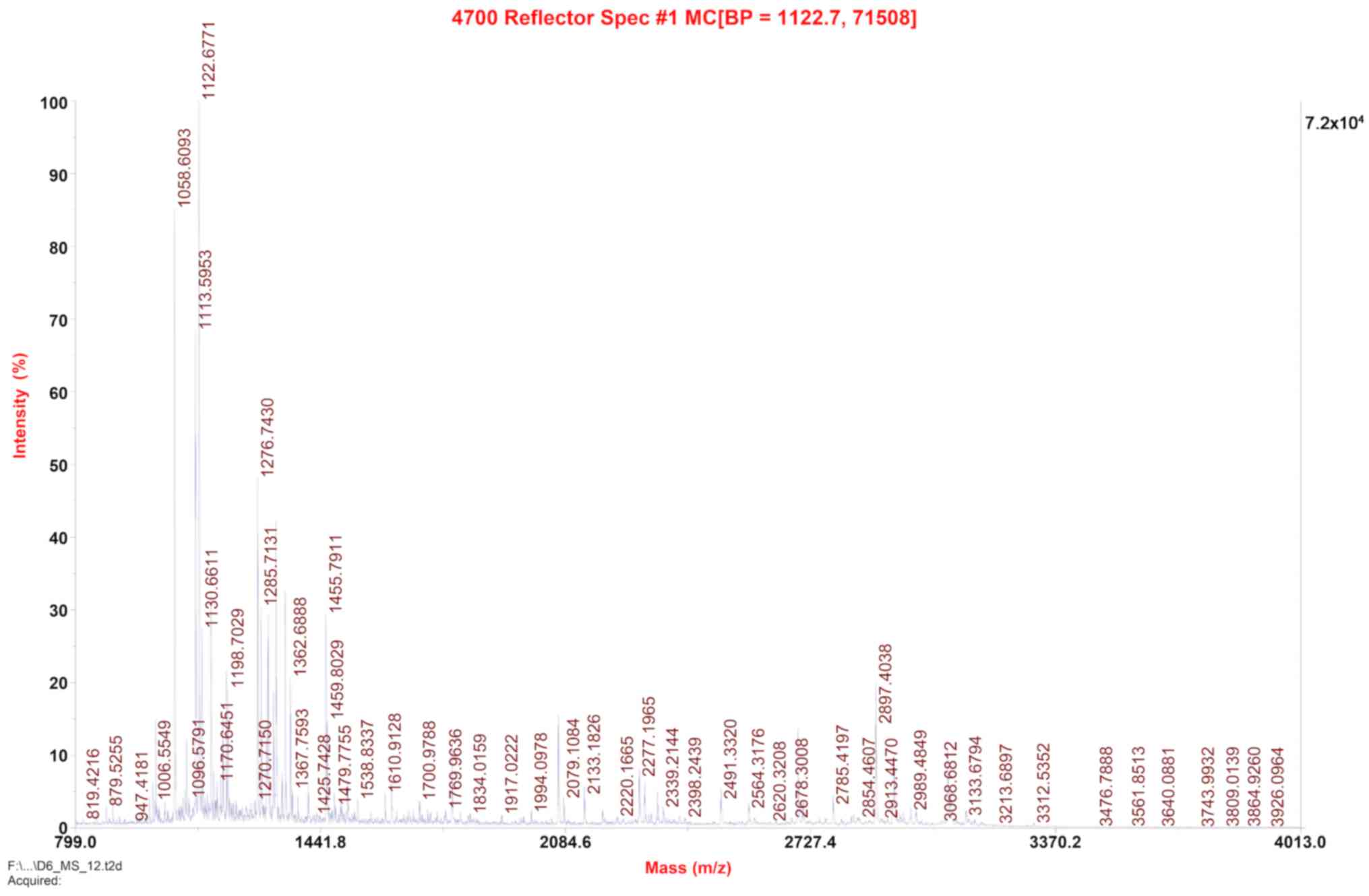
Positron flight mass spectrometry was used at m/z of
1122.7. Protein bands on the SDS-PAGE gel were excised and
subjected to MALDI-TOF-TOF MS/MS protein profiling using an ABI
4700 mass spectrometer (Applied Biosystems; Thermo Fisher
Scientific, Inc.).
Cell treatment
HTR8/SVneo cells were seeded, at a density of 5,000
cells per well, into a 96-well plate with different concentrations
of scFv-RBP4 (0, 1, 10, 100 µg/ml), RBP4 (0, 1, 10, 100 µg/ml) and
anti-hCG (0, 1, 10, 100 µg/ml) along with control HTR8/SVneo cells.
The Cell Counting Kit-8 (Sigma-Aldrich; Merck KGaA) was used to
assay cell survival at 0, 24, 48 and 72 h. The optical density was
measured at 450 nm. These experiments were performed three
times.
Transwell invasion assay
Cell invasion was assessed using Transwell chambers
(24-well inserts; 8 µm-pore size) that had been precoated with
Matrigel (200 µg/ml; BD Biosciences). Briefly, 24 h after
transfection, HTR8-SVneo cells were trypsinized and seeded into the
upper chambers (5×105 cells/chamber) in serum-free
medium. Different concentrations of scFv-RBP4 (0, 1, 10, 100
µg/ml), RBP4 (0, 1, 10, 100 µg/ml) and anti-hCG (0, 1, 10, 100
µg/ml) were used. The lower chambers were filled with RPMI-1640
medium containing 10% FBS as a chemoattractant. The plates were
incubated for 48 h, at which point the cells on the upper surface
of the chambers were gently removed. The cells that had invaded to
the surface of the lower chambers were fixed in paraformaldehyde
(4%) at room temperature for 20 min and stained with crystal violet
(0.1% g/ml) at room temperature for 30 min; the number of cells in
four randomly selected fields of view were imaged using a light
microscope (magnification, ×10) and were counted.
Statistical analysis
Statistical analysis was performed using SPSS 20.0
software (IBM Corp.). Normally distributed data were described as
the mean ± standard deviation, and non-normally distributed data
were described as the median (interquartile range). The data were
tested >3 times for normality using a Shapiro-Wilk test and
analyzed using t-tests, one-way ANOVA, a multiple comparison post
hoc test (least significant difference) and nonparametric tests
(Mood's median test). P<0.05 was considered to indicate a
statistically significant difference.
Results
Construction and purification of the
scFv-RBP4 fusion protein
The full-length RBP4 fusion protein is 453 amino
acids. The target sequence is variable light chain + variable heavy
chain + RBP4. The scFv-RBP4 protein length=453; molecular
weight=50,242.7 kDa. The last residues of scFv were
GGGGSGGGGSGGGGS. The first residues of the RBP4 protein were GGG
SGG GGS (Table I).
 | Table I.Construction and purification of the
scFv-RBP4 fusion protein. |
Table I.
Construction and purification of the
scFv-RBP4 fusion protein.
| Chain | Protein sequence |
|---|
| VL | DIVMSQSPSS LAVSVGEKVT
MTCKSSQSLL YSSNQMNYLA WYQQKPGQSP KLLIYWASTRESGVPDRFTG SGSGTDFTLT
ISSVEAEDLA VYYCQQYHSY PFTFGSGTKL EIKRAGGGGSG |
| VH | GGGGSGGGGSEVNLEESGGG
LVQPGGSMKL SCVASGFTFS NYWMNWVRQS PEKGLEWVA DIRLKSNNYA TLYAESVKGR
FTISRDDSKS SVYLQMNNL RAEDTGIYYC TRGAYYRYDY AMDYWGQGTS VTVSS |
| RBP4 | GGSGGGGSER DCRVSSFRVK
ENFDKARFSG TWYAMAKKDP EGLFLQDNIV AEFSVDETGQ MSATAKGRVR LLNNWDVCAD
MVGTFTDTED PAKFKMKYWG VASFLQKGND DHWIVDTDYD TYAVQYSCRL LNLDGTCADS
YSFVFSRDPN GLPPEAQKIV RQRQEELCLA RQYRLIVHNG YCDGRSERNLL |
Verification of the transfection
plasmid
The expression plasmid was digested and separated on
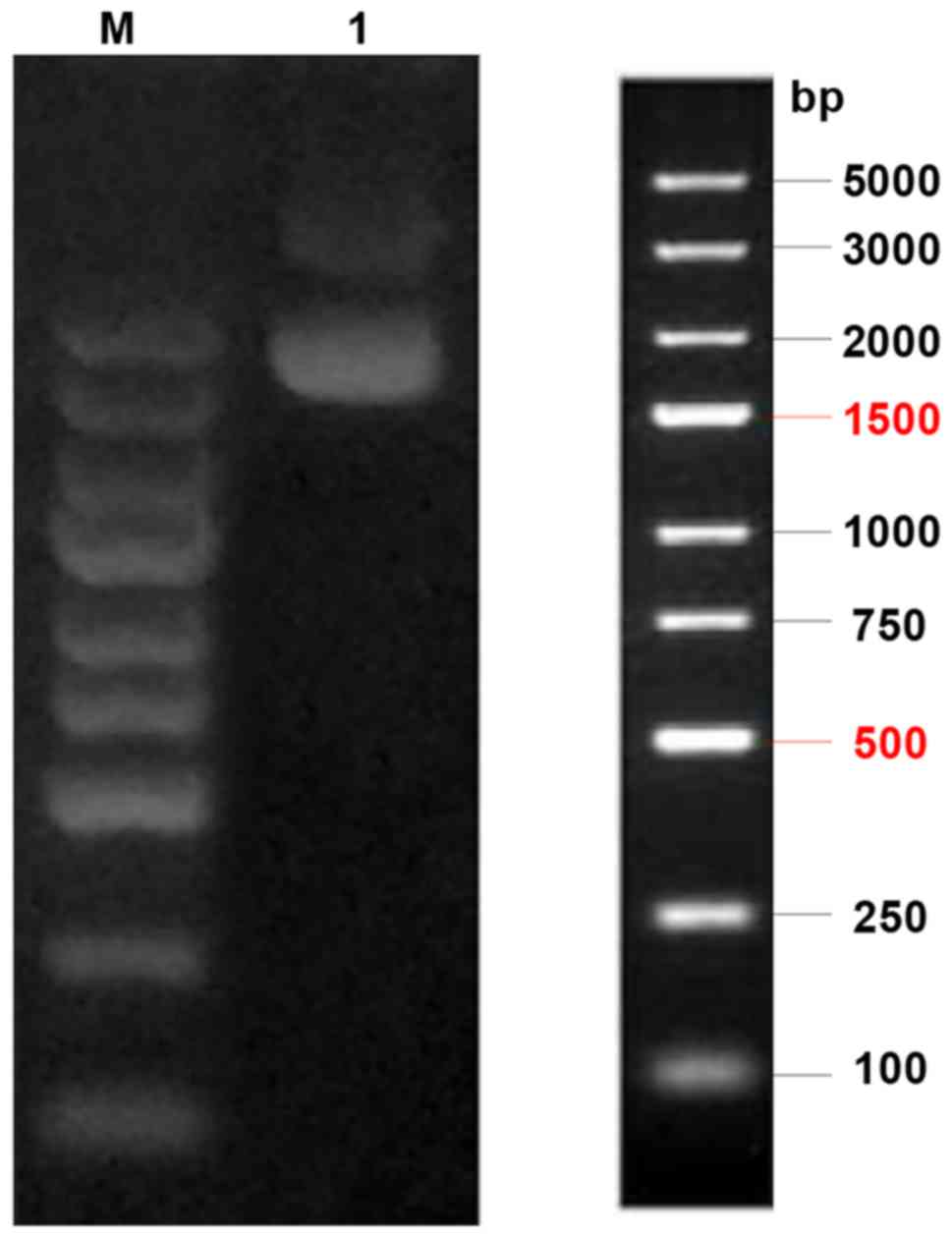
a 1% agarose gel. The electrophoresis result showed that the final
location on the gel strip was as predicted, as shown in Fig. 1.
scFv-RBP4 protein purification
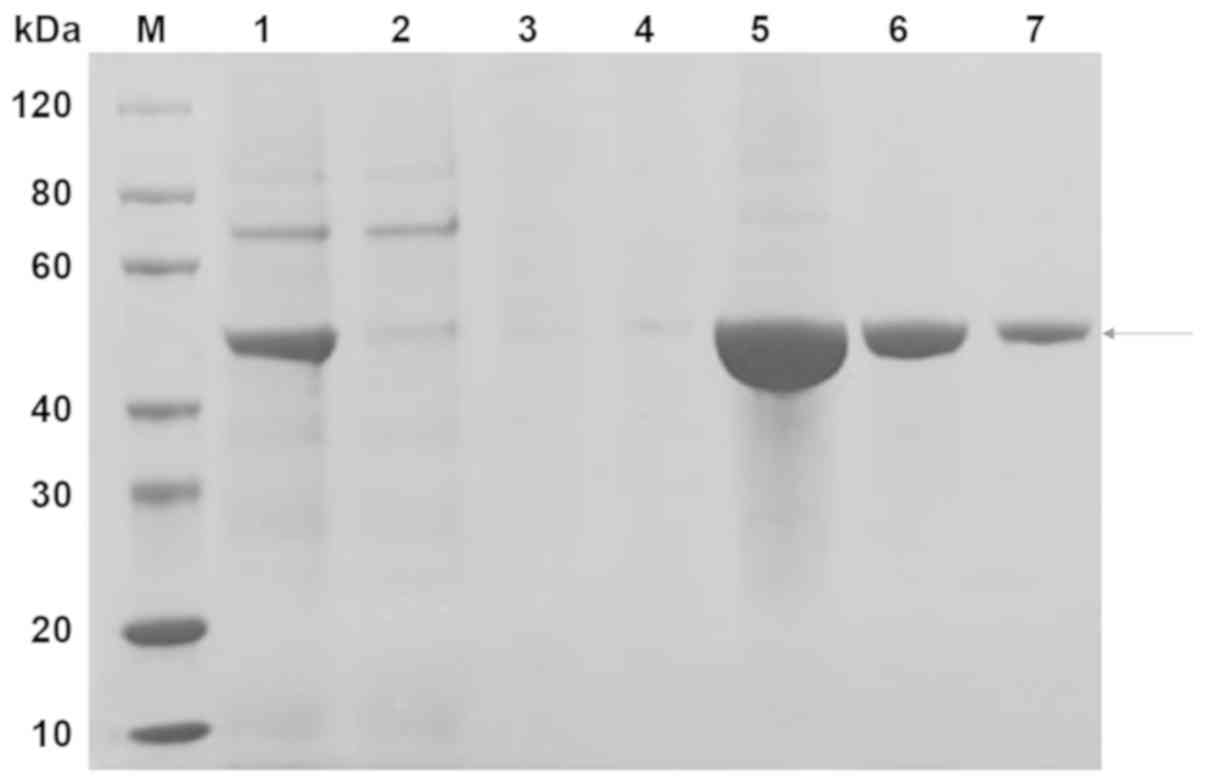
The scFv-RBP4 protein was primarily present in the
fraction eluted with 300 mM imidazole (Fig. 2; lanes 5–7). The desired band was
between the 40 and 60 kDa protein markers and had a molecular
weight of approximately 50 kDa.
Concentration of scFv-RBP4
Based on the Bradford method of protein
concentration determination and using BSA as a standard, the final
concentration of the scFv-RBP4 fusion protein was 1.04 mg/ml, and
the purity of the protein was >90%.
Identification of fusion proteins
using MALDI-TOF
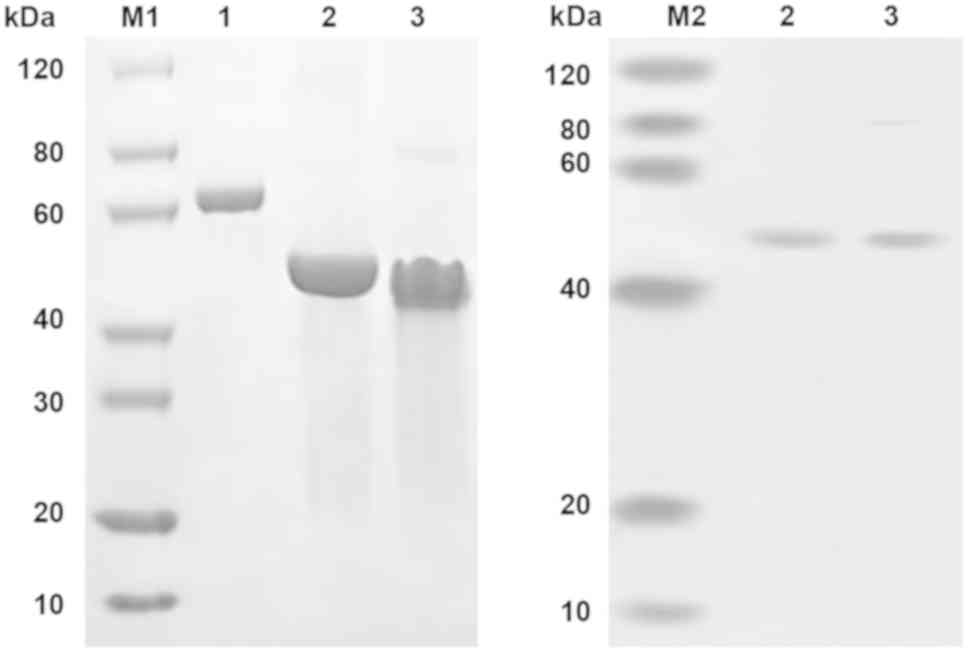
The protein bands corresponding to the predicted
size of scFv-RBP4 on the SDS-PAGE gel (Fig. 3) were excised and subjected to
MALDI-TOF-TOF MS/MS protein profiling using an ABI 4700 mass
spectrometer. The resulting sequence was consistent with the
theoretical sequence of scFv-RBP4 (Fig. 4) [protein score=417; protein score
confidence interval (%)=100].
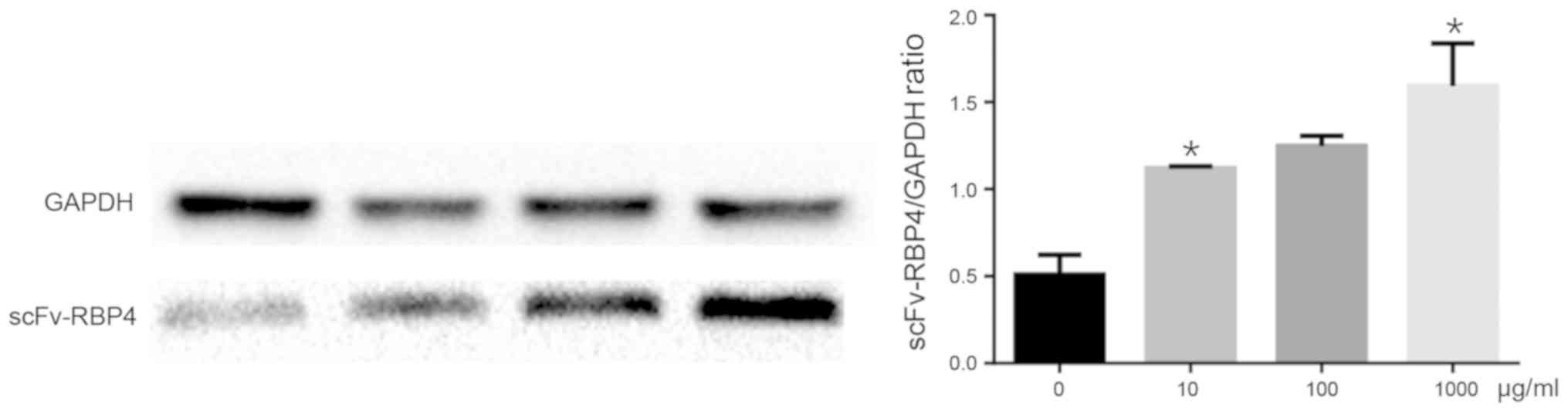
Verification of expression of
scFv-RBP4 in trophoblast cells with different concentrations of
RBP4 by western blot analysis
HBR8/SVneo cells were transfected with different
concentrations of scFv-RBP4 fusion protein. The expression of the
fusion protein was determined by western bolt analysis. The results
showed that the expression of RBP4 was higher when a greater amount
of plasmid was transfected into the cells (Fig. 5).
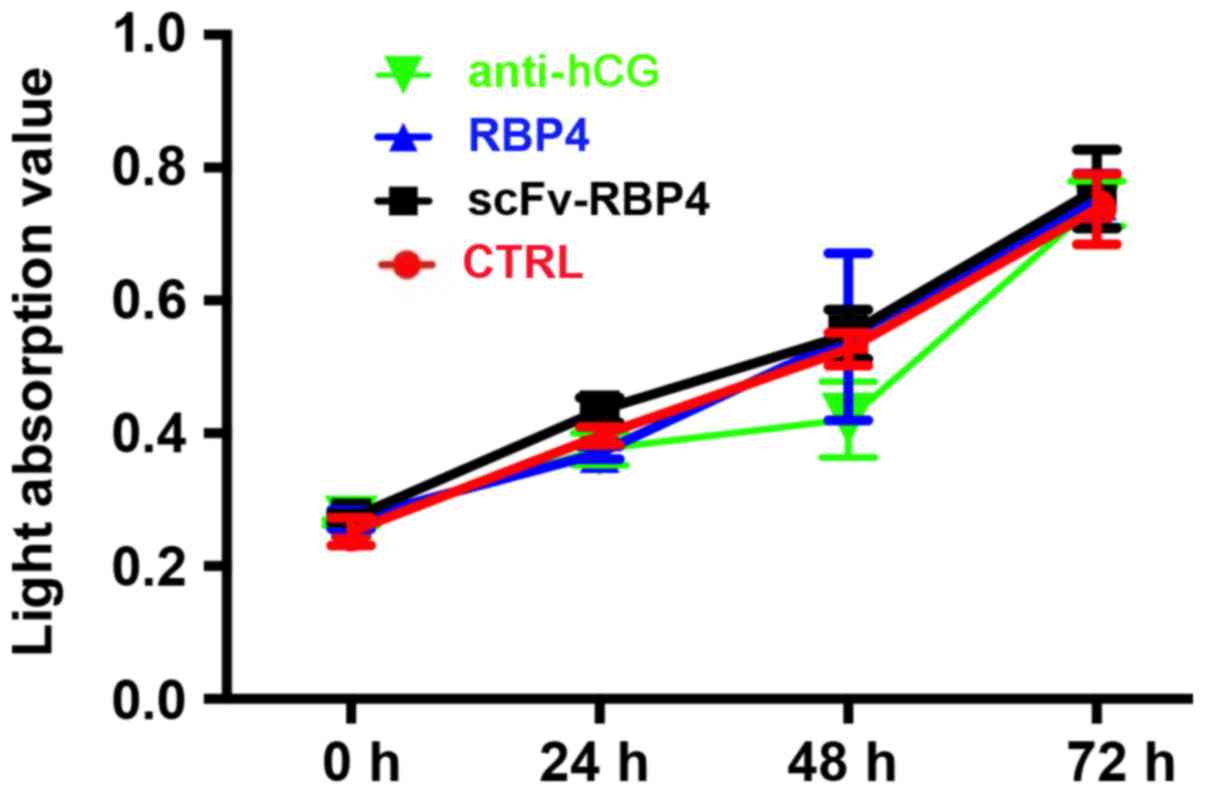
scFv-RBP4 does not increase the
proliferation of HTR8/SVneo cells in vitro
The effect of scFv-RBP4 expression on the biological
behaviors of trophoblast cells was investigated. scFv-RBP4 did not
increase the proliferation of HTR8/SVneo cells (Fig. 6).
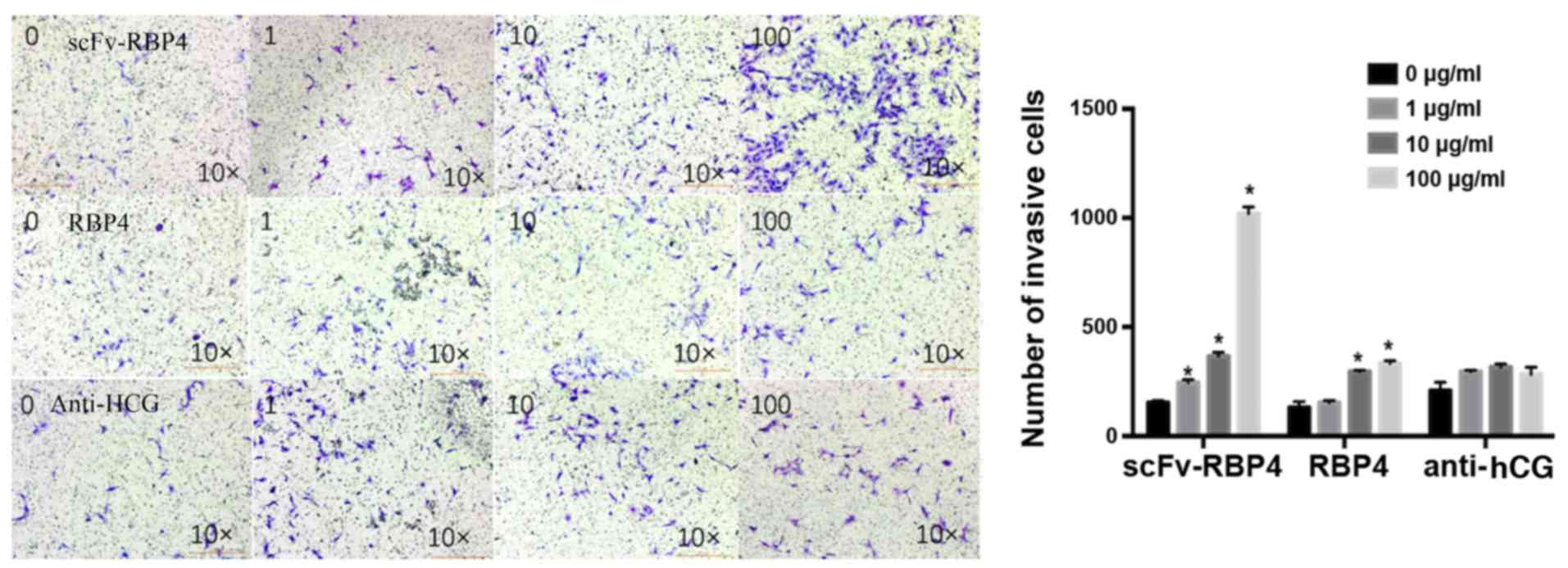
Invasive ability of HTR8/SVneo
cells
Cell invasion assays were employed to investigate
the effect of scFv-RBP4 expression on the invasive ability of
HTR8/SVneo cells (Fig. 7). The
results showed that expression of scFv-RBP4 increased the invasive
ability of HTR8/SVneo cells. With a higher expression of scFv-RBP4
the invasive ability of HTR8/SVneo cells was increased. The
invasiveness of HTR8/SVneo cells did not change significantly in
the hCG group; however, the invasive ability of the cells was
increased with the expression of RBP4 protein alone (10 and 100
µg/l).
Discussion
PE is a pregnancy-related disorder. The pathogenesis
of PE is associated with a variety of factors (2). Unfortunately, the pathophysiology of
this multisystemic disorder, which is characterized by an abnormal
vascular response to placentation, remains unclear. The placenta is
believed to be the primary cause of the disorder (10). During normal pregnancy, in the
early stages of placental development, extravillous
cytotrophoblasts of fetal origin invade the uterine spiral arteries
of the decidua and myometrium. The spiral arteries lose their
endothelium and most of their muscle fibers, transforming these
arteries from small, high-resistance vessels to high-caliber
capacitance vessels capable of providing adequate placental
perfusion to sustain the growing fetus. The invasive
cytotrophoblasts replace the endothelial layer of the maternal
spiral arteries. In PE, this transformation is incomplete, with
almost no intravascular extravillous cytotrophoblastic invasion
(11,12). Therefore, the invasive capability
of trophoblasts is an important factor in the occurrence of PE.
The development of during pregnancy PE can be
life-threatening for the mother and the unborn child, leading to an
increase in morbidity and mortality for both (13). In the case of the mother, PE may
cause cardiovascular disease, including chronic hypertension,
ischemic heart disease and stroke, later in life (14). For the child, preeclamptic
pregnancies can lead to an increased risk of stroke, coronary heart
disease and metabolic syndrome in adulthood (15). To date, delivery is the only
curative treatment for PE (13).
The criteria that define PE have changed little over
the past decade. The criteria include the following: Onset at
>20 weeks of gestation; 24 h proteinuria ≥300 mg/day or, if not
available, a protein concentration ≥30 mg (≥1 on dipstick) with a
minimum of two random urine samples collected at least 4–6 h, but
no more than 7 days, apart; a systolic blood pressure >140 mm Hg
or a diastolic blood pressure ≥90 mm Hg measured twice at least 4–6
h, but less than 7 days, apart using an appropriate cuff; and the
disappearance of all these abnormalities before the end of the 6th
postpartum week (16).
A single-chain antibody (single-chain fragment,
scFv) is a protein obtained via genetic engineering using a peptide
linker sequence to combine the heavy-chain and light-chain variable
regions of an immunoglobulin, thereby producing a single
recombinant protein that has a complete antigen binding site. These
small, genetically engineered antibody fragments, which are less
immunogenic, can be expressed in bacteria and can be genetically
engineered to construct fusion proteins linked to other effector
molecules. There has been extensive application of scFv molecules
in the penetration of tumor tissues (17).
RBP4, previously believed to only be a specific
carrier for vitamin A and primarily produced by the liver (6), has recently been added to the rapidly
expanding family of adipocyte factors (18). RBP4 is widely distributed in human
blood, cerebrospinal fluid, urine and other bodily fluids. Chen
et al (19) showed that
RBP4 is not only a carrier of retinol but is also a circulating
cytokine. Yang et al (18)
reported that RBP4 is an adipocyte-derived signal that may
contribute to the pathogenesis of type 2 diabetes. Lowering RBP4
may be a new strategy for treating type 2 diabetes. A previous
study found that the concentration of RBP4 was significantly lower
in women with severe PE than in women with a healthy pregnancy
(20).
At present, the most important clinical application
of scFv fusion proteins is in immune targeting. An scFv antibody
fragment has reduced non-specific binding, allowing a greater
concentration of scFv to reach a tumor and other regions. Because
an scFv antibody fragment is regarded as the ideal carrier for
target-guided drugs, these constructs have been widely used to
treat tumor cells, thrombolysis and other clinical diseases. The
role of RBP4 in the pathogenesis of PE, improvements in
trophoblastic infiltration and the shallow placental bed have
rarely been reported. In this present study, an anti-hCG scFv-RBP4
fusion protein was constructed in vitro. The fusion protein
components were independent of each other. RBP4 function was not
affected by the scFv domain of anti-hCG, which can be used to
target placental tissue and improve the local concentration of RBP4
at the placenta.
In conclusion, this present study showed that the
scFv-RBP4 fusion protein was involved in the invasiveness of
trophoblastic cells. scFv-RBP4 and RBP4 increased the invasive
ability of HTR8/SVneo trophoblastic cells. Although the invasion
produced by scFv-RBP4 expression may be significant, anti-hCG did
not change the properties of trophoblasts. Decreased expression
levels of RBP4 in the placenta may contribute to the development of
PE by reducing the invasive ability of trophoblasts. These data
suggest that the scFv-RBP4 fusion protein promotes the invasiveness
of trophoblasts.
The advantages of fusion protein technology have not
yet been demonstrated in the field of obstetrics. Therefore,
further studies using this approach are required for the
development of treatment strategies for PE.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the National
Natural Science Foundation of China (grant no. 81571455) and by the
Sino-RUS Cooperation Funds (grant no. 2015DFR31070).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
All authors made substantial contributions to study
conception and design, and the acquisition or analysis and
interpretation of data. All authors were involved in drafting the
manuscript or revising it critically for important intellectual
content. All authors have given final approval of the version to be
published. All authors have agreed to be accountable for all
aspects of the work in ensuring that questions related to the
accuracy or integrity of any part of the work are appropriately
investigated and resolved. ZZ and CL conceived and designed the
experiments. HL and TL performed the experiments. ZZ and GC
analyzed the data. ZZ and CL interpreted the data and HL wrote the
first draft of the manuscript. HL developed the structure and
arguments for the paper. HL, CL and ZZ made critical revisions. All
authors have reviewed and approved the final manuscript.
Ethics approval and consent to
participate
The research protocol was conducted in accordance
with the guidelines of the World Medical Association's Declaration
of Helsinki and was performed following approval from the Medical
Ethics Committee (11-S-59) of Beijing Chao-Yang Hospital, Capital
Medical University.
Patient consent for publication
All patients enrolled in the study provided written
informed consent before inclusion in the study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Poon LC and Nicolaides KH: Early
prediction of preeclampsia. Obstet Gynecol Int. 2014:2973972014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Redman CW and Sargent IL: Latest advances
in understanding preeclampsia. Science. 308:1592–1594. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rana S, Lemoine E, Granger J and
Karumanchi SA: Preeclampsia. Circ Res. 124:1094–1112. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ball E, Bulmer JN, Ayis S, Lyall F and
Robson SC: Late sporadic miscarriage is associated with
abnormalities in spiral artery transformation and trophoblast
invasion. J Pathol. 208:535–542. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goldman-Wohl D and Yagel S: Regulation of
trophoblast invasion: From normal implantation to pre-eclampsia.
Mol Cell Endocrinol. 187:233–238. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zanotti G and Berni R: Plasma
retinol-binding protein: Structure and interactions with retinol,
retinoids, and transthyretin. Vitam Horm. 69:271–295. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Christou GA, Tselepis AD and Kiortsis DN:
The metabolic role of retinol binding protein 4: An update. Horm
Metab Res. 44:6–14. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kotnik P, Fischer-Posovszky P and Wabitsch
M: RBP4: A controversial adipokine. Eur J Endocrinol. 165:703–711.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grönwall C and Ståhl S: Engineered
affinity proteins-generation and applications. J Biotechnol.
140:254–269. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Uzan J, Carbonnel M, Piconne O, Asmar R
and Ayoubi JM: Pre-eclampsia: Pathophysiology, diagnosis, and
management. Vasc Health Risk Manag. 7:467–474. 2011.PubMed/NCBI
|
|
11
|
North RA, Ferrier C, Long D, Townend K and
Kincaid-Smith P: Uterine artery Doppler flow velocity waveforms in
the second trimester for the prediction of preeclampsia and fetal
growth retardation. Obstet Gynecol. 83:378–386. 1994.PubMed/NCBI
|
|
12
|
Meekins JW, Pijnenborg R, Hanssens M,
McFadyen IR and van Asshe A: A study of placental bed spiral
arteries and trophoblast invasion in normal and severe
pre-eclamptic pregnancies. Br J Obstet Gynaecol. 101:669–674. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sibai B, Dekker G and Kupferminc M:
Pre-eclampsia. Lancet. 365:785–799. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang A, Rana S and Karumanchi SA:
Preeclampsia: The role of angiogenic factors in its pathogenesis.
Physiology (Bethesda). 24:147–158. 2009.PubMed/NCBI
|
|
15
|
Meads CA, Cnossen JS, Meher S,
Juarez-Garcia A, ter Riet G, Duley L, Roberts TE, Mol BW van der
Post JA, Leeflang MM, et al: Methods of prediction and prevention
of pre-eclampsia: Systematic reviews of accuracy and effectiveness
literature with economic modelling. Health Technol Assess.
12:iii–iv, 1-270. 2008. View
Article : Google Scholar
|
|
16
|
Schroeder BM; American College of
Obstetricians Gynecologists, : ACOG practice bulletin on diagnosing
and managing preeclampsia and eclampsia. American College of
Obstetricians and Gynecologists. Am Fam Physician. 66:330–331.
2002.PubMed/NCBI
|
|
17
|
Laginha KM, Moase EH, Yu N, Huang A and
Allen TM: Bioavailability and therapeutic efficacy of HER2
scFv-targeted liposomal doxorubicin in a murine model of
HER2-overexpressing breast cancer. J Drug Target. 16:605–610. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang Q, Graham TE, Mody N, Preitner F,
Peroni OD, Zabolotny JM, Kotani K, Quadro L and Kahn BB: Serum
retinol binding protein 4 contributes to insulin resistance in
obesity and type 2 diabetes. Nature. 436:356–362. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen CH, Hsieh TJ, Lin KD, Lin HY, Lee MY,
Hung WW, Hsiao PJ and Shin SJ: Increased unbound retinol-binding
protein 4 concentration induces apoptosis through receptor-mediated
signaling. J Biol Chem. 287:9694–9707. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu Q, Liu C, Liu Y, Zhang N, Deng H and
Zhang Z: Serum markers of pre-eclampsia identified on proteomics. J
Obstet Gynaecol Res. 42:1111–1118. 2016. View Article : Google Scholar : PubMed/NCBI
|