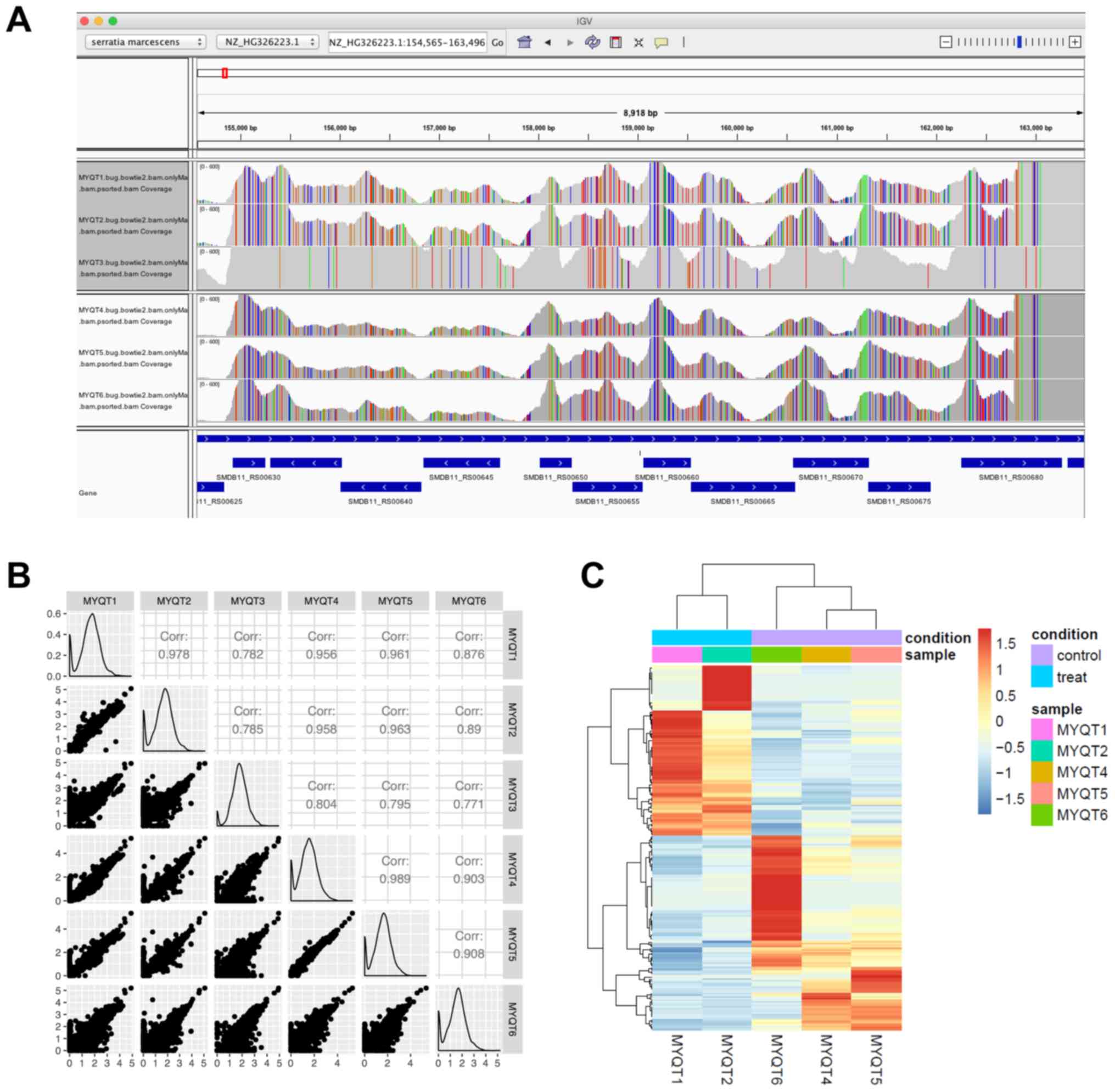
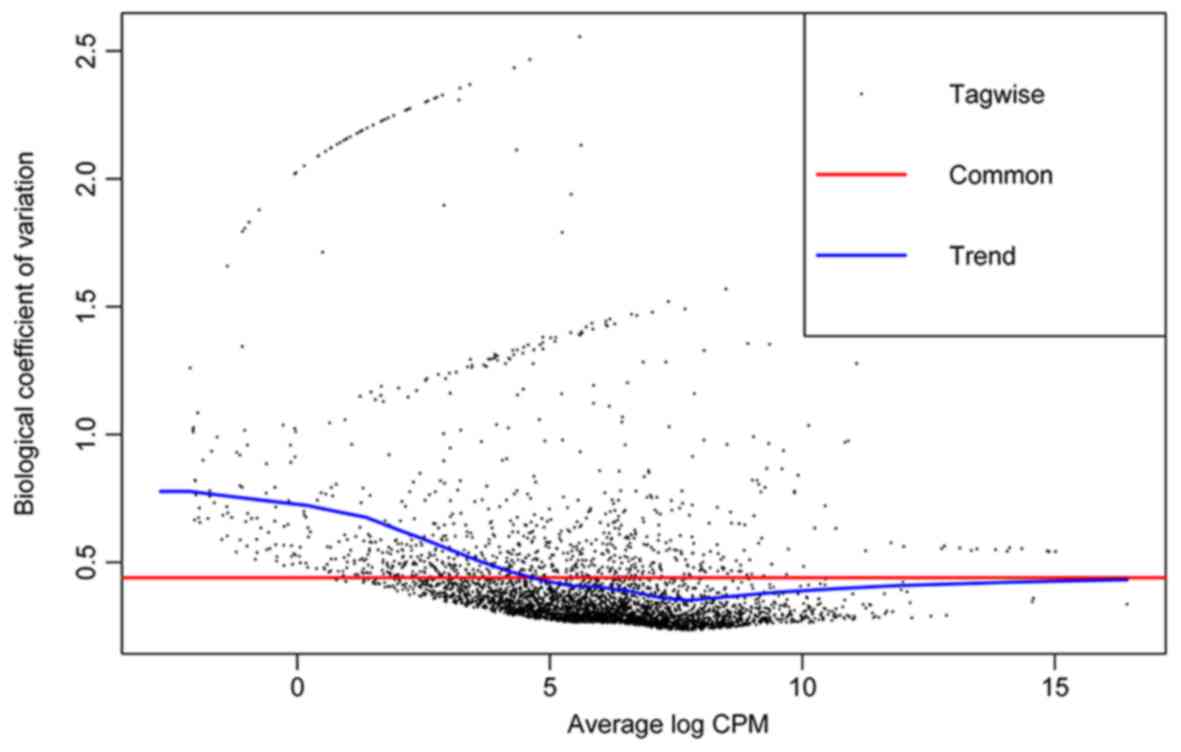
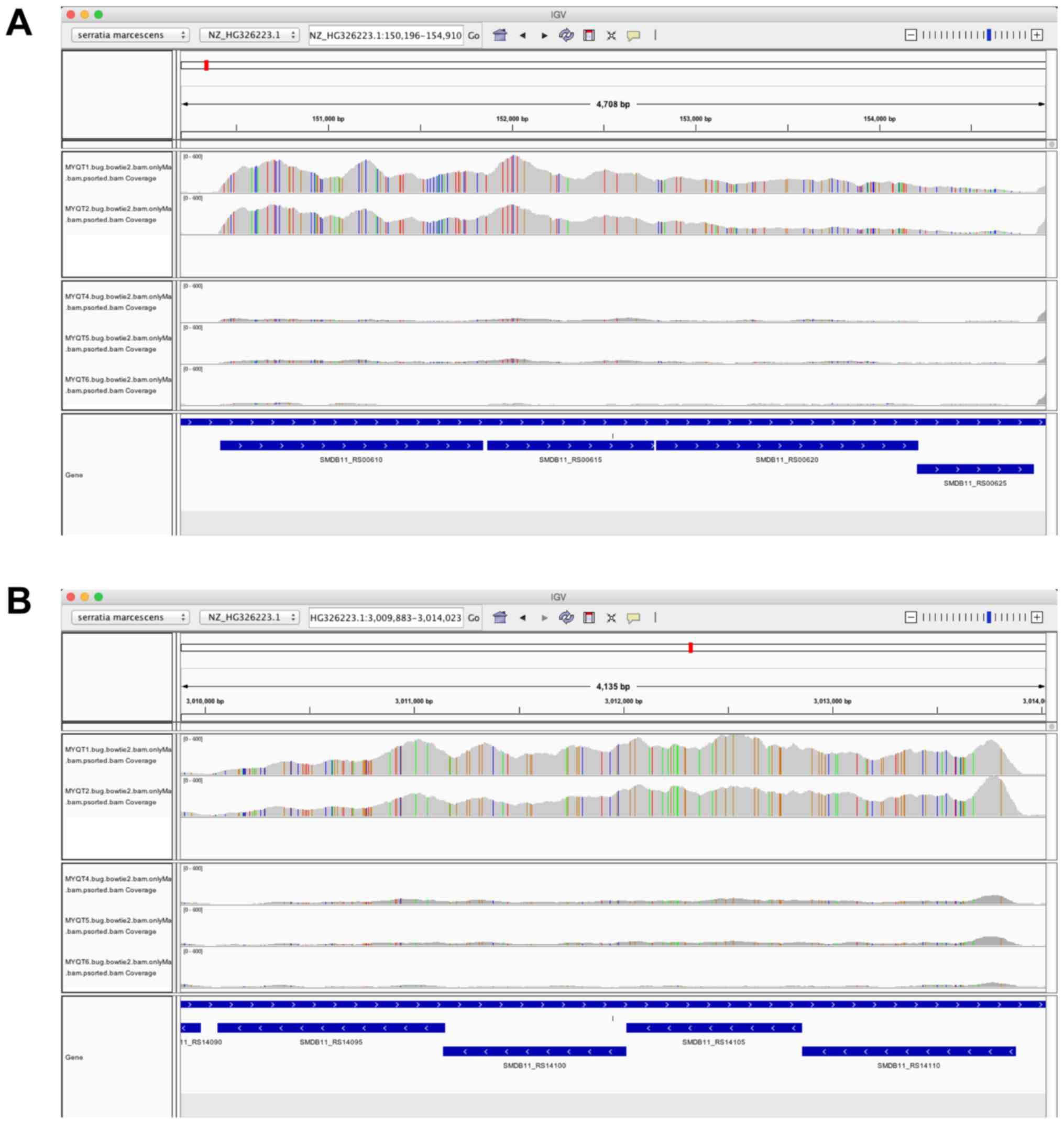
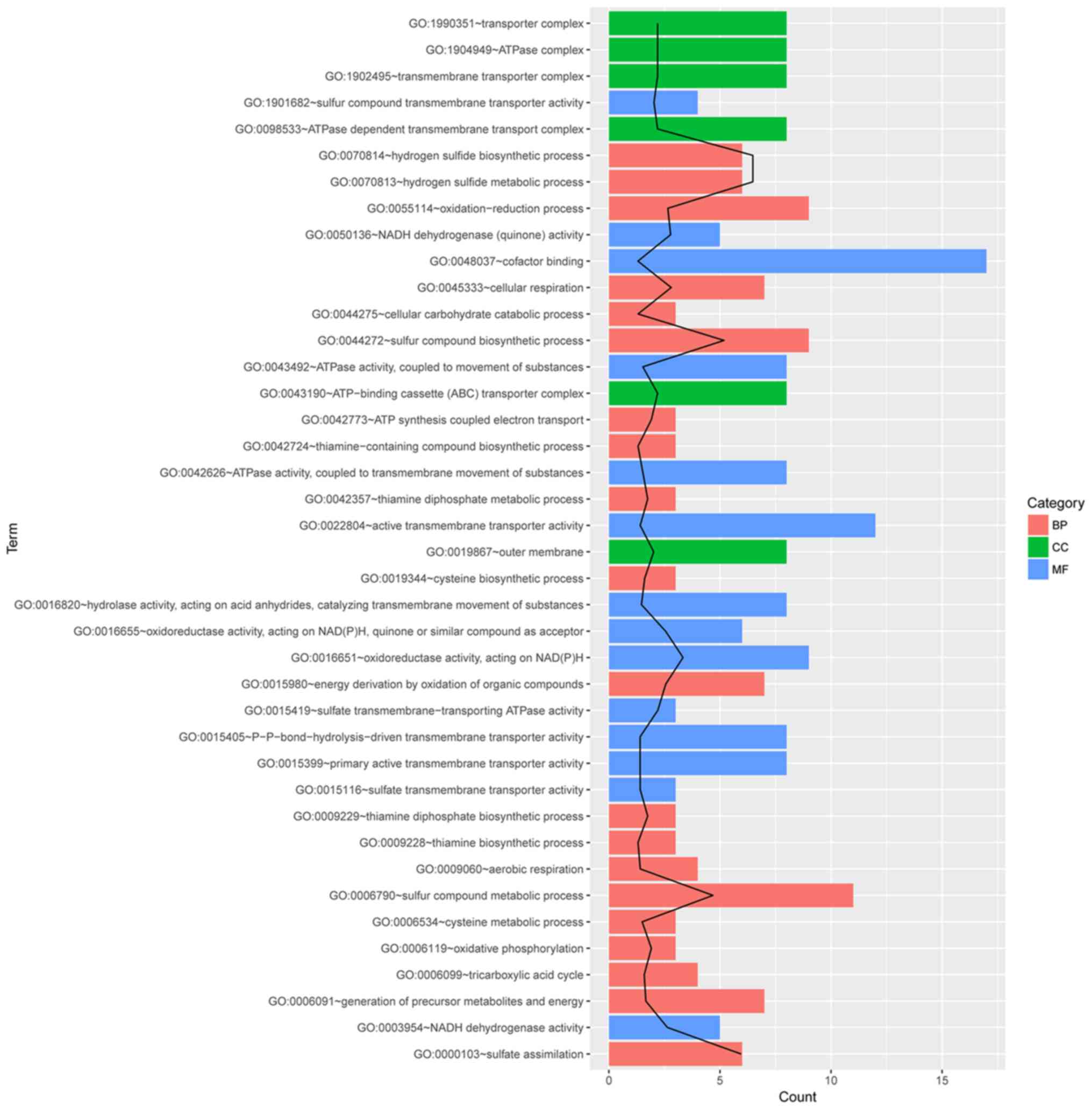
|
1
|
Courtney CM, Goodman SM, McDaniel JA,
Madinger NE, Chatterjee A and Nagpal P: Photoexcited quantum dots
for killing multidrug-resistant bacteria. Nat Mater. 15:529–534.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Iguchi A, Nagaya Y, Pradel E, Ooka T,
Ogura Y, Katsura K, Kurokawa K, Oshima K, Hattori M, Parkhill J, et
al: Genome evolution and plasticity of Serratia marcescens,
an important multidrug-resistant nosocomial pathogen. Genome Biol
Evol. 6:2096–2110. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Blair JM, Webber MA, Baylay AJ, Ogbolu DO
and Piddock LJ: Molecular mechanisms of antibiotic resistance. Nat
Rev Microbiol. 13:42–51. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alekshun MN and Levy SB: Molecular
mechanisms of antibacterial multidrug resistance. Cell.
128:1037–1050. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma XJ, Yang HF, Liu YY, Mei Q, Ye Y, Li
HR, Cheng J and Li JB: The emergence of the 16S rRNA
methyltransferase RmtB in a multidrug-resistant Serratia
marcescens isolate in China. Ann Lab Med. 35:172–174. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Su W, Zhu Y, Deng N and Li L:
Imipenem-resistance in Serratia marcescens is mediated by
plasmid expression of KPC-2. Eur Rev Med Pharmacol Sci.
21:1690–1694. 2017.PubMed/NCBI
|
|
7
|
Olaitan AO, Morand S and Rolain JM:
Mechanisms of polymyxin resistance: Acquired and intrinsic
resistance in bacteria. Front Microbiol. 5:6432014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rabbani B, Nakaoka H, Akhondzadeh S, Tekin
M and Mahdieh N: Next generation sequencing: Implications in
personalized medicine and pharmacogenomics. Mol Biosyst.
12:1818–1830. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Crofts TS, Gasparrini AJ and Dantas G:
Next-generation approaches to understand and combat the antibiotic
resistome. Nat Rev Microbiol. 15:422–434. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Crowley E, Bird P, Fisher K, Goetz K,
Boyle M, Benzinger MJ Jr, Juenger M, Agin J, Goins D and Johnson R:
Evaluation of the VITEK 2 Gram-negative (GN) microbial
identification test card: Collaborative study. J AOAC Int.
95:778–785. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McClure R, Balasubramanian D, Sun Y,
Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK and Tjaden B:
Computational analysis of bacterial RNA-Seq data. Nucleic Acids
Res. 41:e1402013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Robinson JT, Thorvaldsdóttir H, Winckler
W, Guttman M, Lander ES, Getz G and Mesirov JP: Integrative
genomics viewer. Nat Biotechnol. 29:24–26. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Quinlan AR and Hall IM: BEDTools: A
flexible suite of utilities for comparing genomic features.
Bioinformatics. 26:841–842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Robinson MD and Oshlack A: A scaling
normalization method for differential expression analysis of
RNA-seq data. Genome Biol. 11:R252010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gene Ontology Consortium: Gene ontology
consortium: Going forward. Nucleic Acids Res 43 (Database Issue).
D1049–D1056. 2015. View Article : Google Scholar
|
|
19
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res 43
(Database Issue). D447–D452. 2015. View Article : Google Scholar
|
|
20
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Opsahl T, Agneessens F and Skvoretz J:
Node centrality in weighted networks: Generalizing degree and
shortest paths. Soc Netw. 32:245–251. 2010. View Article : Google Scholar
|
|
22
|
Cukierski WJ and Foran DJ: Using
betweenness centrality to identify manifold shortcuts. Proc IEEE
Int Conf Data Min. 2008:949–958. 2008.PubMed/NCBI
|
|
23
|
Du Y, Gao C, Chen X, Hu Y, Sadiq R and
Deng Y: A new closeness centrality measure via effective distance
in complex networks. Chaos. 25:0331122015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of protein interaction networks. Biosystems. 127:67–72. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Safari-Alighiarloo N, Taghizadeh M,
Rezaei-Tavirani M, Goliaei B and Peyvandi AA: Protein-protein
interaction networks (PPI) and complex diseases. Gastroenterol
Hepatol Bed Bench. 7:17–31. 2014.PubMed/NCBI
|
|
26
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sankaran B, Bonnett SA, Shah K, Gabriel S,
Reddy R, Schimmel P, Rodionov DA, de Crécy-Lagard V, Helmann JD,
Iwata-Reuyl D and Swairjo MA: Zinc-independent folate biosynthesis:
Genetic, biochemical, and structural investigations reveal new
metal dependence for GTP cyclohydrolase IB. J Bacteriol.
191:6936–6949. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rebelo J, Auerbach G, Bader G, Bracher A,
Nar H, Hösl C, Schramek N, Kaiser J, Bacher A, Huber R and Fischer
M: Biosynthesis of pteridines. Reaction mechanism of GTP
cyclohydrolase I. J Mol Biol. 326:503–516. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Babitzke P, Gollnick P and Yanofsky C: The
mtrAB operon of Bacillus subtilis encodes GTP cyclohydrolase I
(MtrA), an enzyme involved in folic acid biosynthesis, and MtrB, a
regulator of tryptophan biosynthesis. J Bacteriol. 174:2059–2064.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bermingham A and Derrick JP: The folic
acid biosynthesis pathway in bacteria: Evaluation of potential for
antibacterial drug discovery. Bioessays. 24:637–648. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rengarajan J, Sassetti CM, Naroditskaya V,
Sloutsky A, Bloom BR and Rubin EJ: The folate pathway is a target
for resistance to the drug para-aminosalicylic acid (PAS) in
mycobacteria. Mol Microbiol. 53:275–282. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pan FL and Coote JG: Glutamine synthetase
and glutamate synthase activities during growth and sporulation in
Bacillus subtilis. J Gen Microbiol. 112:373–377. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Harth G, Zamecnik PC, Tang JY, Tabatadze D
and Horwitz MA: Treatment of Mycobacterium tuberculosis with
antisense oligonucleotides to glutamine synthetase mRNA inhibits
glutamine synthetase activity, formation of the
poly-L-glutamate/glutamine cell wall structure, and bacterial
replication. Proc Natl Acad Sci USA. 97:418–423. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Harth G and Horwitz MA: An inhibitor of
exported Mycobacterium tuberculosis glutamine synthetase
selectively blocks the growth of pathogenic mycobacteria in axenic
culture and in human monocytes: Extracellular proteins as potential
novel drug targets. J Exp Med. 189:1425–1436. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Utaida S, Dunman PM, Macapagal D, Murphy
E, Projan SJ, Singh VK, Jayaswal RK and Wilkinson BJ: Genome-wide
transcriptional profiling of the response of Staphylococcus aureus
to cell-wall-active antibiotics reveals a cell-wall-stress
stimulon. Microbiology. 149:2719–2732. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schnell R, Sandalova T, Hellman U,
Lindqvist Y and Schneider G: Siroheme- and [Fe4-S4]-dependent NirA
from Mycobacterium tuberculosis is a sulfite reductase with a
covalent Cys-Tyr bond in the active site. J Biol Chem.
280:27319–27328. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bhattacharyya S, Saha S, Giri K, Lanza IR,
Nair KS, Jennings NB, Rodriguez-Aguayo C, Lopez-Berestein G, Basal
E, Weaver AL, et al: Cystathionine beta-synthase (CBS) contributes
to advanced ovarian cancer progression and drug resistance. PLoS
One. 8:e791672013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bisht R, Katiyar A, Singh R and Mittal P:
Antibiotic resistance-A global issue of concern. Asian J Pharm Clin
Res. 2:34–39. 2009.
|
|
39
|
Bhave DP, Muse WB III and Carroll KS: Drug
targets in mycobacterial sulfur metabolism. Infect Disord Drug
Targets. 7:140–158. 2007. View Article : Google Scholar : PubMed/NCBI
|