Introduction
Necrosis of the femoral head (NFH) is a common
disease of the hip, with a high incidence among elderly patients;
the most common clinical symptom is severe pain (1). The pathological characteristics of
NFH include reduced blood supply to the hip, collapse of the
femoral head and microfracture accumulation without sustained
remodeling (2,3). Early clinical symptoms may involve
pain, ultimately leading to loss of movement in the hip (4). NFH is frequently treated by total hip
arthroplasty in the end-stage of hip arthritis (5); however, the pathogenesis and
molecular mechanisms underlying NFH remain unclear.
Numerous studies have focused on the etiology of
NFH. Huang et al (6)
reported that fibroblast growth factor 2 (FGF2) and family with
sequence similarity 201 member A were associated with the
development of NFH, and that insulin-like growth factor 1, SOX9 and
collagen type II α1 may also affect the pathogenesis of NFH. The
signal transducer and activator of transcription (STAT)1-caspase 3
pathway upregulated the expression of caspase 3, resulting in
apoptosis in NFH (7). Tian et
al (8) demonstrated that NFH
was associated with the Toll-like receptor 4 signaling pathway,
which may serve an important role in the pathogenesis of
osteonecrosis. MicroRNAs (miRs) are also notable diagnostic markers
and therapeutic targets of NFH. In a study by Li et al
(9), has-miR-195-5p exhibited
notable downregulation during the collapse of osteonecrotic femoral
heads, suggesting that the collapse may be associated with the
downregulation of miR-195-5p. Wei et al (10) revealed that the long non-coding RNA
Hox antisense intergenic RNA may inhibit miR-17-5p to regulate
osteogenic differentiation and proliferation in the osteonecrosis
of the femoral head. Ma et al (11) revealed that Runt-related
transcription factor 2 and transcription factor Sp7 were
downregulated in a rat model of NFH, whereas AJ18 was
upregulated.
Microarray analysis using high-throughput platforms
is a promising and efficient tool for the investigation of the
molecular mechanisms of disease, and the identification of useful
biomarkers for the diagnosis and prognosis of disease. Lin and Lin
(12) reported 215 differentially
expressed genes (DEGs) based on gene expression profiles generated
from 3 steroid-induced samples from a rat model of NFH and 3 normal
rat samples. Tong et al (13) revealed 190 DEGs in a rat model of
NFH, 52 of which were upregulated and 138 downregulated. Biological
functions identified from enrichment analysis of DEGs included
signal transduction, apoptosis, extracellular matrix (ECM),
angiogenesis and oxidative stress.
In the present study, DEGs were identified in
patients with NFH, and associated pathways were analyzed to
identify the underlying molecular mechanisms of this disease. Gene
expression profiles in the cartilage of patients with NFH and
healthy individuals were acquired from the National Center of
Biotechnology Information (NCBI) Gene Expression Omnibus (GEO)
database and compared. The GSE74089 microarray dataset was analyzed
using R software, Bioconductor packages and the Database for
Annotation, Visualization and Integrated Discovery (DAVID) 6.8
online resource. A protein-protein interaction (PPI) network of
DEGs was then constructed in order to analyze the putative hub
genes of NFH.
Materials and methods
Agilent microarray data processing and
gene expression profile mining
The microarray data of GSE74089 (14) in NFH was obtained from the NCBI GEO
database (15). GSE74089 contained
data from cartilage samples from patients with NFH and healthy
controls. The microarray data of GSE74089 were obtained using
GPL13497 (Agilent-026652 Whole Human Genome Microarray 4×44K v2;
Agilent Technologies, Inc., Santa Clara, CA, USA); the data were
based on cartilage samples from 4 patients with NFH and 4
controls.
The pre-processing of gene expression profile data
was performed using R software (version 3.4.0; https://www.r-project.org) and Bioconductor packages
3.8 (https://www.bioconductor.org/) for
data analysis. Via the Agilent platform, R software was used to
analyze the pre-processing and normalization of Series Matrix Files
(.TXT files). The parameters used in the R software included robust
multi-array average (for background correction), quantiles (for
normalization), perfect match (PM)-only (PM correction) and median
polish (as a summary measure).
Identification of DEGs
The Linear Models for Microarray Data 3.8 (LIMMA,
http://www.bioconductor.org/packages/release/bioc/html/limma.html)
package (16) in Bioconductor was
employed to evaluate DEGs by comparing the expression values in the
cartilage of patients with NFH and controls. The corresponding
P-value of gene symbols following a t-test was defined as the
adjusted P-value; log2 fold change >2 and P<0.01 were
considered to be the cut-off criteria for DEGs.
Enrichment analysis of DEGs
DAVID 6.8 was employed for enrichment analysis, in
order to investigate DEGs at the molecular and functional level
(17,18). DAVID is a gene functional
classification tool, which provides typical batch annotation and
gene-Gene Ontology (GO) term enrichment analysis to highlight the
most relevant GO terms associated with a specific gene list. DAVID
was employed for GO function and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analyses of DEGs in NFH (19–21).
GO terms included ‘cellular component (CC)’, ‘molecular function
(MF)’ and ‘biological process (BP)’; P<0.05 was set as the
cut-off for enrichment analysis.
Analysis of PPI networks
Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING; http://string-db.org) (22,23)
is a database that predicts the PPIs of DEGs. According to the
STRING database, PPIs of DEGs with a score (median confidence) of
>0.4 were selected, and Cytoscape (Version 3.4.0, available
online: http://www.cytoscape.org/) was used to
analyze the PPI network (24).
Hub-proteins are significant nodes of protein interaction within
the PPI network (25). To
characterize hubs in the PPI network of DEGs, betweenness
centrality, degree centrality, Maximum Neighborhood Component (MNC)
centrality and stress centrality were evaluated.
Results
Identification of DEGs in NFH
To identify DEGs in the cartilage of NFH and healthy
patients, the transcription profile data of GSE74089 were obtained
from the NCBI GEO database based on 4 patients with NFH and 4
healthy controls. According to the cut-off criteria, 1,191 DEGs
were identified in the cartilage of patients with NFH compared with
the controls, including 448 downregulated and 743 upregulated DEGs.
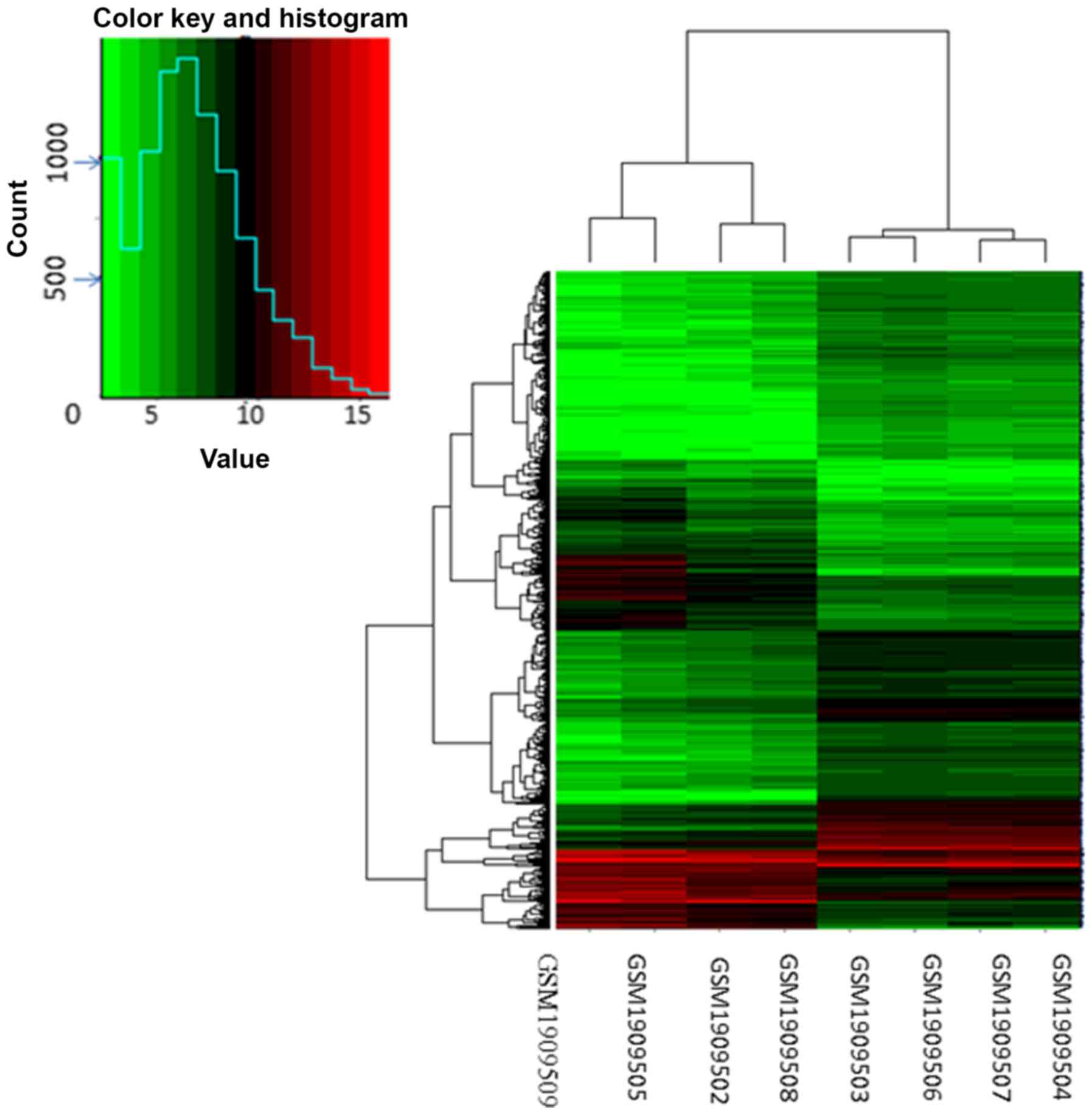
DEGs in NFH samples were identified using hierarchical cluster
analysis of the data (Fig. 1).
Enrichment analysis of DEGs
GO functional enrichment analysis revealed that the
BPs of upregulated DEGs included ‘ECM organization’, ‘in
utero embryonic development’, ‘collagen catabolic processes’,
‘collagen fibril organization’ and ‘angiogenesis’ (Table I). CC analysis revealed that
upregulated DEGs were primarily enriched in ‘proteinaceous ECM’,
‘collagen trimer’ and ‘ECM’. The MFs of upregulated DEGs were
demonstrated to include ‘protein binding’, ‘platelet-derived growth
factor binding’, ‘ubiquitin-protein transferase activity’, ‘ECM
structural constituent’ and ‘oxidoreductase activity’. The BPs of
downregulated DEGs included ‘antigen processing and the
presentation of peptide or polysaccharide antigen via major
histocompatibility complex (MHC) class II’, and ‘immunoglobulin
(Ig) production associated with Ig-mediated immune response’, and
‘antigen processing and presentation of exogenous peptide antigen
via MHC class II’ (Table II).
Additionally, CC enrichment analysis revealed downregulated DEGs to
be mainly associated with ‘MHC class II protein complex’, ‘integral
component of the lumenal side of the endoplasmic reticulum
membrane’ and ‘endocytic vesicle membrane’. The MFs of
downregulated DEGs were demonstrated to included ‘MHC class II
receptor activity’, ‘peptide antigen binding’, ‘MHC class II
protein complex binding’, ‘monooxygenase activity’ and
‘N,N-dimethylaniline monooxygenase activity’. According to KEGG
pathway enrichment analysis, the downregulated DEGs were mainly
enriched in pathways involved in staphylococcus aureus
infection, asthma and graft-versus-host disease. The upregulated
DEGs were mainly enriched in pathways involved in focal adhesion,
phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) signaling,
pathways in cancer and ECM-receptor interactions (Table III).
 | Table I.Top 5 terms identified by GO
enrichment analysis for the upregulated DEGs in necrosis of the
femoral head samples. |
Table I.
Top 5 terms identified by GO
enrichment analysis for the upregulated DEGs in necrosis of the
femoral head samples.
| Category | Term | Description | Count | P-value | Genes |
|---|
| BP | GO:0030574 | Collagen catabolic
process | 14 |
2.87×10−7 | COL18A1, COL13A1,
COL3A1, COL15A1, COL2A1, MMP13, COL5A2, … |
|
| GO:0030198 | Extracellular
matrix organization | 24 |
4.46×10−7 | COL18A1, PXDN,
COL13A1, LUM, COL3A1, CCDC80, DAG1, … |
|
| GO:0030199 | Collagen fibril
organization | 11 |
6.94×10−7 | LUM, TGFBR1,
COL3A1, COL1A2, COL2A1, COL1A1, LOX, LOXL2, … |
|
| GO:0001701 | In utero embryonic
development | 23 |
7.58×10−7 | SRSF1, MAFF, CCM2,
BMP2, ADAM10, TGFBR1, GJA1, UBR3, … |
|
| GO:0001525 | Angiogenesis | 23 |
1.38×10−5 | PRKCA, SLC12A6,
COL18A1, PTGS2, COL15A1, RORA, ECM1, THY1, … |
| CC | GO:0031012 | Extracellular
matrix | 37 |
3.37×10−11 | ASPN, PXDN, LTBP1,
LUM, IGFBP7, COL3A1, POSTN, COL2A1, … |
|
| GO:0005578 | Proteinaceous
extracellular matrix | 30 |
4.00×10−8 | ASPN, CTHRC1, PXDN,
LTBP1, AMTN, MAMDC2, LUM, POSTN, … |
|
| GO:0005581 | Collagen
trimer | 17 |
7.43×10−8 | COL18A1, CTHRC1,
COL13A1, COL3A1, COL15A1, COL2A1, MMP13, … |
|
| GO:0070062 | Extracellular
exosome | 141 |
1.05×10−6 | S100A4, TSPO,
RARRES1, FSTL1, AQP1, PNP, OGN, HMCN1, DYSF, … |
|
| GO:0005615 | Extracellular
space | 80 |
1.20×10−6 | S100A4, COPA,
CTHRC1, PXDN, IGFBP7, FSTL1, POSTN, IL11, OGN, … |
| MF |
|
| GO:0005515 | Protein
binding | 361 |
2.83×10−7 | S100A4, XRCC4,
LTBP1, PTGS2, SNIP1, LEMD3, PTPN21, FSTL1, … |
|
| GO:0048407 | Platelet-derived
growth factor binding | 6 |
1.80×10−5 | COL3A1, COL1A2,
COL6A1, COL2A1, COL1A1, COL5A1 |
|
| GO:0004842 | Ubiquitin-protein
transferase activity | 28 |
2.64×10−5 | KBTBD13, FEM1B,
KLHL2, LNX1, ZYG11B, FBXW7, UBE2D2, KLHL24, … |
|
| GO:0005201 | Extracellular
matrix structural constituent | 11 |
8.76×10−5 | PXDN, LUM, COL3A1,
COL1A2, COL15A1, COL2A1, VCAN, COL1A1, … |
|
| GO:0016641 | Oxidoreductase
activity | 4 |
3.79×10−4 | LOXL4, LOXL3, LOX,
LOXL2 |
 | Table II.Top 5 terms identified by GO
enrichment analysis for the downregulated DEGs in necrosis of the
femoral head samples. |
Table II.
Top 5 terms identified by GO
enrichment analysis for the downregulated DEGs in necrosis of the
femoral head samples.
| Category | Term | Description | Count | P-value | Genes |
|---|
| BP | GO:0002504 | Antigen processing
and presentation of peptide or polysaccharide antigen via MHC class
II | 9 |
8.90×10−11 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, HLA-DRB4, HLA-DRB5, … |
|
| GO:0002381 | Immunoglobulin
production involved in immunoglobulin mediated immune response | 5 |
3.28×10−7 | GAPT, HLA-DQB1,
HLA-DRB1, HLA-DRB4, HLA-DRB5 |
|
| GO:0019882 | Antigen processing
and presentation | 9 |
2.62×10−6 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, HLA-DRB4, HLA-DRB5, … |
|
| GO:0019886 | Antigen processing
and presentation of exogenous peptide antigen via MHC class II | 10 |
1.76×10−5 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, SPTBN2, HLA-DRB4, … |
|
| GO:0060333 |
Interferon-γ-mediated signaling
pathway | 9 |
1.83×10−5 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, HLA-DRB4, HLA-DRB5, … |
| CC | GO:0042613 | MHC class II
protein complex | 9 |
1.58×10−9 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, HLA-DRB4, HLA-DRB5, … |
|
| GO:0071556 | Integral component
of lumenal side of endoplasmic reticulum membrane | 8 |
4.17×10−7 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, HLA-DRB4, SPPL2B, … |
|
| GO:0030666 | Endocytic vesicle
membrane | 9 |
1.49×10−5 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, WNT3A, HLA-DRB4, … |
|
| GO:0012507 | ER to Golgi
transport vesicle membrane | 8 |
2.56×10−5 | HLA-DQB1, HLA-DRB1,
FOLR1, HLA-DRB3, HLA-DRB4, … |
|
| GO:0030658 | Transport vesicle
membrane | 7 |
3.89×10−5 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, HLA-DRB4, HLA-DRB5, … |
| MF | GO:0032395 | MHC class II
receptor activity | 7 |
7.34×10−8 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, HLA-DRB4, HLA-DOA, … |
|
| GO:0042605 | Peptide antigen
binding | 7 |
4.63×10−6 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, HLA-DRB4, HLA-DRB5, … |
|
| GO:0023026 | MHC class II
protein complex binding | 4 | 0.001983 | HLA-DRB1, HLA-DMB,
HLA-DOA, HLA-DRA |
|
| GO:0004497 | Monooxygenase
activity | 6 | 0.00239 | FMO1, FMO2, FMO6P,
CYP4F22, CYP2E1, CYP4B1 |
|
| GO:0004499 | N,N-dimethylaniline
monooxygenase activity | 3 | 0.003718 | FMO1, FMO2,
FMO6P |
 | Table III.Results of KEGG pathway enrichment
analysis of differentially expressed genes. |
Table III.
Results of KEGG pathway enrichment
analysis of differentially expressed genes.
| Category | Term | Description | Count | P-value | Genes |
|---|
| Upregulated
genes | hsa04510 | Focal adhesion | 28 |
1.44×10−8 | COL3A1, COL2A1,
ITGB8, SOS2, COL6A3, PPP1R12A, COL6A1, PDGFC, … |
|
| hsa04151 | PI3K-Akt signaling
pathway | 33 |
2.29×10−6 | PPP2R3A, STK11,
COL3A1, COL2A1, GNG12, PKN3, ITGB8, SOS2, … |
|
| hsa04512 | ECM-receptor
interaction | 14 |
2.24×10−5 | COL3A1, DAG1,
COL2A1, COL5A2, COL5A1, ITGB8, COL6A3, COL1A2, … |
|
| hsa04974 | Protein digestion
and absorption | 14 |
2.54×10−5 | COL18A1, COL13A1,
SLC16A10, COL3A1, COL15A1, COL2A1, COL5A2, … |
|
| hsa05200 | Pathways in
cancer | 31 |
1.96×10−4 | GNAI3, ADCY7,
PTGS2, EGLN3, BDKRB1, GNG12, GLI3, MMP1, GLI1, … |
| Downregulated
genes | hsa05150 | Staphylococcus
aureus infection | 15 |
3.71×10−14 | HLA-DQB1, C3AR1,
HLA-DRB1, C4B, HLA-DRB3, HLA-DMB, ITGAM, … |
|
| hsa05310 | Asthma | 9 |
1.12×10−8 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, HLA-DRB4, HLA-DRB5, HLA-DMB, … |
|
| hsa05332 | Graft-versus-host
disease | 9 |
2.56×10−8 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, HLA-DRB4, HLA-DRB5, HLA-DMB, … |
|
| hsa05330 | Allograft
rejection | 9 |
6.76×10−8 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, HLA-DRB4, HLA-DRB5, HLA-DMB, … |
|
| hsa04940 | Type I diabetes
mellitus | 9 |
1.94×10−7 | HLA-DQB1, HLA-DRB1,
HLA-DRB3, HLA-DRB4, HLA-DRB5, HLA-DMB, … |
PPI network analysis is important for understanding
the biological responses of NFH. The STRING online database and
Cytoscape software were employed to analyze the identified DEGs.
The top 20 upregulated genes were evaluated for MNC centrality,
betweenness centrality, stress centrality and degree centrality in
the PPI network (Table IV). Based
on various centrality, vascular endothelial growth factor A (VEGFA)
was the most notable gene in the PPI network. Jun proto-oncogene
(JUN), cyclin D1 (CCND1), FGF2, HECT domain and ankyrin
repeat-containing E3 ubiquitin protein ligase 1 (HACE1), protein
kinase Cα (PRKCA), bone morphogenetic protein (BMP) 2 and
prostaglandin-endoperoxide synthase 2 (PTGS2) were within the top 5
genes of at least one of the centrality rankings. The significant
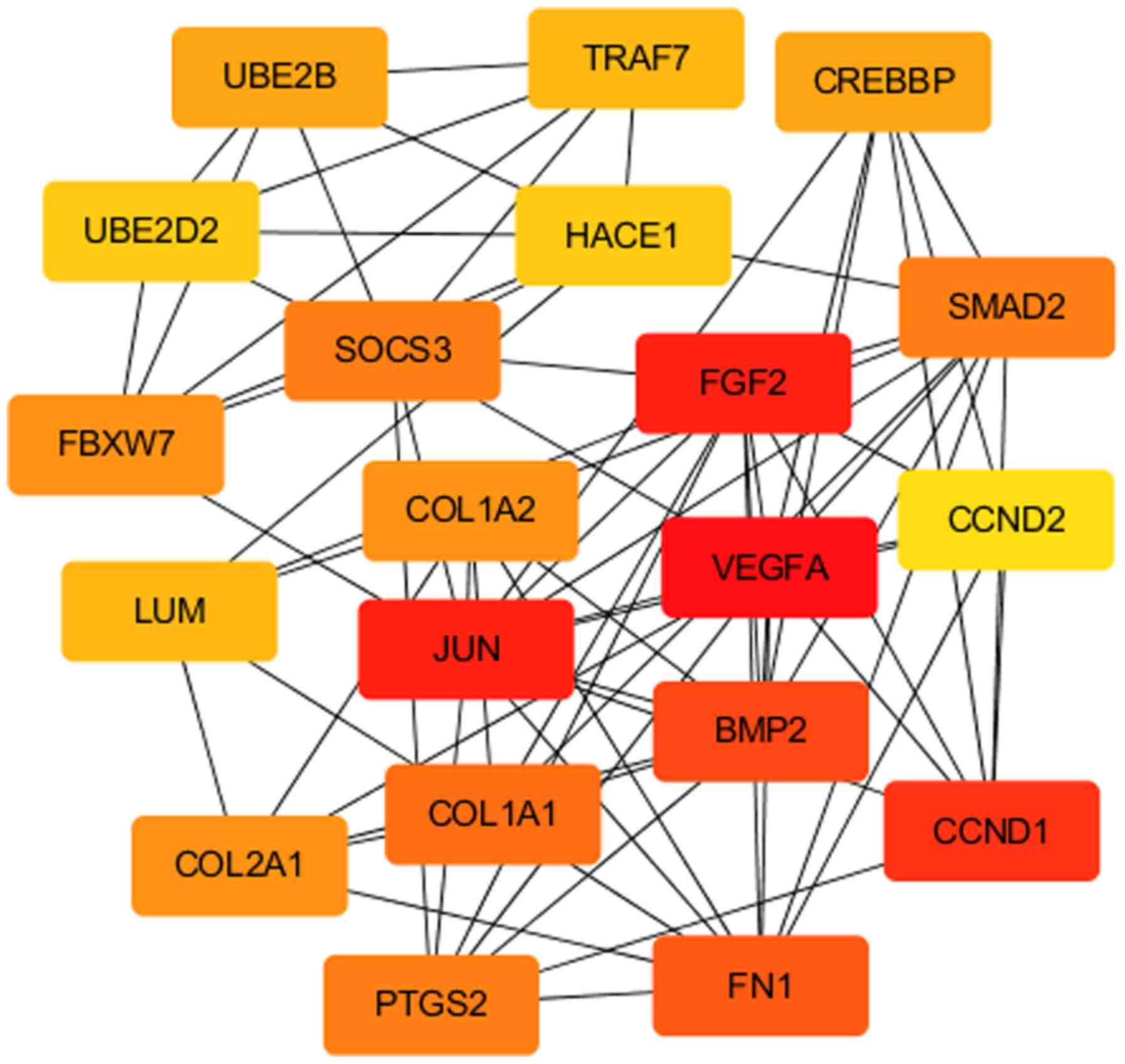
network module generated based on MNC centrality is presented in
Fig. 2. The top 15 downregulated
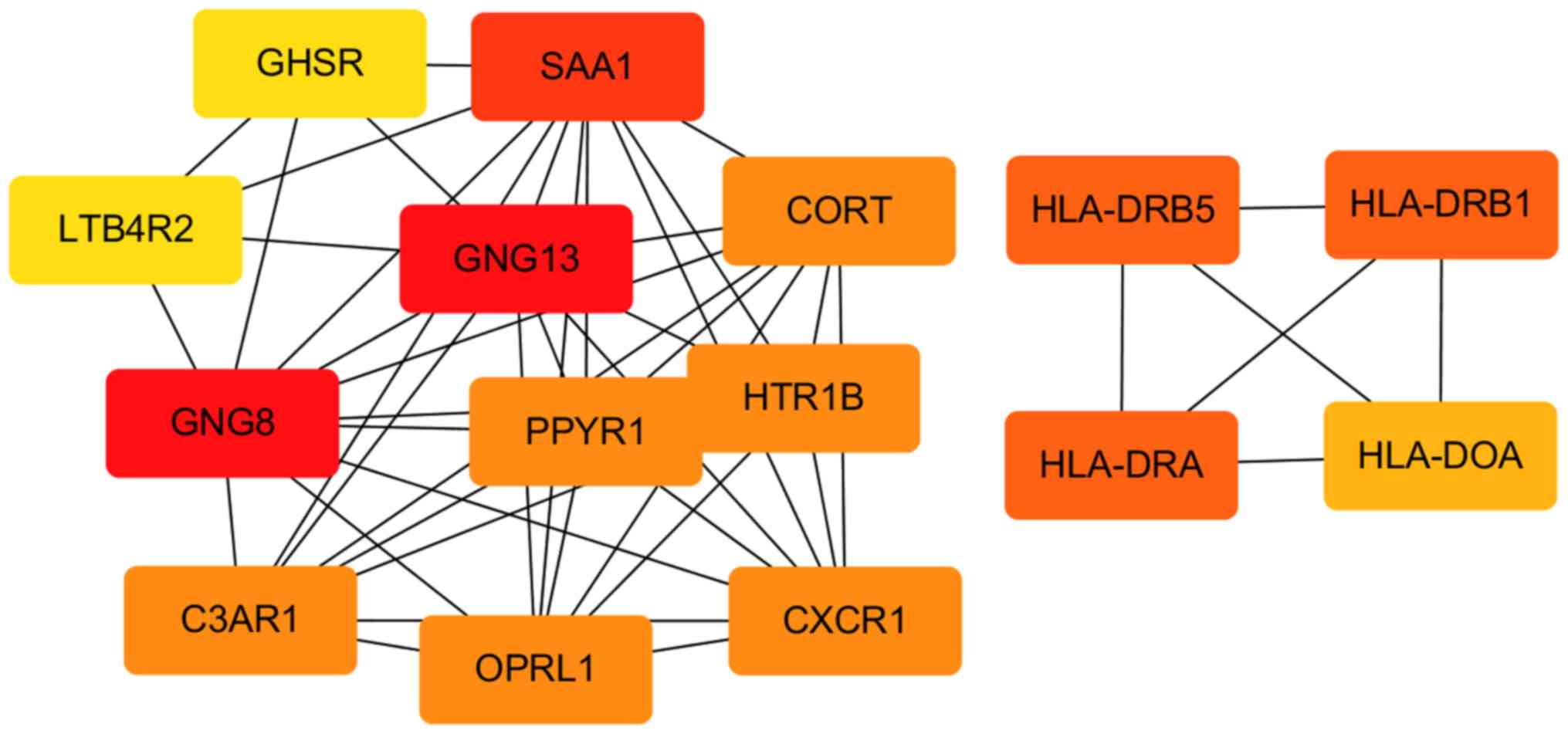
DEGs were analyzed in the PPI network (Table V). Guanine nucleotide-binding
protein, γ13 (GNG13) was the most common gene based on various
centrality. According to the MNC centrality, the significant
network module is presented in Fig.
3.
 | Table IV.Evaluation of the top 20 upregulated
DEGs of the protein-protein interaction network by MNC centrality,
betweenness centrality, stress centrality and degree
centrality. |
Table IV.
Evaluation of the top 20 upregulated
DEGs of the protein-protein interaction network by MNC centrality,
betweenness centrality, stress centrality and degree
centrality.
| Rank | MNC centrality | Betweenness
centrality | Stress
centrality | Degree
centrality |
|---|
| 1 | VEGFA | VEGFA | VEGFA | VEGFA |
| 2 | JUN | CCND1 | JUN | JUN |
| 3 | FGF2 | JUN | CCND1 | FGF2 |
| 4 | CCND1 | PRKCA | FGF2 | CCND1 |
| 5 | BMP2 | PTGS2 | HACE1 | HACE1 |
| 6 | FN1 | FGF2 | PTGS2 | BMP2 |
| 7 | COL1A1 | HACE1 | PRKCA | FN1 |
| 8 | PTGS2 | CREBBP | SMAD2 | PTGS2 |
| 9 | SMAD2 | SMAD2 | FN1 | COL1A1 |
| 10 | SOCS3 | FN1 | BMP2 | SMAD2 |
| 11 | COL2A1 | BMP2 | CREBBP | CREBBP |
| 12 | FBXW7 | CD55 | LUM | SOCS3 |
| 13 | COL1A2 | SOCS3 | SOCS3 | COL2A1 |
| 14 | CREBBP | TJP1 | JAK2 | FBXW7 |
| 15 | UBE2B | LUM | CDK9 | PRKCA |
| 16 | LUM | JAK2 | NT5E | UBE2B |
| 17 | TRAF7 | NOTCH3 | TGFBR1 | LUM |
| 18 | HACE1 | TGFBR1 | NOTCH3 | UBE2N |
| 19 | UBE2D2 | NT5E | GNAI3 | COL1A2 |
| 20 | GNAI3 | CDK9 | CD55 | GNAI3 |
 | Table V.Evaluation of the top 15
downregulated DEGs of the protein-protein interaction network by
MNC centrality, betweenness centrality, stress centrality and
degree centrality. |
Table V.
Evaluation of the top 15
downregulated DEGs of the protein-protein interaction network by
MNC centrality, betweenness centrality, stress centrality and
degree centrality.
| Rank | MNC centrality | Betweenness
centrality | Stress
centrality | Degree
centrality |
|---|
| 1 | GNG13 | GNG13 | PTAFR | GNG13 |
| 2 | GNG8 | PTAFR | GNG13 | GNG8 |
| 3 | SAA1 | CTSH | CTSH | SAA1 |
| 4 | CXCR1 | SFTPB | HLA-DRA | PTAFR |
| 5 | OPRL1 | HLA-DRA | HLA-DRB5 | HLA-DRA |
| 6 | C3AR1 | HLA-DRB5 | HLA-DRB1 | HLA-DRB5 |
| 7 | CORT | HLA-DRB1 | SFTPB | HLA-DRB1 |
| 8 | PPYR1 | TYROBP | GNG8 | CXCR1 |
| 9 | HTR1B | GNG8 | SAA1 | TYROBP |
| 10 | PTAFR | C1QB | SPTBN2 | OPRL1 |
| 11 | XCR1 | SPTBN2 | TYROBP | C3AR1 |
| 12 | GHSR | SAA1 | XCR1 | CORT |
| 13 | LTB4R2 | ITGAM | C1QB | PPYR1 |
| 14 | HLA-DRA | XCR1 | ITGAM | HTR1B |
| 15 | HLA-DRB5 | CD163 | CD163 | XCR1 |
Discussion
NFH is a destructive bone disease, mainly induced by
disruption of the blood supply and the dysfunction of the
coagulation and fibrinolysis systems, which lead to the collapse of
the femoral head (26,27). Various molecular and genetic
studies have investigated the causes of the disease (28,29);
however, the exact pathogenic mechanisms remain unclear. A
genome-wide approach was employed to investigate differential gene
expression in cartilage samples from patients with NFH and healthy
controls. GSE74089 was analyzed to identify potentially important
genes in NFH using bioinformatics analysis. The roles and
interactions of the identified DEGs in NFH were also determined. A
total of 1,191 DEGs were identified in cartilage samples from
patients with NFH compared with the control, 448 of which were
downregulated and 743 of which were upregulated. These DEGs may
serve as important biomarkers with mechanistic relevance to the
pathogenesis and progression of NFH.
The DEGs reported in the present study could aid the
identification of novel molecules or pathways involved in NFH that
may serve as targets in the diagnosis and treatment of the disease,
and provide novel insight into its pathogenesis. Upregulated DEGs
were involved in the organization of the ECM, collagen catabolism
and fibril organization, in utero embryonic development and
angiogenesis. The genes were mainly enriched in proteinaceous ECM,
ECM and collagen trimers for CC enrichment. Liu et al
(14), the study in which the
GSE47089 microarray data were obtained, reported that DEGs in NFH
were enriched in growth factors, cytokines, and proteins involved
in cell cycle, platelet-derived growth factor binding, the ECM and
apoptosis; these DEGs were enriched in collagen, the ECM and
extracellular regions and platelet-derived growth factor binding.
These findings were consistent with the results of the present
study. Upregulated DEGs were mainly involved in PI3K-Akt signaling
pathway, focal adhesion and ECM-receptor interactions.
Downregulated DEGs were mainly enriched in pathways involved in
staphylococcus aureus infection, asthma and
graft-versus-host disease. Focal adhesion kinase (FAK) is a
non-receptor tyrosine kinase associated with a number of different
signaling proteins (30). Zhang
et al (31) reported that
CXC chemokine ligand 13 (CXCL13)/CXC chemokine receptor 5
(CXCR5)/FAK signaling is involved in the differentiation and
trafficking of bone marrow stromal cells (BMSCs) in NFH;
CXCL13/CXCR5 signaling was proposed to induce the phosphorylation
of FAK via the mitogen-activated protein kinase (MAPK) pathway
(32). The PI3K/Akt pathway is
associated with various fundamental cellular processes, including
survival, proliferation, growth and differentiation (33). Xue et al (34) reported that Salidroside alleviated
the dexamethasone-induced apoptosis of osteoblasts by
downregulating caspase-3 and activating the PI3K/Akt signaling
pathway in osteoblasts. In addition, the osteogenic
differentiation-inducing and bone regenerative properties of
graphene-incorporated poly(lactic-co-glycolic acid) were mediated
via the activation of the PI3K/Akt/glycogen synthase kinase
3β/β-catenin signaling pathway (35).
PPI networks of DEGs were constructed using STRING
and Cytoscape to investigate the associations between important
proteins identified by GO enrichment and pathway analyses (36). The upregulated DEGs included VEGFA,
JUN, CCND1, FGF2, HACE1, PRKCA, BMP2 and PTGS2. In the early stages
of a rabbit model of NFH, BMP and VEGF were co-expressed using an
adeno-associated virus (AAV) vector; the AAV–VEGF/BMP vector
increased the bone repair capacity of the femoral head by inducing
angiogenesis and improving bone quality (37). VEGF-expressing transgenic
autologous BMSCs improved bone reconstruction and blood vessel
regeneration in a canine model of NFH (38). Adenovirus-mediated expression of
BMP2 and basic FGF in BMSCs in combination with a demineralized
bone matrix improved bone formation in a dog model of NFH (39). CCND1 is an important regulatory
factor of the cell cycle and is a frequently used biomarker for the
diagnosis and prognosis of human primary tumors (40). Phosphorylated CCND1 is associated
with the development of osteosarcoma, which may occur via
MAPK-induced expression of CCND1, and the continuous proliferation
of tumor cells (41).
Downregulated DEGs identified in the present study
included GNG13, platelet-activating factor receptor (PAFR), GNG8,
serum amyloid A1 and cathepsin H. PPI analysis identified GNG13 as
a central downregulated DEG. GNG13 is a divergent member of the GNG
subunit γ family and contains a C-terminal NPW tripeptide (42). It is a component of the gustducin
G-protein heterotrimer involved in bitter and sweet taste reception
in taste bud cells (42). PAF is a
potent phospholipid regulator of inflammation; PAFR is expressed on
plasma and nuclear membranes in various cell types, and binds to
PAF and oxidized phospholipids (43). Activation of PAFR in macrophages
induces an anti-inflammatory phenotype (44). PAF activates PAFR, which may serve
an important role in the malignant development of esophageal
squamous cell carcinoma by stimulating PI3K/AKT activation, and
promoting disease progression and metastasis via the initiation of
a forward feedback loop between PAFR and STAT3 (45).
There were certain limitations to the present study.
Gene expression data were obtained from only a single dataset
containing 4 patients with NFH and 4 controls. The use of
additional datasets with increased sample sizes in future studies
would increase the accuracy and reliability of identified DEGs.
Additionally, the potential role of DEGs and PPIs identified in the
hip cartilage of patients with NFH require further investigation
in vivo and in vitro; for example, the effects of
silencing or upregulating DEGs in cellular or in vivo models
could be determined. Furthermore, the altered expression of genes
and proteins identified by microarray analysis should be
investigated via reverse transcription-quantitative polymerase
chain reaction and histological analyses of samples from patients
with NFH. These experiments may validate the diagnostic and
prognostic potential of identified DEGs for the disease.
In conclusion, following integrated bioinformatical
analysis of the gene expression profiles of cartilage from patients
with NFH and controls, 1,191 DEGs were identified in necrotic
samples, 743 of which were upregulated and 448 were downregulated.
DEG enrichment analysis identified molecules and pathways, which
may provide novel insight into the pathogenesis of NFH. PPI network
analysis identified VEGFA, JUN, CCND1, FGF2, HACE1, PRKCA, BMP2 and
PTGS2, and GNG13 as central upregulated and downregulated DEGs,
respectively. These DEGs and signaling pathways may serve as
biomarkers and targets in the treatment of NFH; however, further
investigation is required.
Acknowledgements
Not applicable.
Funding
This study was supported by Clinical Support
Foundation of Chinese PLA General Hospital (grant nos.
2017FC-TSYS-3015 and 2017FC-TSYS-2043), Nursery Foundation of
Chinese PLA General Hospital (grant no. 17KMM12), Beijing Natural
Science Foundation (grant no. 7192196) and National Natural Science
Foundation of China (grant no. 81702169).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
WCL made substantial contributions towards the
conception and design of the study, and experiments. DLB and YX
performed the major bioinformatics analysis of the gene database.
RJX and WBH performed the analysis of DEGs. WCL drafted the
manuscript, aggregated the figures and discussed the results. RJX
contributed to the revision of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NFH
|
necrosis of the femoral head
|
|
NCBI
|
National Center of Biotechnology
Information
|
|
GEO
|
gene expression omnibus
|
|
DEGs
|
differentially expressed genes
|
|
GO
|
Gene Ontology
|
|
STAT
|
signal transducer and activator of
transcription
|
|
PPI
|
protein-protein interaction
|
|
DAVID
|
Database for Annotation, Visualization
and Integrated Discovery
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
BP
|
biological process
|
|
CC
|
cellular component
|
|
MF
|
molecular function
|
|
STRING
|
Search Tool for the Retrieval of
Interacting Genes/Proteins
|
|
VEGFA
|
vascular endothelial growth factor
A
|
|
GNG13
|
guanine nucleotide binding protein,
γ13
|
|
BMP
|
bone morphogenetic protein
|
|
AAV
|
adeno-associated virus
|
|
MAPK
|
mitogen-activated protein kinase
|
|
PAF
|
platelet activating factor
|
|
PAFR
|
platelet-activating factor
receptor
|
References
|
1
|
Zhang QY, Li ZR, Gao FQ and Sun W:
Pericollapse stage of osteonecrosis of the femoral Head: A last
chance for joint preservation. Chin Med J (Engl). 131:2589–2598.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hauzeur JP, Malaise M and de Maertelaer V:
A prospective cohort study of the clinical presentation of
non-traumatic osteonecrosis of the femoral head: Spine and knee
symptoms as clinical presentation of hip osteonecrosis. Int Orthop.
40:1347–1351. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moya-Angeler J, Gianakos AL, Villa JC, Ni
A and Lane JM: Current concepts on osteonecrosis of the femoral
head. World J Orthop. 6:590–601. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shah KN, Racine J, Jones LC and Aaron RK:
Pathophysiology and risk factors for osteonecrosis. Curr Rev
Musculoskelet Med. 8:201–209. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Al-Khateeb H, Kwok IH, Hanna SA, Sewell MD
and Hashemi-Nejad A: Custom cementless THA in patients with
legg-calve-perthes disease. J Arthroplasty. 29:792–796. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang G, Zhao G, Xia J, Wei Y, Chen F,
Chen J and Shi J: FGF2 and FAM201A affect the development of
osteonecrosis of the femoral head after femoral neck fracture.
Gene. 652:39–47. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu X, Wen H, Hu Y, Yu H, Zhang Y, Chen C
and Pan X: STAT1-caspase 3 pathway in the apoptotic process
associated with steroid-induced necrosis of the femoral head. J Mol
Histol. 45:473–485. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tian L, Wen Q, Dang X, You W, Fan L and
Wang K: Immune response associated with Toll-like receptor 4
signaling pathway leads to steroid-induced femoral head
osteonecrosis. BMC Musculoskelet Disord. 15:182014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li P, Zhai P, Ye Z, Deng P, Fan Y, Zeng Y,
Pang Z, Zeng J, Li J and Feng W: Differential expression of
miR-195-5p in collapse of steroid-induced osteonecrosis of the
femoral head. Oncotarget. 8:42638–42647. 2017.PubMed/NCBI
|
|
10
|
Wei B, Wei W, Zhao B, Guo X and Liu S:
Long non-coding RNA HOTAIR inhibits miR-17-5p to regulate
osteogenic differentiation and proliferation in non-traumatic
osteonecrosis of femoral head. PLoS One. 12:e01690972017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma XL, Liu ZP, Ma JX, Han C and Zang JC:
Dynamic expression of Runx2, Osterix and AJ18 in the femoral head
of steroid-induced osteonecrosis in rats. Orthop Surg. 2:278–284.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin Z and Lin Y: Identification of
potential crucial genes associated with steroid-induced necrosis of
femoral head based on gene expression profile. Gene. 627:322–326.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tong P, Wu C, Jin H, Mao Q, Yu N, Holz JD,
Shan L, Liu H and Xiao L: Gene expression profile of
steroid-induced necrosis of femoral head of rats. Calcif Tissue
Int. 89:271–284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu R, Liu Q, Wang K, Dang X and Zhang F:
Comparative analysis of gene expression profiles in normal hip
human cartilage and cartilage from patients with necrosis of the
femoral head. Arthritis Res Ther. 18:982016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jensen LJ, Kuhn M, Stark M, Chaffron S,
Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et
al: STRING 8-a global view on proteins and their functional
interactions in 630 organisms. Nucleic Acids Res. 37:D412–D416.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bertolazzi P, Bock ME and Guerra C: On the
functional and structural characterization of hubs in
protein-protein interaction networks. Biotechnol Adv. 31:274–286.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Z, Yang B, Weng X, Tse G, Chan MTV and
Wu WKK: Emerging roles of MicroRNAs in osteonecrosis of the femoral
head. Cell Prolif. 51:2018. View Article : Google Scholar :
|
|
27
|
Cohen-Rosenblum A and Cui Q: Osteonecrosis
of the femoral Head. Orthop Clin North Am. 50:139–149. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wei B and Wei W: Identification of
aberrantly expressed of serum microRNAs in patients with
hormone-induced non-traumatic osteonecrosis of the femoral head.
Biomed Pharmacother. 75:191–195. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song Y, Du ZW, Yang QW, Ren M, Wang QY,
Wang A, Chen GY, Zhao HY, Yu T and Zhang GZ: Association of genes
variants in RANKL/RANK/OPG signaling pathway with the development
of osteonecrosis of the femoral head in Chinese population. Int J
Med Sci. 14:690–697. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peng X and Guan JL: Focal adhesion kinase:
From in vitro studies to functional analyses in vivo. Curr Protein
Pept Sci. 12:52–67. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y, Ma C, Yu Y, Liu M and Yi C: Are
CXCL13/CXCR5/FAK critical regulators of MSCs migration and
differentiation? Med Hypotheses. 84:213–215. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
MacDonald RJ and Yen A: CXCR5
overexpression in HL-60 cells enhances chemotaxis toward CXCL13
without anticipated interaction partners or enhanced MAPK
signaling. In Vitro Cell Dev Biol Anim. 54:725–735. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gu YX, Du J, Si MS, Mo JJ, Qiao SC and Lai
HC: The roles of PI3K/Akt signaling pathway in regulating MC3T3-E1
preosteoblast proliferation and differentiation on SLA and SLActive
titanium surfaces. J Biomed Mater Res A. 101:748–754. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xue XH, Feng ZH, Li ZX and Pan XY:
Salidroside inhibits steroid-induced avascular necrosis of the
femoral head via the PI3K/Akt signaling pathway: In vitro
and in vivo studies. Mol Med Rep. 17:3751–3757.
2018.PubMed/NCBI
|
|
35
|
Wu X, Zheng S, Ye Y, Wu Y, Lin K and Su J:
Enhanced osteogenic differentiation and bone regeneration of
poly(lactic-co-glycolic acid) by graphene via activation of
PI3K/Akt/GSK-3β/β-catenin signal circuit. Biomater Sci.
6:1147–1158. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Aki T, Hashimoto K, Ogasawara M and Itoi
E: A whole-genome transcriptome analysis of articular chondrocytes
in secondary osteoarthritis of the hip. PLoS One. 13:e01997342018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang C, Ma J, Li M, Li XH, Dang XQ and
Wang KZ: Repair effect of coexpression of the hVEGF and hBMP genes
via an adeno-associated virus vector in a rabbit model of early
steroid-induced avascular necrosis of the femoral head. Transl Res.
166:269–280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hang D, Wang Q, Guo C, Chen Z and Yan Z:
Treatment of osteonecrosis of the femoral head with VEGF165
transgenic bone marrow mesenchymal stem cells in mongrel dogs.
Cells Tissues Organs. 195:495–506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Peng WX and Wang L: Adenovirus-mediated
expression of BMP-2 and BFGF in bone marrow mesenchymal stem cells
combined with demineralized bone matrix for repair of femoral head
osteonecrosis in beagle dogs. Cell Physiol Biochem. 43:1648–1662.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu J and Lin DI: Oncogenic c-terminal
cyclin D1 (CCND1) mutations are enriched in endometrioid
endometrial adenocarcinomas. PLoS One. 13:e01996882018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu J, Cui LL, Yuan J, Wang Y and Song S:
Clinical significance of the phosphorylation of MAPK and protein
expression of cyclin D1 in human osteosarcoma tissues. Mol Med Rep.
15:2303–2307. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Blake BL, Wing MR, Zhou JY, Lei Q,
Hillmann JR, Behe CI, Morris RA, Harden TK, Bayliss DA, Miller RJ
and Siderovski DP: G beta association and effector interaction
selectivities of the divergent G gamma subunit G gamma(13). J Biol
Chem. 276:49267–49274. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
da Silva Junior IA, de Sousa Andrade LN,
Jancar S and Chammas R: Platelet activating factor receptor
antagonists improve the efficacy of experimental chemo- and
radiotherapy. Clinics (Sao Paulo). 73 (Suppl 1):e792s2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Filgueiras LR, Koga MM, Quaresma PG,
Ishizuka EK, Montes MB, Prada PO, Saad MJ, Jancar S and Rios FJ:
PAFR in adipose tissue macrophages is associated with
anti-inflammatory phenotype and metabolic homoeostasis. Clin Sci
(Lond). 130:601–612. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen J, Lan T, Zhang W, Dong L, Kang N,
Zhang S, Fu M, Liu B, Liu K, Zhang C, et al: Platelet-activating
factor receptor-mediated PI3K/AKT activation contributes to the
malignant development of esophageal squamous cell carcinoma.
Oncogene. 34:5114–5127. 2015. View Article : Google Scholar : PubMed/NCBI
|