Introduction
Congenital heart disease (CHD) is the most common
type of developmental abnormality at birth, with an incidence of
~1% of live births worldwide (1,2).
Furthermore, it is the leading non-infectious cause of mortality in
newborns (3). The most common
types of congenital heart defects, including ventricular septal
defect (VSD) and atrial septal defect (ASD), account for a high
proportion of total congenital heart disease (4). Therefore, it is urgent to determine
the pathogenesis of these congenital cardiovascular diseases. VSD
and ASD are anatomically characterized by an interatrial and
ventricular septum that is defective or absent, causing the blood
to flow directly between the atria and ventricles of the heart
(5).
Cardiogenesis from the early embryo to formation of
the fully functional four-chambered heart is a highly dynamic and
complex process that requires numerous factors (6); cardiac transcription factors are
considered to be the leading contributors to the normal development
of the embryonic heart and include the GATA family (7). In addition, an increasing number of
studies have reported that genetic risk factors may disrupt the
biological process of heart development and subsequently lead to
CHD (8,9). Several genes that are essential for
heart development have been identified in the occurrence of CHD,
including GATA-binding protein 4 (GATA4), T-box
transcription factor 5 (TBX5) and NK2 homeobox 5 (7,10,11).
GATA4 belongs to a family of DNA-binding
proteins with conserved zinc finger domains that can specifically
bind the consensus DNA sequence GATA motif present in the promoter
of several target genes involved in cardiogenesis, such as atrial
natriuretic factor (ANF) (12,13),
and can interact with other transcriptional factors. GATA4
serves an important role in heart development, including in the
proliferation of cardiomyocytes, endocardial cushion formation,
development of the right ventricle and septation of the outflow
tract. A previous study reported that the GATA4
transcription factor is required for ventral morphogenesis and
heart tube formation in mice via knockout of the GATA4 gene
(14). Furthermore, mice
homozygous for the GATA4 G295S mutant allele exhibit normal
ventral body patterning and heart looping, but have a thin
ventricular myocardium, single ventricular chamber and lethality at
embryonic day 11.5 (15). The
importance of GATA4 in cardiac development in other
organisms, such as chicks, flies and fish, has also been
demonstrated (16). These findings
indicate that the GATA4 transcription factor may be closely
associated with cardiac development in humans and other animals.
Based on our previous study, we have established a mouse model of
the GATA4 p.H435Y mutation and propagated it successfully
for further research (17). To
date, numerous mutations in the GATA4 gene have been
reported in patients with CHD.
Our previous study reported a heterozygous
GATA4 c.1306C>T (pH436Y) mutation in four Chinese infants
with congenital heart defects (18). In the present study, further
functional analysis of the GATA4 H436Y mutation was
performed in vitro, and the molecular mechanism underlying
the effect of this mutation on gene function was explored.
Materials and methods
GATA4 amino acid sequence conservation
and mutation prediction
In our previous study (18), a heterozygous GATA4
c.1306C>T (p.H436Y) mutation was detected in four infants with
sporadic cardiac septal defects via sequencing of all exons and
flanking intron sequences. Conservation of the amino acids was
estimated by aligning genes from various species using National
Center for Biotechnology Information Blast (blast.ncbi.nlm.nih.gov/Blast.cgi).
PolyPhen-2 (genetics.bwh.harvard.edu/pph2), SIFT (sift.jcvi.org) and Mutation Taster (www.mutationtaster.org) programs were used to predict
the disease-causing potential of the mutation.
Plasmid construction and site-directed
mutagenesis
A wild-type GATA4 expression plasmid was
constructed by cloning the entire human GATA4 cDNA
(accession no: NM_002052) into pcDNA3.1 (+) expression vector with
a C-terminal flag-tag (Youbio). A point mutation was introduced
into the wild-type GATA4-pcDNA3.1 plasmid using the
KOD-plus-mutagenesis kit (cat. no. SMK-101; Toyobo Life Science),
according to manufacturer's protocol, and confirmed by Sanger
sequencing (19). The reporter
plasmid, ANF-luciferase (ANF-luc), was constructed as previously
described (20,21).
Cell culture, transfection and
luciferase reporter assay
HeLa cells, originally purchased from the Cell Bank
of type culture collection of the Chinese Academy of Sciences, were
cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.) at 37°C in an atmosphere
containing 5% CO2. Cells were seeded in 12-well plates
at a density of 1–4×105 cells/well at 24 h prior to
transient transfection using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.). For co-transfection
luciferase assays, 2.5 µg pcDNA3.1, 2.5 µg wild-type
GATA4-pcDNA3.1 or 2.5 µg mutant GATA4-pcDNA3.1 were
co-transfected with 2.5 µg ANF reporter plasmid. The pRL-TK plasmid
(Promega Corporation) was co-transfected with the plasmids
mentioned previously to normalize the luciferase activity.
Luciferase activity was measured at 48 h after transient
transfection; three independent experiments were performed in
duplicate with the ANF-luc reporters. The firefly luciferase
activity was normalized to Renilla luciferase activity, and
fold activation of wild-type GATA4 and mutant GATA4
luciferase activities were calculated with respect to the pcDNA3.1
value.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from HeLa cells transfected
with wild-type GATA4-pcDNA3.1 and mutant
GATA4-pcDNA3.1 expression plasmids using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) 24 h after
transfection. RNA samples were reverse transcribed into cDNA using
a PrimeScript RT kit (cat. no. #RR036A; Takara Biotechnology Co.,
Ltd.), according to manufacturer's protocol. Subsequently, relative
quantification was performed using the 2−ΔΔCq method
(22) and the TB Green system kit
(Toyob Life Science), according to manufacturer's protocol. The
primer sequences were synthesized by Generay Biotech Co., Ltd. as
follows: GATA4, forward, 5′-GTCACACATGCTTCCAGGTAATG-3′ and
reverse, 5′-GGGAACGGTAAATGGCTCTCTA-3′; GAPDH forward,
5′-GTCACACATGCTTCCAGGTAATG-3′ and reverse,
5′-GGGAACGGTAAATGGCTCTCTA-3′. The PCR thermal cycling conditions
were as follows: 95°C for 60 sec followed by 40 cycles of
amplification at 95°C for 30 sec, and annealing and extension at
60°C for 30 sec; extension at 60°C for 5 min. All reactions were
performed in triplicate, and GAPDH was used as an internal
control to normalize expression levels.
Western blot analysis
HeLa cells were transfected with wild-type
GATA4-pcDNA3.1 or mutant GATA4-pcDNA3.1 expression
plasmids, and whole cell extracts were obtained using RIPA lysis
buffer (Thermo Fisher Scientific, Inc.) containing 1X protease
inhibitor cocktail. The protein concentrations were detected using
a bicinchoninic acid (BCA) protein assay kit (Takara Biotechnology
Co., Ltd.) according to the manufacturer's protocol. Subsequently,
~10 µg protein extracts were separated using 10% SDS-PAGE gels and
transferred onto nitrocellulose membranes. The membranes were
blocked with PBS with 1% Tween-20 (PBST) containing 5% BSA for 2 h
at room temperature and then probed with primary antibodies against
GATA4 (dilution 1:10,000; cat. no. ab124265; Abcam) and
GAPDH (dilution 1:3,000; cat. no. ab9482; Abcam) at 4°C
overnight. The antigen-antibody complex was then incubated with
horseradish peroxidase-conjugated anti-rabbit secondary antibody
(dilution 1:3,000; cat. no. M21002; Abmart) or anti-mouse secondary
antibody (dilution 1:3,000; cat. no. M21001; Abmart) for 1 h at
room temperature. Blots were visualized using an ImageQuant LAS
4000 (GE Healthcare). GAPDH served as a loading control.
Immunofluorescence and subcellular
localization
HeLa cells were seeded onto 20 mm glass-bottom cell
culture dishes (NEST Scientific) at a density of 0.6×105
cells/ml 24 h before transfection with wild-type
GATA4-pcDNA3.1 or mutant GATA4-pcDNA3.1 expression
plasmids using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.). HeLa cells were fixed with 3.7%
formaldehyde/PBS for 20 min at room temperature and permeabilized
with 0.1% Triton X-100/PBS for 1 h at room temperature 48 h after
transfection. Subsequently, cells were incubated with a primary
antibody against GATA4 (dilution 1:800; cat. no. ab124265;
Abcam) overnight at 4°C and then detected using anti-rabbit
fluorescein isothiocyanate-conjugated secondary antibody (dilution:
1:500, cat. no. ab150080; Abcam) at room temperature for 2 h.
Nuclear staining was performed with a 1:1,000 dilution of DAPI at
room temperature for 20 min. The cells were observed under a Leica
TCS SP8 Laser Scanning Confocal microscope (Leica Microsystems
GmbH).
Electrophoretic mobility shift assay
(EMSA)
The enhancer region of the heart and neural crest
derivatives expressed 2 (HAND2) gene contains two conserved
consensus binding sites for GATA factors (23). The biotin-labeled oligonucleotide
corresponding to the conserved GATA-binding sites at −3,039 and
−3,140 (i.e. G1: 5′-TGATAA-3′) of the HAND2 gene was
synthesized by Generay Biotech Co., Ltd., as follows: Forward
5′-GCAGTTAACTGATAATGACACTGTG-3′ and reverse
5′-CACAGTGTCATTATCAGTTAACTGC-3′. An unlabeled oligonucleotide with
the same sequence was used as the competitor. Oligonucleotide pairs
were annealed into double strands; the DNA-binding ability was
detected by EMSA using a scientific light-shift EMSA kit (cat. no.
20148; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocols. HeLa cells were harvested 48 h after
transfection with wild-type GATA4-pcDNA3.1 or mutant
GATA4-pcDNA3.1 expression plasmids Whole cell extracts were
prepared using RIPA lysis buffer (Thermo Fisher Scientific, Inc.).
Protein concentrations were determined using a BCA protein assay
kit. Whole cell extracts (10 µg) were incubated with 20 fmol of
biotin-labelled probe in binding buffer containing poly (dI-dC),
50% glycerol and 1% NP-40 (included with EMSA kit) for 30 min at
room temperature. A 200-fold excess of unlabeled probe was added to
the reaction for competition experiments to confirm the specificity
of the binding. Supershift analysis was performed by adding 1 µl
neat GATA4 antibody (cat. no. ab124265; Abcam) to the whole
cell extracts for 20 min prior to the addition of the labelled
probe. Protein-DNA complexes were separated from the free probe by
6% polyacrylamide gel electrophoresis. The DNA-protein complexes
were analyzed using GE ImageQuant LAS4000 mini (GE Healthcare).
Statistical analysis
All data are presented as the mean ± standard
deviation of three independent experiments. Differences between
multiple groups were analyzed using one-way ANOVA and the least
significant difference post hoc test. Statistical analysis was
performed using SPSS software v20.0 (IBM Corp.). P<0.05 was
considered to indicate a statistically significant difference.
Results
Effects of the GATA4 mutation on gene
transcription activity
Our previous study (18) reported a heterozygous GATA4
c.1306C>T (p.H436Y) mutation in four Chinese children with
congenital heart defects, which was located on exon 7. This
mutation exhibits conserved evolution, and was predicted to be
deleterious and disease causing, as determined using SIFT,
Polyphen-2 and Mutation Taster. In order to confirm whether the
GATA4 H436Y mutation affects the functional activity of
GATA4, the present study used ANF-luc in HeLa cells, as
described previously (24). As
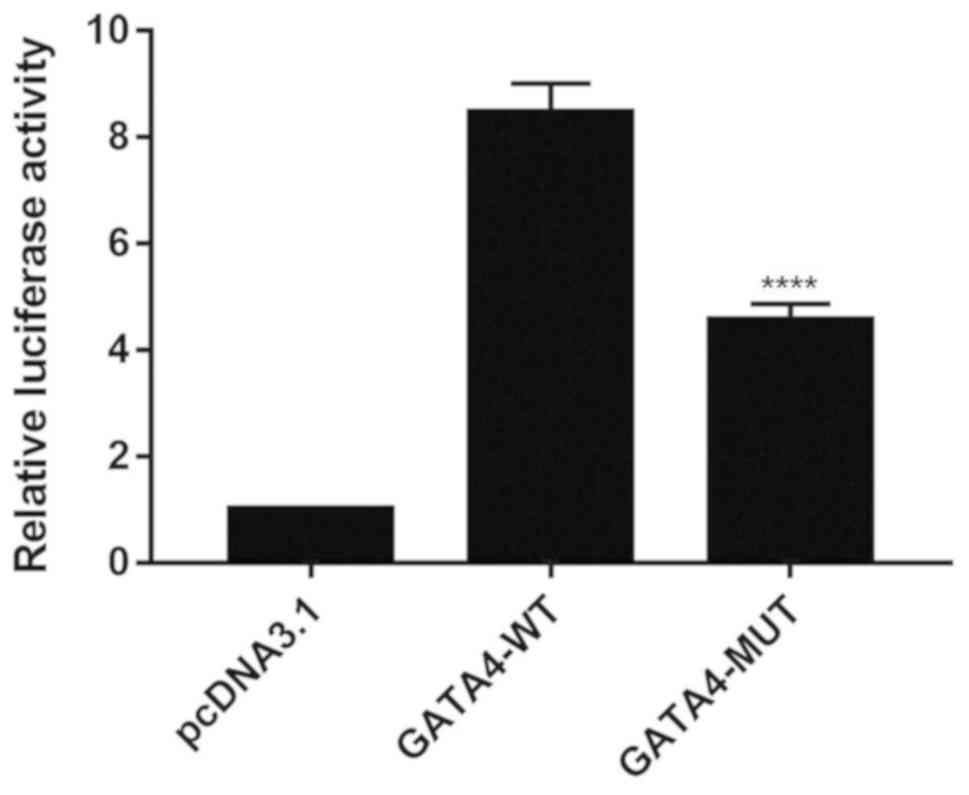
shown in Fig. 1, the GATA4
mutation significantly reduced reporter gene transcription activity
compared with the wild-type controls (P<0.01), thus suggesting
that the mutant GATA4 significantly diminished the
transcriptional activity of GATA4.
Expression of wild-type and mutant
GATA4 in HeLa cells
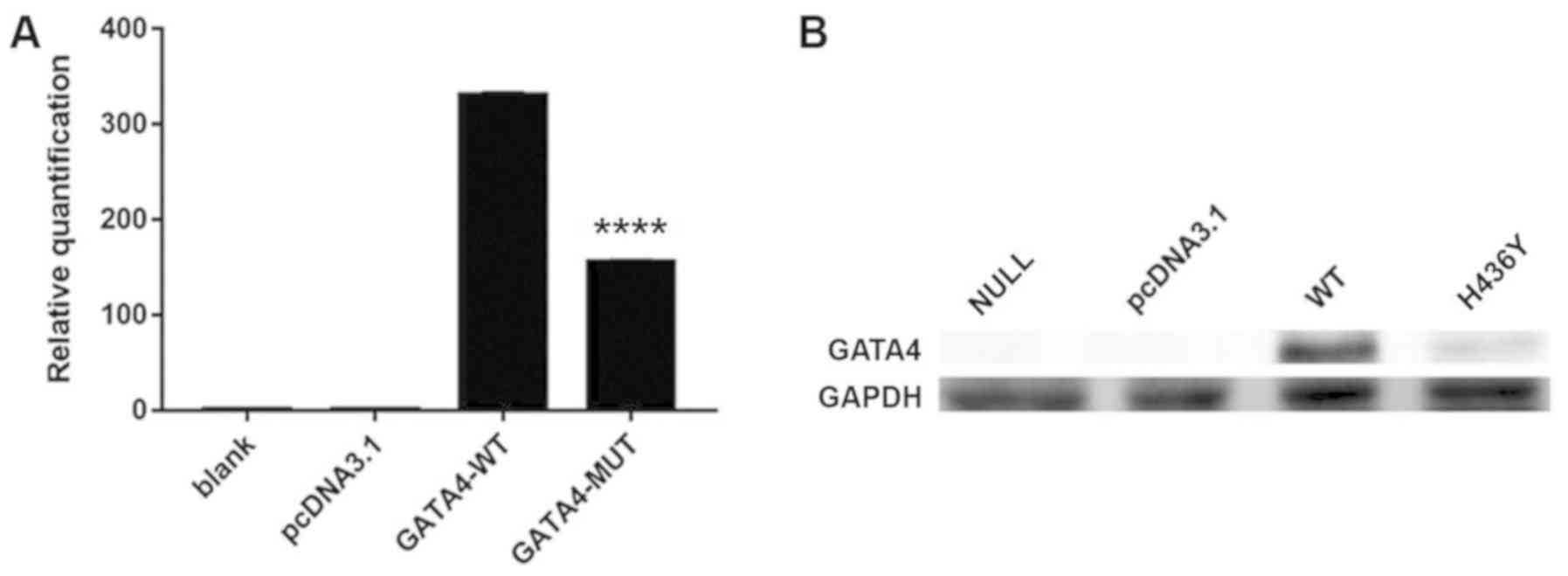
RT-qPCR was performed to measure the mRNA expression
levels of wild-type and mutant GATA4 following extraction of
total RNA from HeLa cells. The results of RT-qPCR revealed that the
mRNA expression levels of GATA4 were significantly lower in
the mutant group compared with in the wild-type group (Fig. 2A; P<0.01). The present study
also performed western blot analysis to detect the protein
expression levels of GATA4 in the wild-type and mutant
groups; the results demonstrated that the GATA4 mutation
significantly reduced GATA4 protein expression (Fig. 2B). These results are consistent
with those of RT-qPCR, in which the GATA4 mutation reduced
transcriptional activity of the gene.
Subcellular localization of the
wild-type and mutant GATA4 proteins
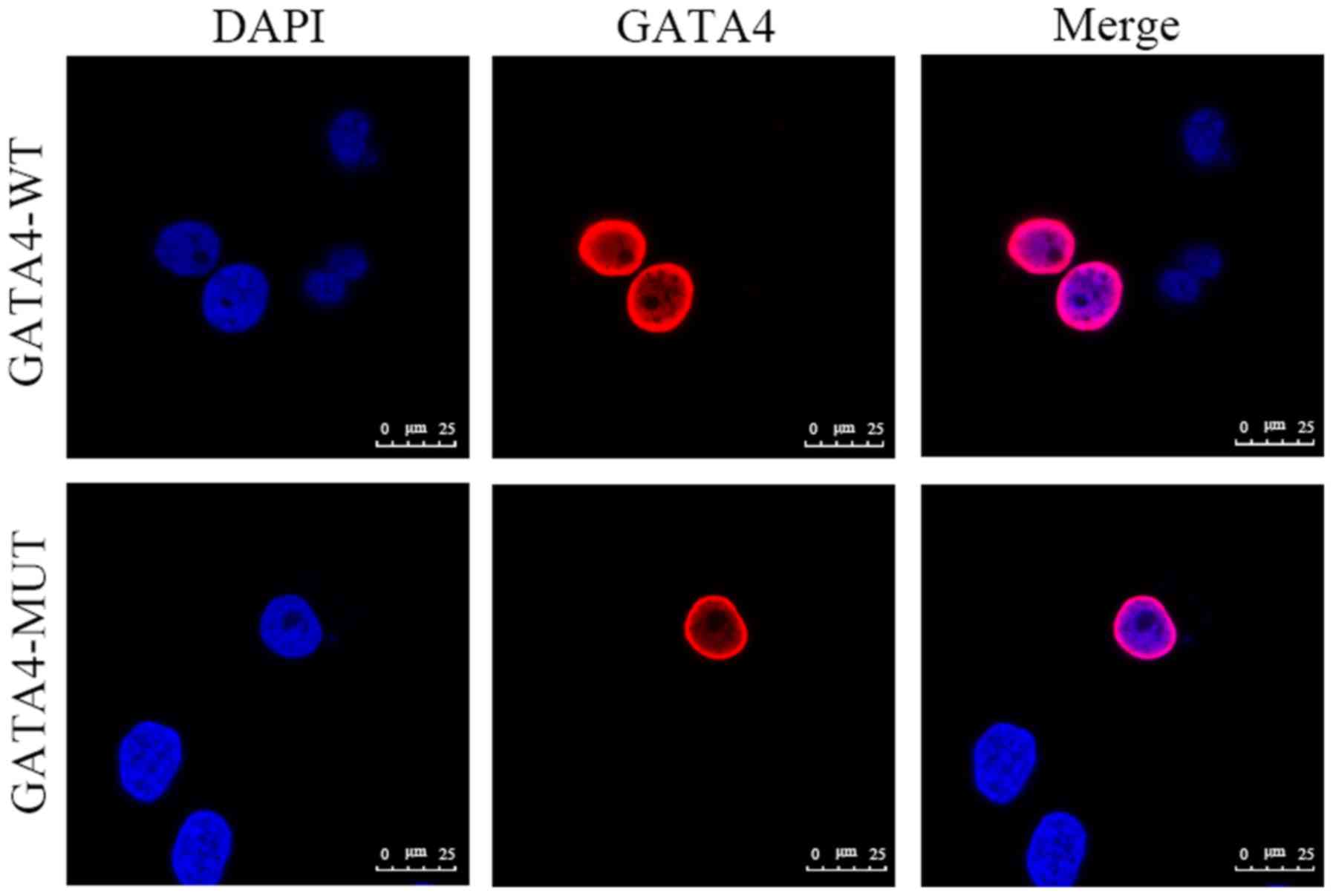
GATA4 is a nuclear transcription factor that
is localized in the nucleus. To determine whether the GATA4
c.1306C>T (p.H436Y) mutation altered distribution of the
GATA4 protein in cells, the present study performed
immunofluorescence analysis in HeLa cells, in order to detect its
cellular localization. As shown in Fig. 3, although the protein expression of
mutant GATA4 was reduced compared with the wild-type, the
protein localization of mutant GATA4 was similar to
wild-type GATA4, with both localized in the nucleus, thus
suggesting that the GATA4 mutation did not influence the
protein subcellular localization.
Effects of the GATA4 mutation on
DNA-binding affinity
A previous study (25) demonstrated that GATA4 could
interact with the cardiac-expressed basic helix-loop-helix
transcription factor HAND2 gene to regulate transcription of
the downstream gene by binding to the conserved GATA-binding sites
on the HAND2 gene (G1: 5′-TGATAA-3′; G2: 5′-CTATCT-3′;
Fig. 4A). To determine whether the
GATA4 mutant (c.1306C>T; p.H436Y) affects the binding
ability of the GATA4 protein to the conserved GATA-binding
site in the promoter of the HAND2 gene, the present study
performed EMSA using wild-type and mutant GATA4 proteins
from transfected HeLa cells, with a biotin-labeled probe. As shown
in Fig. 4B (lane 2), the wild-type
GATA4 protein could bind to the conserved GATA-binding site.
In order to confirm the binding specificity of the GATA4
protein to the conserved GATA-binding site on the HAND2
gene, an unlabeled probe at 100X was used to compete with the
biotin-labeled probe at 1X bound to the wild-type GATA4
protein. As shown in Fig. 4B (lane
3), the protein/DNA complex could compete with an excessive amount
of unlabeled probe. However, when the equivalent amount of mutant
GATA4 protein was added, the DNA/protein band showed a
lighter band than the GATA4-WT group but a stronger band
than the competitor group (Fig.
4B, lane 4), thus suggesting that the GATA4 mutation
reduces DNA-binding affinity. In addition, supershift analysis was
conducted to prove that binding was caused by the GATA4
protein (Fig. 4B, lane 5). These
results indicated that the mutant GATA4 protein decreased
the ability to bind the conserved GATA-binding site on the
HAND2 gene, which may contribute to abnormal expression of
the HAND2 gene.
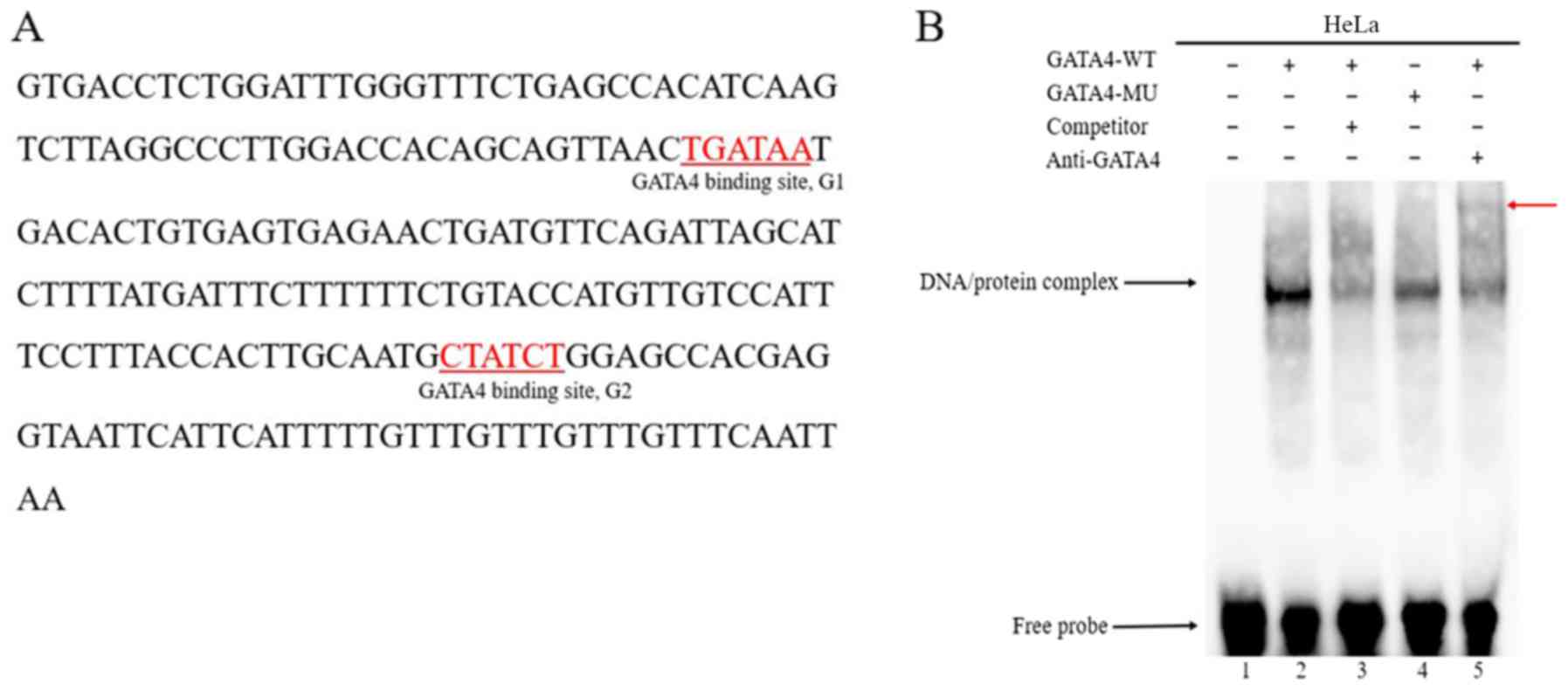 | Figure 4.GATA4 binds to the conserved
GATA-binding sites on the HAND2 gene. (A) Enhancer region of
the HAND2 gene contains two conserved GATA-binding sites
(underlined sequence, transcription factor binding sequence). (B)
EMSA results revealed that the MU GATA4 protein exhibited
decreased DNA-binding affinity. Lane 1, labeled probe; lane 2,
protein from HeLa cells transfected with GATA4 (WT) +
labeled probe; lane 3, protein from HeLa cells transfected with
GATA4 (WT) + unlabeled competitor probe + labeled probe;
lane 4, protein from HeLa cells transfected with GATA4 (MU)
+ labeled probe; lane 5, protein from HeLa cells transfected with
GATA4 (WT) + labeled probe + anti-GATA4. The specific
DNA/protein complexes are indicated by arrows. A supershift,
indicated by the red arrow, revealed that the GATA4 antibody
could specifically bind with whole cell lysate, which was
transfected with the GATA4 plasmid. EMSA, electrophoretic
mobility shift assay; GATA4, GATA-binding factor 4;
HAND2, heart and neural crest derivatives expressed 2; MU,
mutant; WT, wild-type. |
Discussion
The GATA4 gene serves an important role in
cardiac development, and numerous mutations in this gene have
previously been reported in congenital heart defects. For example,
a heterozygous G296S missense mutation in GATA4 results in
reduced DNA-binding affinity and transcriptional activity of
GATA4. Furthermore, the GATA4 mutation abrogates a
physical interaction between GATA4 and TBX5 (15). GATA4 R311W resides in the
nuclear localization signal domain (NLS), and the mutant protein
does not alter its intracellular distribution; however, the
mutation reduces the ability of GATA4 to activate its downstream
target gene (26). Furthermore,
the GATA4 K300T mutation may impair cardiogenesis by
impeding the GATA4-DNA-binding process and the transcription of
GATA4 target genes (27). In our
previous study, a heterozygous missense mutation, GATA4
c.1306C>T (p.H436Y), was identified in four children with
sporadic cardiac septal defects, including two VSDs, one VSD
associated with ASD, and one VSD associated with an ASD and patent
foramen ovale (18).
It has been reported that GATA4 is an upstream
transcriptional regulator of ANF and HAND2 (12), and that it regulates their protein
expression. GATA4 can interact with the HAND2 gene to
regulate transcription of the downstream gene by binding to the
conserved GATA-binding sites on the promoter region of the
HAND2 gene. In this study, two different methods (ANF
luciferase assay and HAND2 EMSA assay) were used to
determine the functional consequences of the GATA4 H436Y
mutation, in order to obtain more realistic and reliable
results.
A previous study demonstrated that GATA4 is a
transcriptional activator of numerous genes expressed during
cardiac development, including the ANF gene (23). Therefore, the functional
characteristics of this mutation could be analyzed by investigating
the transcriptional activity of the ANF promoter in HeLa
cells expressing GATA4. In the present study, the functional
effects of the GATA4 mutation were studied by ANF-luc
assays; the results revealed that the GATA4 c.1306C>T,
p.H436Y mutation was associated with decreased transcriptional
activity. Furthermore, the present study performed RT-qPCR and
western blotting to explore the expression of GATA4 at the
mRNA and protein levels, and revealed that the mutation induced
decreases in the protein and mRNA expression levels of
GATA4. These results indicated that the haploinsufficiency
or dominant-negative effect resulting from the GATA4
mutation may be a pathogenic mechanism underlying congenital heart
defects.
As reported previously, the human GATA4 gene
is located on chromosome 8p23.1-p22 and consists of sevens exons,
encoding a protein containing 442 amino acids (25). The GATA4 protein is comprised of
two transcriptional activation domains [(TAD)-1, amino acids 1–74;
TAD2, amino acids 130–177], two highly conserved zinc finger
domains [(ZF)-1, amino acids 215–240; ZF2, amino acids 270–294],
and one NLS (amino acids 254–32) (28). Additionally, the results revealed
that subcellular localization of GATA4 was not affected by
the GATA4 mutations analyzed in the present study, which may
be associated with the fact that the mutation is absent in the NLS
region, not affecting the nuclear distribution of GATA4.
Notably, the present study demonstrated that the
DNA-binding affinity was weakened by the mutation, as determined
using EMSA, although the GATA4 p.H436Y mutation is not
located in the ZF2 domain, which is essential for DNA sequence
recognition and binding to the consensus motif (29). The GATA4 C-finger domain
interacts with the basic helix-loop-helix domain of HAND2 to
synergistically activate the expression of cardiac-specific genes,
including ANF and the brain type natriuretic peptide
(12). In the mutation
investigated in the present study, the amino acid at site 436 of
the GATA4 protein was changed from histidine to tyrosine.
The substitution of polar positively charged histidine to neutral
tyrosine may alter the structure and charge of the residue. From
the EMSA results, it may be hypothesized that the significantly
decreased affinity of GATA4 to HAND2 caused by the
p.H436Y mutation may be associated with an alteration in the
three-dimensional structure of the mutated protein, obstructing its
interaction with the DNA, further affecting the subsequent
malfunction in transcriptional regulation, which may lead to the
occurrence of CHD. This finding is consistent with previous studies
(7,30).
A limitation of the study is that the HeLa cell line
was used instead of a cardiac cell line for in vitro
experiments. Although cardiac cell lines could be used to perform
the in vitro experiments, endogenous GATA4 gene
expression in cardiac cells may interfere with the results of the
gene mutation studies. Notably, the HeLa cell line has a strong
proliferative ability and is easy to culture for experimental
research. The endogenous GATA4 gene expression was very low
and had little effect on the experimental results of transfection
with wild-type and mutant GATA4 plasmids in the HeLa cell
line. Considering these advantages, the in vitro experiments
were performed using the HeLa cell line instead of a cardiac cell
line in the present study. In additions, further studies using
additional cell lines, including cardiac cell lines, should be
conducted in future to further the work presented in the present
study.
Similar to previous studies (27,31),
congenital heart defects were observed in four patients bearing the
same GATA4 mutation in our previous study (18), indicating that the GATA4
mutation (c.1306C>T; p.H436Y) may be closely associated with the
occurrence of VSD. However, congenital heart defects are
multifactorial, and both genetic and environmental factors serve an
important role in their occurrence. The same CHD phenotype can be
caused by different mutations, and the same mutation may lead to
different phenotypes in different patients (32). The occurrence of CHD is a complex
process, involving genetic and environmental factors, epigenetic
regulation and many other factors (33). In conclusion, the results of the
present study may broaden the spectrum of known mutations in the
GATA4 gene associated with congenital heart defects, and
could provide novel insights into the mechanism underlying CHD. In
addition, these findings may contribute to the future development
of genetic diagnostic techniques and therapies.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National Key
Research and Development Program of China (grant no.
2016YFC1000500), The Anhui Natural Science Foundation (grant no.
21608085MH196) and The National Natural Science Foundation of China
(grant nos. 8137019, 81570282 and 81570283).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WS and MC were responsible for study design and
revision of the manuscript. GH conceived and designed the study. TF
and YJZ performed the research and analyzed the data. YLZ wrote the
manuscript. AX and QW conducted the statistical analysis. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhao QM, Liu F, Wu L, Ma XJ, Niu C and
Huang GY: Prevalence of congenital heart disease at live birth in
China. J Pediatr. 204:53–58. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
van der Linde D, Konings EE, Slager MA,
Witsenburg M, Helbing WA, Takkenberg JJ and Roos-Hesselink JW:
Birth prevalence of congenital heart disease worldwide a systematic
review and meta-analysis. J Am Coll Cardiol. 58:2241–2247. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yu Z, Xi Y, Ding W, Han S, Cao L, Zhu C,
Wang X and Guo X: Congenital heart disease in a Chinese hospital:
Pre- and postnatal detection, incidence, clinical characteristics
and outcomes. Pediatr Int. 53:1059–1065. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Miranović V: The incidence of congenital
heart disease: Previous findings and perspectives. Srp Arh Celok
Lek. 142:243–248. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marian AJ: Congenital heart disease the
remarkable journey from the ‘post-mortem room’ to adult clinics.
Circ Res. 120:895–897. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Srivastava D and Olson EN: A genetic
blueprint for cardiac development. Nature. 407:221–226. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang YQ, Gharibeh L, Li RG, Xin YF, Wang
J, Liu ZM, Qiu XB, Xu YJ, Xu L, Qu XK, et al: GATA4
loss-of-function mutations underlie familial tetralogy of fallot.
Hum Mutat. 34:1662–1671. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Andersen TA, Troelsen Kde L and Larsen LA:
Of mice and men: Molecular genetics of congenital heart disease.
Cell Mol Life Sci. 71:1327–1352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang X, Li P, Chen S, Xi L, Guo Y, Guo A
and Sun K: Influence of genes and the environment in familial
congenital heart defects. Mol Med Rep. 9:695–700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qu XK, Qiu XB, Yuan F, Wang J, Zhao CM,
Liu XY, Zhang XL, Li RG, Xu YJ, Hou XM, et al: A novel NKX2.5
loss-of-function mutation associated with congenital bicuspid
aortic valve. Am J Cardiol. 114:1891–1895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McCulley DJ and Black BL: Transcription
factor pathways and congenital heart disease. Curr Top Dev Biol.
100:253–277. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dai YS, Cserjesi P, Markham BE and
Molkentin JD: The transcription factors GATA4 and dHAND physically
interact to synergistically activate cardiac gene expression
through a p300-dependent mechanism. J Biol Chem. 277:24390–24398.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Molkentin JD, Kalvakolanu DV and Markham
BE: Transcription factor GATA-4 regulates cardiac muscle-specific
expression of the alpha-myosin heavy-chain gene. Mol Cell Biol.
14:4947–4957. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pikkarainen S, Tokola H, Kerkelä R and
Ruskoaho H: GATA transcription factors in the developing and adult
heart. Cardiovasc Res. 63:196–207. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Misra C, Sachan N, McNally CR, Koenig SN,
Nichols HA, Guggilam A, Lucchesi PA, Pu WT, Srivastava D and Garg
V: Congenital heart disease-causing Gata4 mutation displays
functional deficits in vivo. PLoS Genet. 8:e10026902012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Epstein JA and Parmacek MS: Recent
advances in cardiac development with therapeutic implications for
adult cardiovascular disease. Circulation. 112:592–597. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, Chen M, Fang T, Zhang T and Ni W:
Establishment and verification of a mouse model of Gata4 gene H435Y
mutation. Nan Fang Yi Ke Da Xue Xue Bao. 38:1245–1249. 2018.(In
Chinese). PubMed/NCBI
|
|
18
|
Chen MW, Pang YS, Guo Y, Pan JH, Liu BL,
Shen J and Liu TW: GATA4 mutations in Chinese patients with
congenital cardiac septal defects. Pediatr Cardiol. 31:85–89. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kaetzke A and Eschrich K: Simultaneous
determination of different DNA sequences by mass spectrometric
evaluation of Sanger sequencing reactions. Nucleic Acids Res.
30:e1172002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Seidman CE, Wong DW, Jarcho JA, Bloch KD
and Seidman JG: Cis-acting sequences that modulate atrial
natriuretic factor gene-expression. Proc Natl Acad Sci USA.
85:4104–4108. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sprenkle AB, Murray SF and Glembotski CC:
Involvement of multiple cis-elements in basal-inducible and
alpha-adrenergic agonist-inducible atrial-natriuretic-factor
transcription-roles for serum response elements and an sp-1-like
element. Circ Res. 77:1060–1069. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McFadden DG, Charite J, Richardson JA,
Srivastava D, Firulli AB and Olson EN: A GATA-dependent right
ventricular enhancer controls dHAND transcription in the developing
heart. Development. 127:5331–5341. 2000.PubMed/NCBI
|
|
23
|
Garg V, Kathiriya IS, Barnes R,
Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS,
Hirayama-Yamada K, Joo K, et al: GATA4 mutations cause human
congenital heart defects and reveal an interaction with TBX5.
Nature. 424:443–447. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang E, Sun S, Qiao B, Duan W, Huang G, An
Y, Xu S, Zheng Y, Su Z, Gu X, et al: Identification of functional
mutations in GATA4 in patients with congenital heart disease. PLoS
One. 8:e621382013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
White RA, Dowler LL, Pasztor LM, Gatson
LL, Adkison LR, Angeloni SV and Wilson DB: Assignment of the
transcription factor gata4 gene to human-chromosome-8 and mouse
chromosome-14-GATA4 is a candidate gene for ds (disorganization).
Genomics. 27:20–26. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang X, Wang J, Wang B, Chen S, Fu Q and
Sun K: A novel missense mutation of GATA4 in a chinese family with
congenital heart disease. PLoS One. 11:e01589042016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen J, Qi B, Zhao J, Liu W, Duan R and
Zhang M: A novel mutation of GATA4 (K300T) associated with familial
atrial septal defect. Gene. 575:473–477. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Heineke J, Auger-Messier M, Xu J, Oka T,
Sargent MA, York A, Klevitsky R, Vaikunth S, Duncan SA, Aronow BJ,
et al: Cardiomyocyte GATA4 functions as a stress-responsive
regulator of angiogenesis in the murine heart. J Clin Invest.
117:3198–3210. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Morrisey EE, Ip HS, Tang Z and Parmacek
MS: GATA-4 activates transcription via two novel domains that are
conserved within the GATA-4/5/6 subfamily. J Biol Chem.
272:8515–8524. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nemer G, Fadlalah F, Usta J, Nemer M,
Dbaibo G, Obeid M and Bitar F: A novel mutation in the GATA4 gene
in patients with tetralogy of fallot. Hum Mutat. 27:293–294. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen Y, Mao J, Sun Y, Zhang Q, Cheng HB,
Yan WH, Choy KW and Li H: A novel mutation of GATA4 in a familial
atrial septal defect. Clin Chim Acta. 411:1741–1745. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Moreau JLM, Kesteven S, Martin EMMA, Lau
KS, Yam MX, O'Reilly VC, Del Monte-Nieto G, Baldini A, Feneley MP,
Moon AM, et al: Gene-environment interaction impacts on heart
development and embryo survival. Development. 146:dev1729572019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Thomford NE, Dzobo K, Yao NA, Chimusa E,
Evans J, Okai E, Kruszka P, Muenke M, Awandare G, Wonkam A and
Dandara C: Genomics and epigenomics of congenital heart defects:
Expert review and lessons learned in Africa. OMICS. 22:301–321.
2018. View Article : Google Scholar : PubMed/NCBI
|