Introduction
Liver cancer, including hepatocellular carcinoma and
intrahepatic cholangiocarcinoma, is one of the most common
malignancies worldwide (1).
Additionally, liver cancer is the third leading cause of
cancer-associated mortality worldwide, after lung and gastric
cancer (1). A variety of aberrant
genes and signaling transduction pathways are involved in the
occurrence and development of liver cancer, which contribute to a
complex pathogenesis (2–4). At present, surgery is the main
clinical treatment for liver cancer; however, a lack of early
clinical symptoms makes diagnosis difficult (5,6).
Additionally, despite advances in treatment, such as liver
transplantation and chemotherapeutic interventions, the therapeutic
effects are inadequate due to tumor recurrence and metastasis
(7,8), meaning patients with the disease have
a poor prognosis. Therefore, an in-depth investigation to improve
the understanding of the molecular mechanisms underlying liver
cancer and to identify accurate markers for the prognosis of liver
cancer remains urgent.
Previous studies have found that EPS8 plays an
important role in a wide variety of cancers, including oral cancer
(9), pancreatic cancer (10), non-small cell lung cancer (11), and head and neck squamous cell
carcinomas (12). As such, EPS8 is
proposed to be an important target in multiple cancer types
(13). EPS8 has pro-tumorigenic
potential of in a variety of tumors and specific inhibitors derived
from the nuclear localization signal (NLS) sequence are responsive
to tumor-associated proteins. Chen et al (14) found that the injection of an
EPS8-NLS peptide exerted anti-tumor activity in xenograft tumor
models of acute myeloid leukemia, further indicating the
pro-tumorigenic role of EPS8 and its potential as a therapeutic
target. The EPS8 family of proteins, including EPS8-like (EPS8L) 1,
EPS8L2 and EPS8L3, is a group of important regulators of behavior
in mice and flies (15,16). As the homolog of EPS8, EPS8L3 has
been reported to be overexpressed in liver cancer tissues and
cells. Furthermore, silencing and overexpression of EPS8L3 has been
reported to reduce and increase the rate of proliferation,
respectively; this was found to be dependent on the AKT signaling
pathway, as an inhibition of AKT reverses the effect of EPS8L3
overexpression on proliferation in liver cancer (17). Little is known about the clinical
relevance, significance to prognosis and potential regulatory
effect on invasiveness and trans-migration of EPS8L3 in liver
cancer. Therefore, the aim of the present study was to determine
the potential function of EPS8L3 in liver cancer development by
determining EPS8L3 expression in liver cancer cells, and the
effects of EPS8L3 knockdown on liver cancer cell proliferation and
migration.
Materials and methods
Online database data retrieval
EPS8L3 expression data from liver cancer and
non-tumor liver tissues were obtained from the Oncomine database
(https://www.oncomine.org/resource/login.html). The
clinical data of 338 patients with liver cancer were obtained from
The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/; Table I). The present study is in
accordance with the publication guidelines provided by Oncomine and
TCGA. In order to analyze the role of EPS8L3 in the progression of
liver cancer, all patients with liver cancer were divided into
EPS8L3 high (n=169) or low (n=169) expressing groups, according to
the median value.
 | Table I.Association between the expression of
EPS8L3 and clinicopathological features in 338 patients with liver
cancer. |
Table I.
Association between the expression of
EPS8L3 and clinicopathological features in 338 patients with liver
cancer.
|
| Expression of
EPS8L3 |
|
|---|
|
|
|
|
|---|
| Characteristics | Low | High | P-value |
|---|
| Age |
|
| 0.383 |
|
<60 | 75 | 83 |
|
|
≥60 | 94 | 86 |
|
| Sex |
|
| 0.198 |
|
Female | 48 | 59 |
|
|
Male | 121 | 110 |
|
| Grade |
|
| 0.024 |
| G1 +
G2 | 116 | 96 |
|
| G3 | 53 | 73 |
|
| Clinical stage |
|
| 0.137 |
| I +
II | 131 | 119 |
|
| III +
IV | 38 | 50 |
|
| Tumor stage |
|
| 0.103 |
| T1 +
T2 | 133 | 120 |
|
| T3 +
T4 | 36 | 49 |
|
| Node stage |
|
| 1.000 |
| N0 | 167 | 167 |
|
| N1 | 2 | 2 |
|
| Metastasis
stage |
|
| 0.562 |
| M0 | 167 | 168 |
|
| M1 | 2 | 1 |
|
| Mortality |
|
| 0.011 |
|
Yes | 46 | 68 |
|
| No | 123 | 101 |
|
Cell culture and transfection
The normal human liver cell line HL-7702, and the
liver cancer cell lines HepG2 and HCCLM3 were purchased from the
Type Culture Collection of the Chinese Academy of Sciences. All
cell lines were verified using short tandem repeats profiling. The
cells were cultured in DMEM (Invitrogen; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin and 0.1 mg/ml streptomycin at 37°C with
5% CO2. During the logarithmic growth period, cells were
collected and digested with 0.25% trypsin for 5 min at 37°C to
obtain a single cell suspension. For RNA interference,
5×105 cells were seeded into a 24-well plate the day
before transfection. The following day, transfection of 100 nM
small interfering RNA (siRNA) into liver cancer cells was performed
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
siRNAs were synthesized by Shanghai GenePharma Co, Ltd. The
sequences of the siRNAs were as follows: si-EPS8L3 1,
5′-AAUUCUCGGGGCUCUAAUGGG-3′; si-EPS8L3 1,
5′-UUUUACAAGCUUGAAGAUGCU-3′; si-con, 5′-UUCUCCGAACGUGUCACGU-3′. The
expression of EPS8L3 at the mRNA and protein levels was determined
48 h after transfection using reverse transcription-quantitative
(RT-q)PCR and western blotting. The siRNA whose silencing
efficiency was the greatest was used in the subsequent
experiments.
RNA extraction and RT-qPCR
Total RNA was extracted from cells using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Complementary DNA (cDNA) was synthesized
at 37°C for 15 min and 85°C for 5 sec using PrimeScript™RT reagent
kit with gDNA Eraser (Takara Bio, Inc.). RT-qPCR was performed
using Power SYBR Green PCR Master Mix (Takara Bio, Inc.) on a 7300
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) under the following thermocycling conditions: 94°C for 5 min,
followed by 40 cycles of 94°C for 30 sec, 60°C for 45 sec and 72°C
for 30 sec. GAPDH was used as the endogenous control, and the
primers were as follows: EPS8L3 forward (F),
5′-CCTCATCAAAGGCAGGCTCT-3′ and reverse (R),
5′-GCTCTGAGGTGAGGTTCTGG-3′; GAPDH F, 5′-GGAGCGAGATCCCTCCAAAAT-3′
and R, 5′-GGCTGTTGTCATACTTCTCATGG-3′. The relative expression of
EPS8L3 was normalized to GAPDH using the 2–ΔΔCq method
(18). Each experiment was
performed in triplicate.
Western blot analysis
Cells were harvested 48 h following transfection and
lysed in RIPA buffer (Beyotime Institute of Biotechnology)
supplemented with protease inhibitor (Thermo Fisher Scientific,
Inc.). After measuring protein concentrations using a bicinchoninic
protein assay, proteins (20 µg) were separated on 10–12% SDS-PAGE
gels and then transferred onto PVDF membranes. The membranes were
blocked with 5% skimmed milk for 1 h at room temperature, followed
by incubation with primary antibodies, including EPS8L3 (1:1,000;
cat. no. PA5-49855; Thermo Fisher Scientific, Inc.), phosphorylated
(p)-PI3K (1:1,000; cat. no. 17366; Cell Signaling Technology,
Inc.), PI3K (1:1,000; cat. no. 429; Cell Signaling Technology,
Inc.), p-AKT (1:1,000; cat no. 9271, Cell Signaling Technology,
Inc.), AKT (1:1,000; cat. no. 9272, Cell Signaling Technology,
Inc.) and GAPDH (1:5,000; cat. no. 60004-1-Ig, ProteinTech Group,
Inc.) at 4°C overnight. Next, the membranes were washed with 0.05%
TBS-Tween 20 for 30 min and incubated with anti-rabbit/mouse
horseradish peroxidase-conjugated secondary antibody (1:5,000; cat
nos. SA00001-2 and SA00001-1; ProteinTech Group, Inc) for 1 h at
room temperature. The blots were visualized using a Hypersensitive
ECL chemiluminescence detection kit (ProteinTech Group, Inc).
Signals were analyzed using Quantity One version 4.6 (Bio-Rad
Laboratories, Inc,). The relative expression of each protein was
normalized to GAPDH.
Cell Counting Kit-8 (CCK-8) assay
The effect of EPS8L3 on cell viability was
determined using a CCK-8 assay. Cell suspensions were seeded into
96-well plates at density of 1×103 cells/well and
cultured for 24, 48, 72 or 96 h in a CO2 incubator at
37°C. CCK-8 solution (10 µl/well) was added and the optical density
was measured at 450 nm after incubation at 37°C for 1.5 h.
Wound healing assay
The migration of cells was assessed using wound
healing assays. Cells, at a density of 1×105/well, were
seeded into 6-well plates and grown to 80% confluency. The scratch
test was performed after 24 h. Wounds were scratched in each well
using a 10-µl pipette tip. After washing with PBS to remove
cellular debris, the cells were cultured in medium without FBS.
Wound closure was monitored at 0 and 24 h, and images were captured
at ×100 magnification using an inverted light microscope equipped
with a Nikon camera (Nikon Corporation).
Statistical analysis
Statistical analyses were performed using SPSS 22.0
(IBM Corp.) and GraphPad Prism 7.0 (GraphPad Software, Inc.). All
data were presented as the mean ± standard deviation. Associations
between EPS8L3 expression and clinical pathological parameters were
analyzed using the χ2 test. Differences between multiple
groups were analyzed using ANOVA followed by Tukey's post hoc test.
Survival analysis was performed using the Kaplan-Meier method with
a log-rank test. Univariate and multivariate analyses were
performed using the Cox proportional hazards model to assess
prognostic factors. P<0.05 was considered to indicate a
statistically significant difference.
Results
EPS8L3 is upregulated in human liver
cancer tissues and cell lines
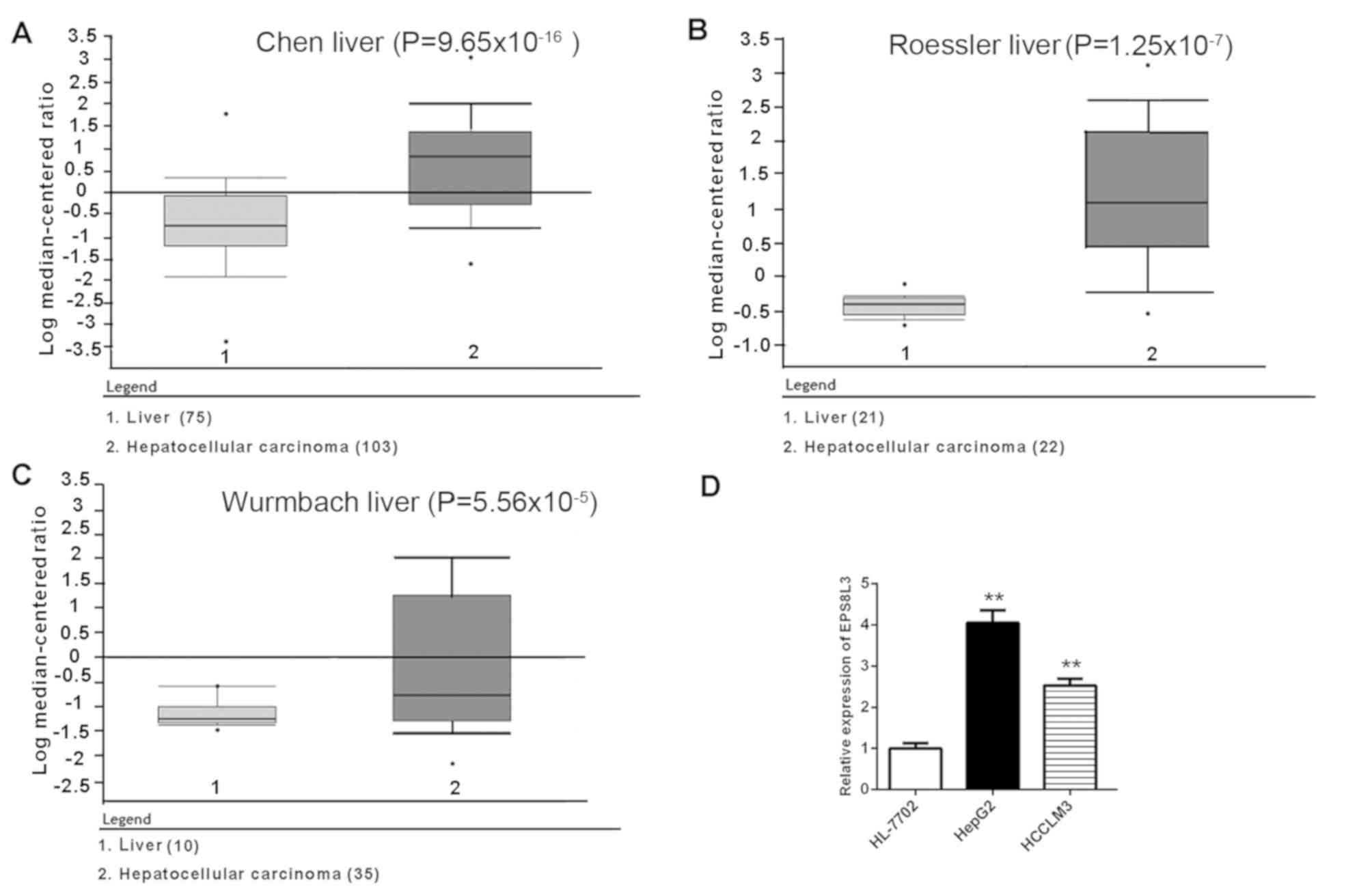
According to data in the Oncomine database, the
expression of EPS8L3 in liver cancer tissues was significantly
upregulated compared with normal liver samples
(P=9.65×10−16 for Chen liver, P=1.25×10−7 for
Roessler liver and P=5.56×10−5 for Wurmbach liver
patient datasets; Fig. 1A-C). To
further investigate the expression profile of EPS8L3, the mRNA
expression levels of EPS8L3 were determined in two liver cancer
cell lines (HepG2 and HCCLM3) and an immortalized human liver cell
line (HL-7702). Consistent with the aforementioned findings in
liver cancer tissue samples, the data revealed that the expression
levels of EPS8L3 were significantly increased in liver cancer cell
lines compared with in the normal liver cell line (P<0.01;
Fig. 1D). Collectively, the
results suggested that EPS8L3 expression was increased in liver
cancer, indicating that it may play an oncogenic role in the
development of liver cancer.
Association between EPS8L3 expression,
clinicopathological features and survival
As presented in Table
I, high EPS8L3 expression was associated with high grade
(P=0.024) and mortality (P=0.011); however, there was no
significant association between EPS8L3 expression and other
clinicopathologic parameters, including age, sex, clinical stage,
tumor stage, node stage and metastasis stage (P>0.05).
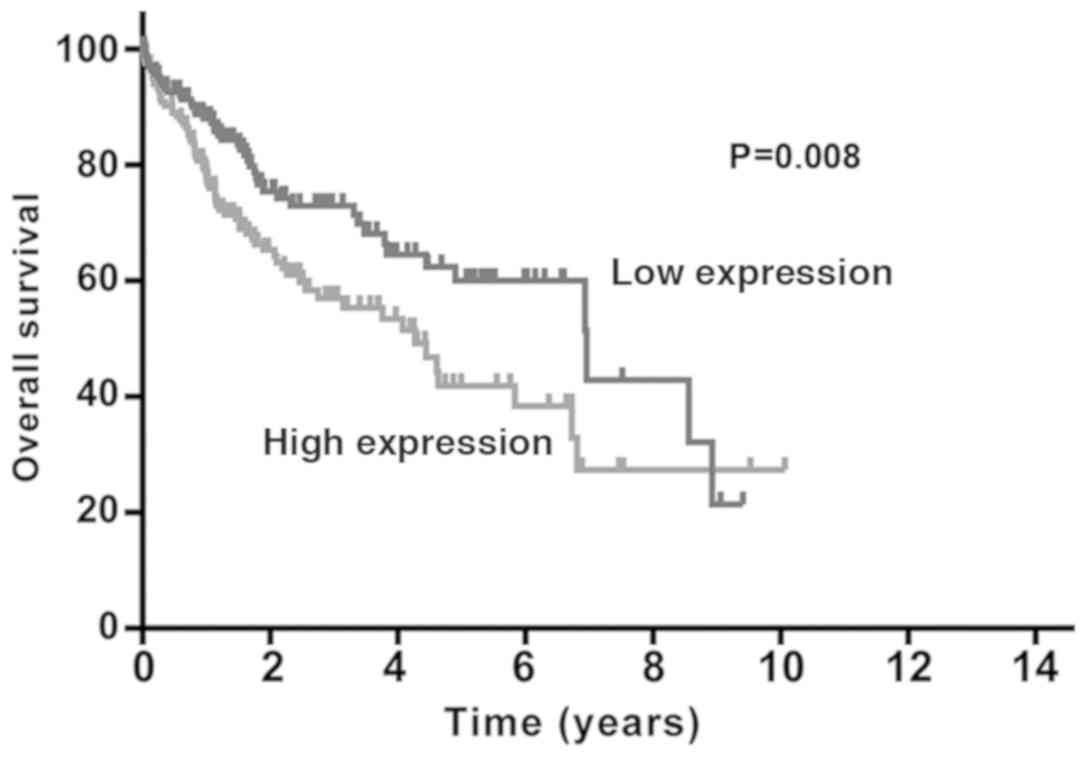
Survival analysis using the Kaplan-Meier method
revealed that patients with liver cancer who exhibited high levels
of EPS8L3 expression had a shorter survival time compared with
those with low levels of EPS8L3 expression (P=0.008; Fig. 2). Furthermore, a Cox proportional
hazards regression model was used for univariate and multivariate
analyses. In univariate analysis, several prognostic factors,
including EPS8L3 expression (P=0.009), clinical stage (P<0.001),
tumor stage (P<0.001) and metastasis stage (P=0.023) were
identified (Table II).
Multivariate analyses revealed that EPS8L3 expression (hazard
ratio=1.58; 95% confidence interval, 1.085–2.301; P=0.017) could be
independent predictor of overall survival (OS).
 | Table II.Univariate and multivariate analyses
of factors associated with survival. |
Table II.
Univariate and multivariate analyses
of factors associated with survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | P-value | HR | 95% CI | P-value | HR | 95% CI |
|---|
| EPS8L3 expression,
high/low |
0.009a | 1.654 | 1.136–2.406 | 0.017 | 1.580 | 1.085–2.301 |
| Clinical stage, I +
II/III + IV |
0.000a | 2.410 | 1.659–3.500 |
|
|
|
| Tumor stage, T1 +
T2/T3 + T4 |
0.000a | 2.429 | 1.671–3.532 |
|
|
|
| Metastasis stage,
M0/M1 |
0.023a | 3.797 | 1.201–12.004 |
|
|
|
| Node stage, N0/N1 +
N2 + N3 | 0.346 | 1.963 | 0.483–7.974 |
|
|
|
| Age,
<60/≥60 | 0.217 | 1.264 | 0.871–1.835 |
|
|
|
| Sex,
female/male | 0.292 | 0.815 | 0.558–1.192 |
|
|
|
| Grade, G1 +
G2/G3 | 0.423 | 1.166 | 0.801–1.697 |
|
|
|
EPS8L3 knockdown inhibits cell
proliferation and migration
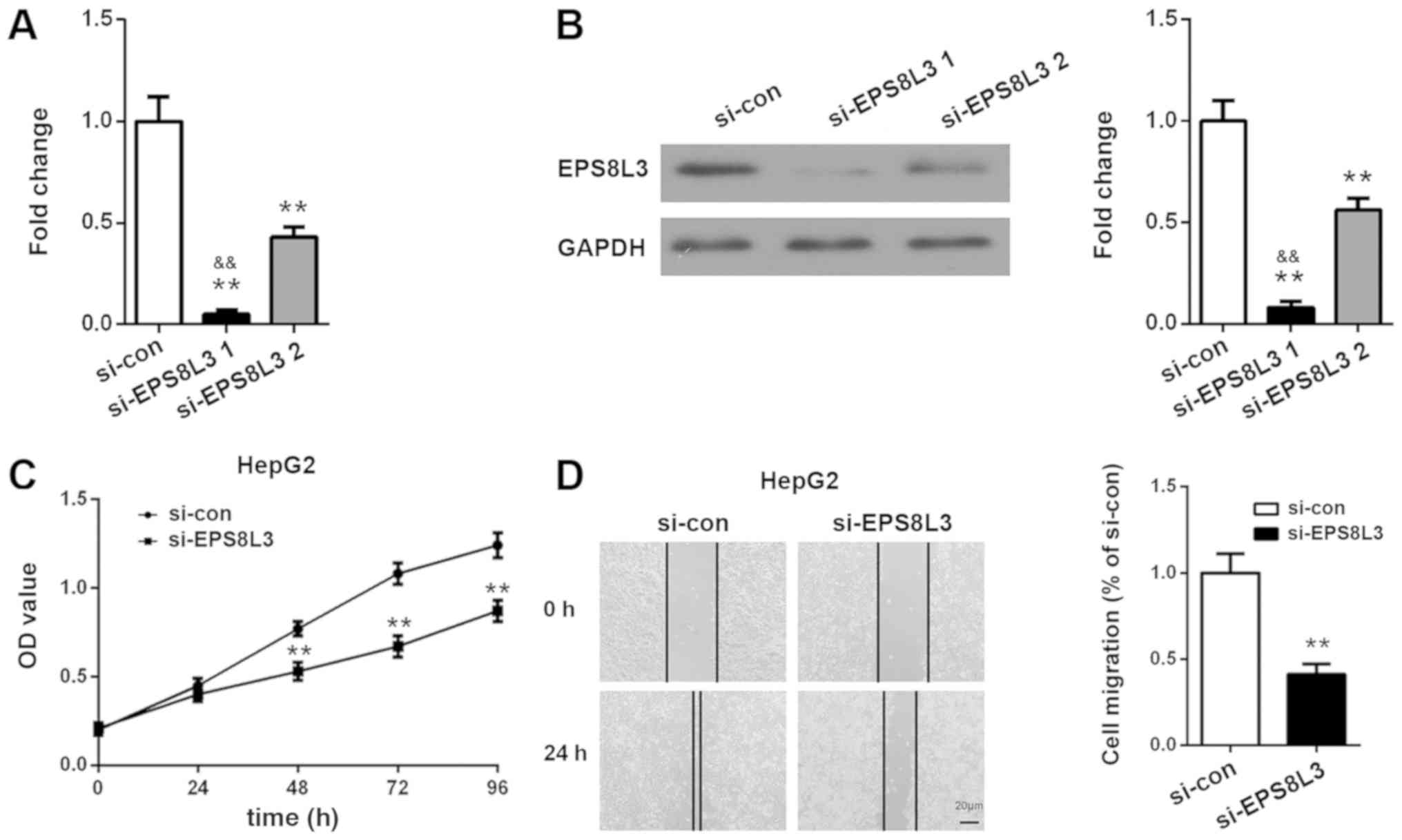
After knocking down EPS8L3 expression in HepG2 cells
using siRNA, the suppressive effects of the siRNAs were determined
at 72 h using RT-qPCR and western blot analysis. As presented in
Fig. 3A and B, EPS8L3 expression
was inhibited significantly following siRNA transfection. EPS8L3
expression was significantly lower following EPS8L3 siRNA-1
transfection compared with EPS8L3 siRNA-2 transfection at the mRNA
and protein levels (P<0.01). Therefore, si-EPS8L3-1, which
achieved a >90% reduction in EPS8L3 expression, was selected for
use in the subsequent experiments. A CCK-8 assay revealed that
EPS8L3 silencing significantly suppressed the rate of proliferation
of the HepG2 cell line at 48, 72 and 96 h (P<0.01; Fig. 3C). Wound healing assays
demonstrated that EPS8L3 knockdown induced a significant reduction
in HepG2 cell migration, as indicated by the greater distance
between the two wound fronts following EPS8L3 depletion compared
with the control siRNA (P<0.01; Fig. 3D). In summary, these results
indicated that the decreased expression of EPS8L3 inhibited the
proliferation and migration of liver cancer cells.
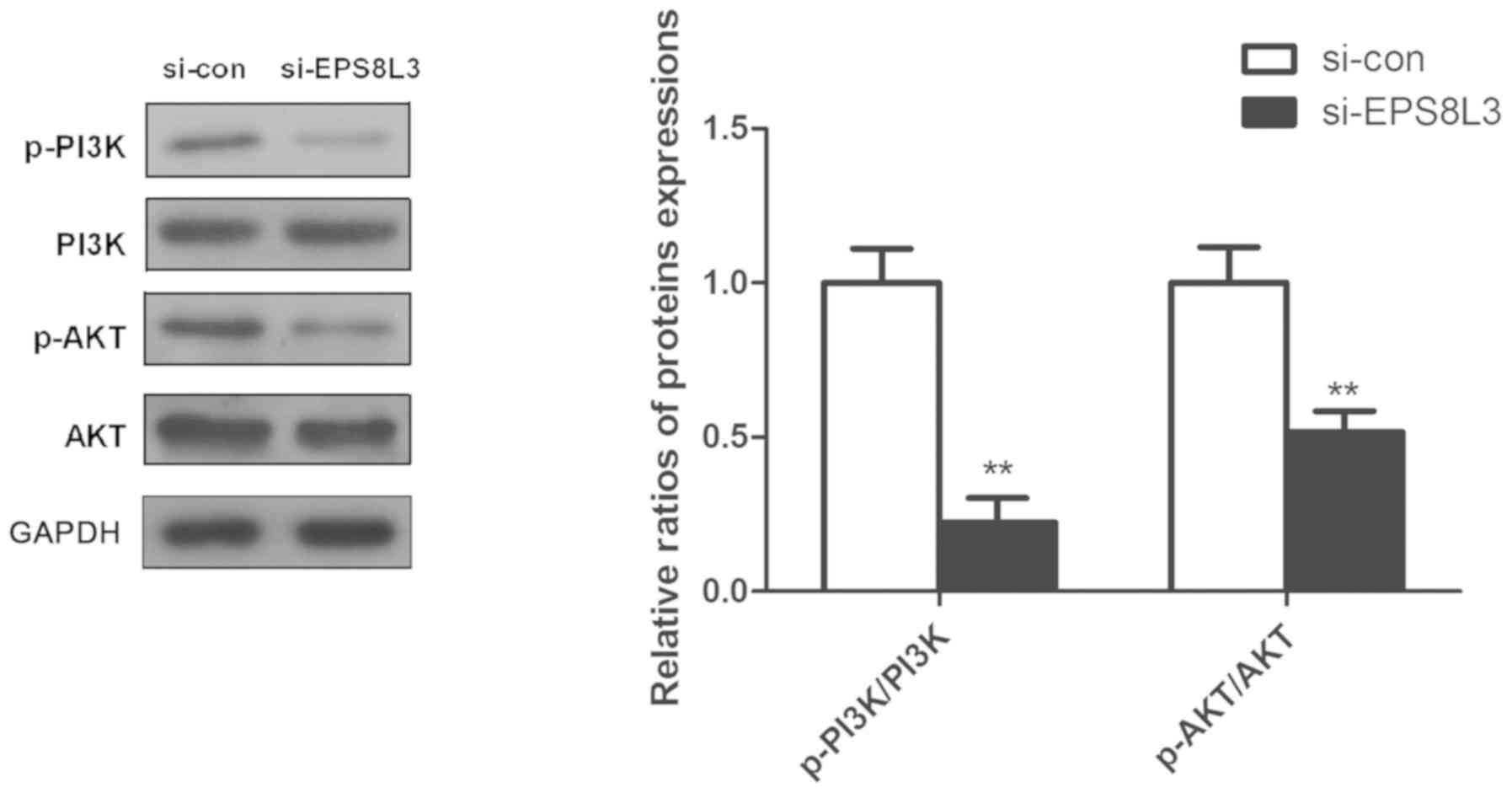
Effects of EPS8L3 on the PI3K/AKT
signaling pathway in liver cancer cells
Previous studies have reported that the PI3K/AKT
signaling pathway plays an important role in the cell survival
(19,20); thus, the expression and
phosphorylation of PI3K and AKT was evaluated to further
investigate the potential role of EPS8L3 in liver cancer. It was
observed that the knockdown of EPS8L3 significantly reduced the
levels of p-PI3K and p-AKT in the HepG2 cell line, as indicated by
the ratio of p-PI3K/PI3K and p-AKT/AKT (Fig. 4; P<0.01).
Discussion
EPS8 serves an important role in regulating the
development and progression of a number of human tumors (21,22);
however, the clinical relevance, significance to prognosis and
potential regulatory effect of EPS8L3 in liver cancer is not fully
understood. In the present study, it was observed that EPS8L3
expression was significantly increased in liver cancer cell lines,
which was consistent with the results from liver cancer tissues
according to data obtained from the Oncomine database. In addition,
the depletion of EPS8 significantly inhibited liver cancer cell
proliferation and migration. Previous studies indicated that EPS8
was overexpressed in various solid tumors, including breast cancer
(23), gliomas (24), squamous carcinoma (25) and ovarian cancer (26). Moreover, EPS8 is an important
regulator of multiple biological functions in tumor cells. It has
been recently been reported that EPS8L3 expression was
significantly elevated in liver cancer tissues and cell lines, and
that EPS8L3 silencing reduced the proliferation of liver cancer
cells (16). The present study
that found a high-level of EPS8L3 was predictive of a poor
prognosis and identified EPS8L3 as an independent prognostic
indicator in patients with liver cancer. The silencing of EPS8L3
reduced liver cancer cell proliferation, these data demonstrated
that EPS8L3 acts as a promotor of liver cancer progression.
EPS8 has been identified as an oncogene (27), and Chen et al (28) reported that the upregulation of
EPS8 expression was associated with shorter OS and disease-free
survival in cervical cancer. Furthermore, He et al (21) revealed that patients with high EPS8
expression exhibited shorter event-free survival, and that EPS8 may
be a valuable clinical biomarker for patients with acute
lymphoblastic leukemia. These previous studies demonstrated the
clinical significance of EPS8; thus, the present study further
explored the potential value of EPS8L3 using data obtained from
TCGA database for 338 patients with liver cancer. The results of
the present study revealed that high EPS8L3 expression
significantly associated with aggressive clinicopathological
features, including higher tumor grade and mortality rate, which
further suggested an oncogenic role for EPS8L3 in the development
of liver cancer. Furthermore, survival analysis suggested that
patients with liver cancer who had a high level of EPS8L3
expression had a significantly lower survival rate than those with
a low EPS8L3 expression level. Therefore, EPS8L3 may be an
independent predictor of prognosis. To the best of our knowledge,
the present study is the first report to reveal the prognostic
value of EPS8L3 for patients with liver cancer.
An increasing volume of evidence has shown that the
abnormal activation of the PI3K/AKT signaling pathway is involved
in the progression of a variety of solid tumors by regulating
proliferation, differentiation, apoptosis and survival, including
in liver cancer (29–33). EPS8 has been reported to be a
signaling molecule involved in the regulation of the PI3K/AKT
signaling pathway (34,35). Additionally, Zeng et al
(17) revealed that an AKT
inhibitor suppressed the effects of EPS8L3 on the proliferation of
EPS8L3-overexpressing cells, suggesting that EPS8L3 may promote
proliferation by hyperactivating the AKT signaling pathway.
Consistent with previous evidence that EPS8 knockdown inactivated
the PI3K/AKT signaling pathway, the present study revealed that
EPS8L3 depletion significantly reduced the levels of phosphorylated
PI3K and AKT in liver cancer cells, indicating that EPS8L3 serves
an important role in tumor development.
Collectively, the present study found that EPS8L3
expression was upregulated in liver cancer tissues and cell lines,
and was associated with aggressive clinicopathological
characteristics, including a higher tumor grade, higher mortality
rate and poor prognosis. Moreover, EPS8L3 inhibited liver cancer
cell proliferation and migration by regulating the PI3K/AKT
signaling pathway. Thus, the results of the present study indicated
that EPS8L3 could act as an oncogene in liver cancer and be a novel
prognostic biomarker for patients with liver cancer. However, to
further verify the findings of the present study, western blot
analysis or immunohistochemistry is required to examine EPS8L3
expression in clinical specimens.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of material and data
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GYC, PL and TH conceived and designed the study. PL,
TH, HSW, YT and YM performed the experiments. XDW and YSX analyzed
the data, revised the figures and drafted the manuscript. All
authors read and approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu M, Liu Z, Zhang A and Li N:
Identification of key genes and pathways in hepatocellular
carcinoma: A preliminary bioinformatics analysis. Medicine
(Baltimore). 98:e142872019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Haines K, Sarabia SF, Alvarez KR,
Tomlinson G, Vasudevan SA, Heczey AA, Roy A, Finegold MJ, Parsons
DW, Plon SE and López-Terrada D: Characterization of pediatric
hepatocellular carcinoma reveals genomic heterogeneity and diverse
signaling pathway activation. Pediatr Blood Cancer. 66:e277452019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu M, Liu Z, Li X, Zhang A, Lin D and Li
N: Analysis of potential key genes in very early hepatocellular
carcinoma. World J Surg Oncol. 17:772019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fu J and Wang H: Precision diagnosis and
treatment of liver cancer in China. Cancer Lett. 412:283–288. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Della Corte C, Triolo M, Iavarone M and
Sangiovanni A: Early diagnosis of liver cancer: An appraisal of
international recommendations and future perspectives. Liver Int.
36:166–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maluccio M and Covey A: Recent progress in
understanding, diagnosing, and treating hepatocellular carcinoma.
CA Cancer J Clin. 62:394–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yeudall A and Patel V: EPS8 signaling as a
therapeutic target in oral cancer. Oral Dis. 24:128–131. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ohshima K, Hatakeyama K, Kanto K, Ide T,
Watanabe Y, Moromizato S, Wakabayashi-Nakao K, Sakura N, Yamaguchi
K and Mochizuki T: Comparative proteomic analysis identifies
exosomal Eps8 protein as a potential metastatic biomarker for
pancreatic cancer. Oncol Rep. 41:1019–1034. 2019.PubMed/NCBI
|
|
11
|
Wen Q, Jiao X, Kuang F, Hou B, Zhu Y, Guo
W, Sun G, Ba Y, Yu D, Wang D, et al: FoxO3a inhibiting expression
of EPS8 to prevent progression of NSCLC: A new negative loop of
EGFR signaling. EBioMedicine. 40:198–209. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nasri E, Wiesen LB, Knapik JA and
Fredenburg KM: Eps8 expression is significantly lower in
p16+ head and neck squamous cell carcinomas (HNSCCs)
compared with p16− HNSCCs. Hum Pathol. 72:45–51. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Q and Yong L: Eps8, a therapeutic
potential target for cancer. Hum Pathol. 82:300–301. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen Y, Xie X, Wu A, Wang L, Hu Y, Zhang H
and Li Y: A synthetic cell-penetrating peptide derived from nuclear
localization signal of EPS8 exerts anticancer activity against
acute myeloid leukemia. J Exp Clin Cancer Res. 37:122018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Offenhäuser N, Castelletti D, Mapelli L,
Soppo BE, Regondi MC, Rossi P, D'Angelo E, Frassoni C, Amadeo A,
Tocchetti A, et al: Increased ethanol resistance and consumption in
Eps8 knockout mice correlates with altered actin dynamics. Cell.
127:213–226. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tocchetti A, Confalonieri S, Scita G, Di
Fiore PP and Betsholtz C: In silico analysis of the EPS8 gene
family: Genomic organization, expression profile, and protein
structure. Genomics. 81:234–244. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zeng CX, Tang LY, Xie CY, Li FX, Zhao JY,
Jiang N, Tong Z, Fu SB, Wen FJ and Feng WS: Overexpression of
EPS8L3 promotes cell proliferation by inhibiting the transactivity
of FOXO1 in HCC. Neoplasma. 65:701–707. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen H, Li H and Chen Q: INPP4B reverses
docetaxel resistance and epithelial-to-mesenchymal transition via
the PI3K/Akt signaling pathway in prostate cancer. Biochem Biophys
Res Commun. 477:467–472. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sheng J, Shen L, Sun L, Zhang X, Cui R and
Wang L: Inhibition of PI3K/mTOR increased the sensitivity of
hepatocellular carcinoma cells to cisplatin via interference with
mitochondrial-lysosomal crosstalk. Cell Prolif. 52:e126092019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He YZ, Liang Z, Wu MR, Wen Q, Deng L, Song
CY, Wu BY, Tu SF, Huang R and Li YH: Overexpression of EPS8 is
associated with poor prognosis in patients with acute lymphoblastic
leukemia. Leuk Res. 39:575–581. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li M, Yang J, Zhang L, Tu S, Zhou X, Tan
Z, Zhou W, He Y and Li Y: A low-molecular-weight compound exerts
anticancer activity against breast and lung cancers by disrupting
EGFR/Eps8 complex formation. J Exp Clin Cancer Res. 38:2112019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen C, Liang Z, Huang W, Li X, Zhou F, Hu
X, Han M, Ding X and Xiang S: Eps8 regulates cellular proliferation
and migration of breast cancer. Int J Oncol. 46:205–214. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ding X, Zhou F, Wang F, Yang Z, Zhou C,
Zhou J, Zhang B, Yang J, Wang G, Wei Z, et al: Eps8 promotes
cellular growth of human malignant gliomas. Oncol Rep. 29:697–703.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schoenherr C, Serrels B, Proby C,
Cunningham DL, Findlay JE, Baillie GS, Heath JK and Frame MC: Eps8
controls Src- and FAK-dependent phenotypes in squamous carcinoma
cells. J Cell Sci. 127:5303–5316. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen H, Wu X, Pan ZK and Huang S:
Integrity of SOS1/EPS8/ABI1 tri-complex determines ovarian cancer
metastasis. Cancer Res. 70:9979–9990. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li YH, Xue TY, He YZ and Du JW: Novel
oncoprotein EPS8: A new target for anticancer therapy. Future
Oncol. 9:1587–1594. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen YJ, Shen MR, Chen YJ, Maa MC and Leu
TH: Eps8 decreases chemosensitivity and affects survival of
cervical cancer patients. Mol Cancer Ther. 7:1376–1385. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu HY, Kim SO, Jin CY, Kim GY, Kim WJ, Yoo
YH and Choi YH: β-lapachone-induced apoptosis of human gastric
carcinoma AGS cells is caspase-dependent and regulated by the
PI3K/Akt pathway. Biomol Ther (Seoul). 22:184–192. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fu YL, Zhang QH, Wang XW and He H:
Antidiabetic drug metformin mitigates ovarian cancer SKOV3 cell
growth by triggering G2/M cell cycle arrest and inhibition of
m-TOR/PI3K/Akt signaling pathway. Eur Rev Med Pharmacol Sci.
21:1169–1175. 2017.PubMed/NCBI
|
|
31
|
Paplomata E and O'Regan R: The
PI3K/AKT/mTOR pathway in breast cancer: Targets, trials and
biomarkers. Ther Adv Med Oncol. 6:154–166. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang H, Cao Y, Chen Y, Li G and Yu H:
Apatinib promotes apoptosis of the SMMC-7721 hepatocellular
carcinoma cell line via the PI3K/Akt pathway. Oncol Lett.
15:5739–5743. 2018.PubMed/NCBI
|
|
33
|
Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T,
Wen H, Yang Y, Wang S, Wang J, et al: Exosomal microRNA-32-5p
induces multidrug resistance in hepatocellular carcinoma via the
PI3K/Akt pathway. J Exp Clin Cancer Res. 37:522018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu M, Shorts-Cary L, Knox AJ,
Kleinsmidt-Demasters B, Lillehei K and Wierman ME: Epidermal growth
factor receptor pathway substrate 8 is overexpressed in human
pituitary tumors: Role in proliferation and survival.
Endocrinology. 150:2064–2071. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang H, Patel V, Miyazaki H, Gutkind JS
and Yeudall WA: Role for EPS8 in squamous carcinogenesis.
Carcinogenesis. 30:165–174. 2009. View Article : Google Scholar : PubMed/NCBI
|