Introduction
The incidence of colorectal cancer is increasing
globally. A total of ~655,000 people succumb to colorectal cancer
each year. The 5-year survival rate of early colorectal cancer is
~90%, but this rate decreases to 15% in metastatic colorectal
cancer (1). Although numerous
protocols for early screening and treatment have been developed in
previous years, there has been no significant increase in the
survival rate of colorectal cancer in the past 20 years (2). In addition, the most common cause of
treatment failure is metastasis. This process is multifaceted,
involving cancer cell invasion, epithelial mesenchymal transition
(EMT), extracellular matrix degradation, angiogenesis and
microenvironment chemotactic action. The abnormal activation or
inactivation of a number of oncogenes or tumor suppressor genes and
signaling pathways are regulated at transcriptional,
post-transcriptional and translational levels, and the regulatory
mechanisms are complex (3).
Colorectal cancer is one of the three most common
fatal types of tumor, with up to 1.03 million cases each
year and 530,000 mortalities each year in developed countries
including Western Europe and the United States of America,.
According to China's Ministry of Health, the incidence of
colorectal cancer is the third-highest incidence of all cancers
(4). Colorectal cancer is a
malignant tumor caused by multiple factors, including individual
genetics and environmental effects. For example, large-scale
genetic studies have identified that the occurrence of colorectal
cancer is associated with multiple genes abnormal expression and
multiple molecular interactions (5–7), but
the studies investigating the molecular mechanism of colorectal
cancer incidence are not exhaustive. Therefore, additional studies
exploring the pathogenesis of colorectal cancer may provide the
theoretical basis for novel clinical anti-tumor drugs (8).
Long non-codingRNAs (lncRNA) are a type of molecule
within the transcriptome in the cell nucleus or cytoplasm, similar
to mRNA, with a length >200 nucleotides. Previous studies have
demonstrated that lncRNA is closely associated with the occurrence
of cancer (9,10). Abnormal lncRNAs may serve key roles
as tumor suppressor genes or oncogenes. Colorectal cancer has one
of the most rapidly increasing morbidity rates and has become a
threat to health and life. (11).
Studies investigating this association between lncRNAs and cancer
have indicated from non-small cell lung cancer genome-wide
microarray data that small nucleolar RNA host gene 6 (SNHG6),
LINC00649, DLX6-AS1 and 3 lncRNA were identified to be abnormally
expressed in patients with lung cancer (9–11),
but no study has demonstrated whether lncRNA SNHG6 serves a
significant role in colorectal cancer.
The normal human E26 transformation specific-1
(ETS1) gene is a proto-oncogene. It has been suggested that the
ETS1 gene is involved in cell growth and extracellular matrix
invasion, which promotes tumor invasion and metastasis (12). Previous studies have indicated that
the expression of ETS1 is increased in a number of malignant tumors
(12,13). The expression of ETS1 and its roles
in tumor invasion and metastasis have attracted widespread
attention.
The present study investigated the role of lncRNA
SNHG6 in the development of colorectal cancer; the results
identified that the expression of lncRNA SNHG6 was inhibited in
tumor cells and tissues, and that the overexpression of lncRNA
SNHG6 in cells may suppress ETS1 expression, inhibit cell
proliferation. In addition, it was confirmed that the lncRNA SNHG6
inhibited ETS1 expression by directly targeting its 3′-untranslated
regions (UTR) and activated the intrinsic invasion pathway through
downregulating expression of phosphoinositide 3-kinase
(PI3K)/protein kinase B (AKT)/mechanistic target of rapamycin
(mTOR).
Materials and methods
Colorectal cancer tissue samples and
the variant choice of SNHG6
A total of 30 colon tumors and adjacent non-tumor
tissues samples were collected from patients with colorectal cancer
from the Cancer Hospital of China Medical University (Shenyang,
China) between April 2017 and September 2017. There were 18 males
and 12 females; the age range was 34–82 years old, with a median
age of 52 years. Patients were diagnosed according to the World
Health Organization colon cancer histology classification and
grading standards 2010 (14). The
colon cancer staging system proposed by the American Cancer
Association (AJCC) has become widely recognized as an independent
indicator that can comprehensively reflect the progress of
malignant tumors and judge prognosis (15). All patients with colon tumors
enrolled in the present study did not receive any treatment prior
to surgery. This was due to the fact that different radiotherapy or
chemotherapy regimens may affect the outcomes of genetic studies of
colon cancer, and it is difficult to standardize the frequency and
intensity of different chemotherapy and radiotherapy treatments
within the patient cohort. The protocol was approved by the Ethics
Committee of Cancer Hospital of China Medical University, and
written informed consent was obtained from all study participants.
SNHG6 has several variants; from previous optimization experiments
(16) it was identified that only
variant 1 successfully completed the editing of the gene, so
variant 1 was used to establish the overexpression model.
Concomitantly, subsequent experiments within the present study
confirmed that this variant was effective.
Cell culture and transfection
The human colon cancer SW480 and HCT-116 cell lines
and normal colonic mucosa NCM460 cell line were purchased from
American Type Culture Collection (Manassas, VA, USA). These cells
were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and
incubated in a humidified atmosphere containing 5% CO2
at 37°C. LncRNA pWPXL-SNHG6 and small interfering (si)-SNHG6 and
si-negative control were purchased from Shanghai GenePharma Co.,
Ltd. (Shanghai, China).
For transfection, cells at a density of
1×105 cells/well were seeded in each well of a 24-well
microplate, grown for 24 h to reach 30–50% confluence and then
incubated with 50 nM LncRNA pWPXL-SNHG6 and small interfering
si-SNHG6 and si-negative control with Lipofectamine®
2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in 100 µl
serum-free DMEM, according to the manufacturer's protocols.
RNA isolation and reverse
transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from SW480 and HCT-116 cell
lines using TRIzol® (Thermo Fisher Scientific, Inc.).
The corresponding RNA was reverse transcribed into cDNA using a
QuantiFast SYBR Green PCR kit (Qiagen GmbH, Hilden, Germany). Then,
RT-qPCR analysis was detected using the SYBR Green qPCR Master Mix
(5 µl; Invitrogen; Thermo Fisher Scientific, Inc.) on a 7500 Fast
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). For
the PCR experiments, the following forward and reverse primers were
used: SNHG6; Forward, 5′-TGCCAGCAGTGACAGCAGCA-3′ and reverse,
5′-TACGGAGGTGGAGTGCCAT-3′; and reference gene GAPDH; Forward,
5′-CAAAGGTGGATCAGATTCAAG-3′ and reverse,
5′-GGTGAGCATTATCACCCAGAA-3′.
The PCR conditions included an initial denaturation
step of 94°C for 2 min, followed by 30 cycles of 94°C for 30 sec,
59°C for 30 sec, 72°C for 2 min and a final elongation step at 72°C
for 10 min. Taq DNA polymerase was purchased from Sigma-Aldrich
(Merck KGaA, Darmstadt, Germany). GAPDH was used as an internal
control to normalize gene expression. The relative gene expression
levels were calculated using the ΔΔCq method (17).
Western blot analysis
Protein samples from cells were homogenized using
radioimmunoprecipitation assay lysis buffer (Invitrogen; Thermo
Fisher Scientific, Inc.). The protein concentrations of the cell
extracts were then measured using Bradford protein dye reagent
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). A total of 30
µg/lane protein was loaded. The proteins lysates were separated on
a 10% SDS-PAGE separation gel and transferred onto a nitrocellulose
membrane by 300 mA for 90 mins, blocked with 5% bovine serum
albumin (Gibco; Thermo Fisher Scientific, Inc.) for 2 h at room
temperature, and incubated with primary antibodies (Abcam, UK):
anti-ETS (1:2,000; ab26096), anti-p-PI3K (1:2,000; ab151549), anti-
PI3K (1:2,000; ab32089) anti p-AKT (1:3,000; ab8932), anti AKT
(1:2,000; ab38449), anti p-mTOR (1:2,000; ab109268), anti mTOR
(1:2,000; ab2732) and anti-GAPDH (1:1,000; ab9485) at 4°C
overnight. Then, membranes were incubated at room temperature for 2
h with anti-IgG conjugated with horseradish peroxidase secondary
antibodies (1:5,000; ab97040) before being visualized using the
SuperSignal West Pico Chemiluminescent Substrate Trial kit (Pierce
Protein Biology; Thermo Fisher Scientific, Inc.). Images were
obtained using the ChemiDoc XRS system with Quantity One software
(Bio-Rad Laboratories, Inc.). Protein expression was analyzed using
BandScan 5.0 software (Glyko, Inc., Novato, CA, USA). All
experiments were repeated three times.
Cell proliferation analysis
Cells proliferation was detected in vitro
using an MTT assay. Briefly, following transfection, HCT-116 cell
lines were seeded at a density of 2,000 cells/well in 96-well
plates and all groups were incubated for 24, 48, 72 or 96 h. MTT
was added into each well (5 mg/ml; Sigma-Aldrich; Merck KGaA), and
culture was maintained at 37°C for 4–6 h. Samples were measured at
570 nm using the MTT cell proliferation kit (Cayman Chemical
Company, Ann Arbor, MI, USA).
Flow cytometry
For the cell apoptosis analysis, HCT116 cells were
harvested 48 h after transfection and immobilized in 70% ethanol at
−20°C for 30 min. Then, the cells were resuspended in 10 ml RNase,
and stained with propidium iodide (PI; 1:200) and Annexin V-FITC
(1:200; BD Pharmingen, San Diego, CA, USA) for 30 min at 37°C. A
total of 500 µl PBS was added and mixed thoroughly. The samples
were then analyzed by analyzed with a FACScan flow cytometer (BD
Biosciences). Apoptotic rate of cells was analyzed by flow
cytometry (BD, Biosciences) using WinMDI software (version 2.9; BD
Biosciences). All experiments were repeated 3 times.
Transwell invasion assays
The pore size in the bottom membrane of the
Transwell chambers (Corning Incorporated, Corning, NY, USA) was 8
µm. The chambers were coated with Matrigel (Sigma-Aldrich; Merck
KGaA) for the determination of cell invasive abilities at room
temperature for 24 h. HCT116 cells were grown to ~80% confluence in
the 24-well Transwell plates. Then, after 24 h incubation at 37°C
in 15% fetal bovine serum medium, the invasion of cells was
measured using light microscopy at a magnification of ×200
(Shanghai Bime Instrument Co., Ltd., Shanghai, China).
Cell scratch assay
HCT116 cells (5×104 cells/well) were
seeded in a 6-well plate (5×104 cells per well). Lines
were then drawn behind the plate with a ruler. Following attachment
of the cells, a scratch was made across the middle of the wells,
the width of a 10 µl pipette tip. Non-adherent cells were removed
by washing with PBS 3 times, and then serum-free medium (Gibco;
Thermo Fisher Scientifc, Inc.) was added. Cells were incubated at
37°C and 5% CO2. Images were captured at 0 and 48 h.
Statistical analysis
Data are presented as the mean ± standard deviation
of 3 independent experiments. The results were analyzed using a
Student t-test or a one-way analysis of variance with Tukey's
post-hoc test. All the data were analyzed using SPSS Statistics
19.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism version 5.0
(GraphPad Software, Inc., La Jolla, CA, USA) with Microsoft Excel
(Microsoft Corporation, Redmond, WA, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
LncRNA SNHG6 is downregulated in colon
cancer tissues and cell lines
A previous study demonstrated that SNHG6 is
upregulated in hepatocellular carcinoma and indicates the
possibility of high recurrence in cancer patients (9). However, the expression of lncRNA
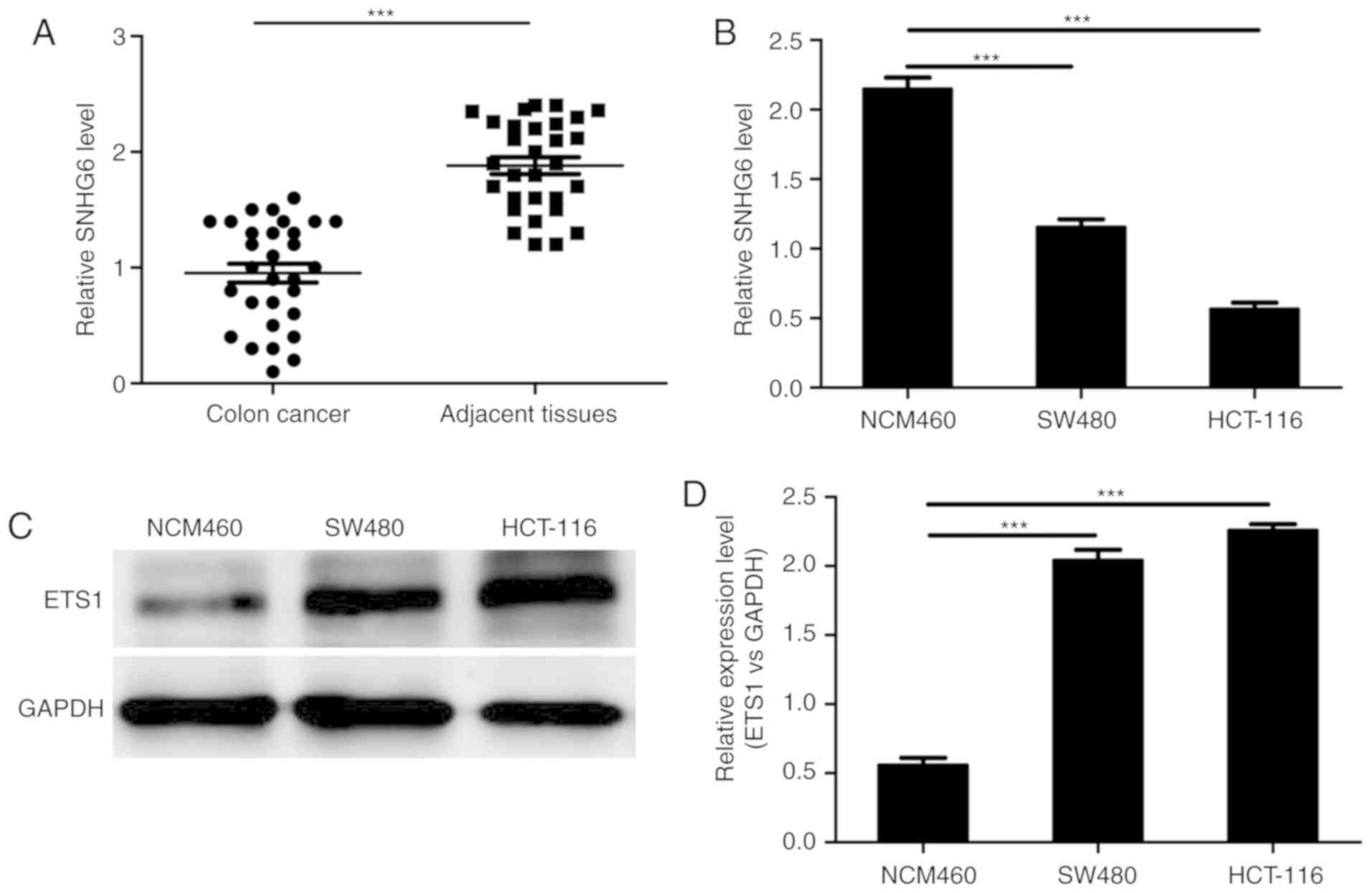
SNHG6 has not been determined in colon cancer. Firstly, the
expression of lncRNA SNHG6 was detected in 30 pairs of colon cancer
and adjacent tissues by RT-qPCR. The RT-qPCR results verified that
the expression of lncRNA SNHG6 was markedly decreased in colon
cancer tissues compared with the normal tissues (Fig. 1A). Furthermore, similar results
were observed when examining colon cancer cell lines, in which the
expression of lncRNA SNHG6 was decreased compared with the normal
colon cell line (Fig. 1B). In
addition, the expression of ETS1, which is a transcription factor,
was detected. The western blot analysis results suggested that ETS1
was highest in colon cancer cell lines (Fig. 1C). These data indicate that lncRNA
SNHG6 may serve a key role in colon cancer progression.
LncRNA SNHG6 represses colon cancer
proliferation
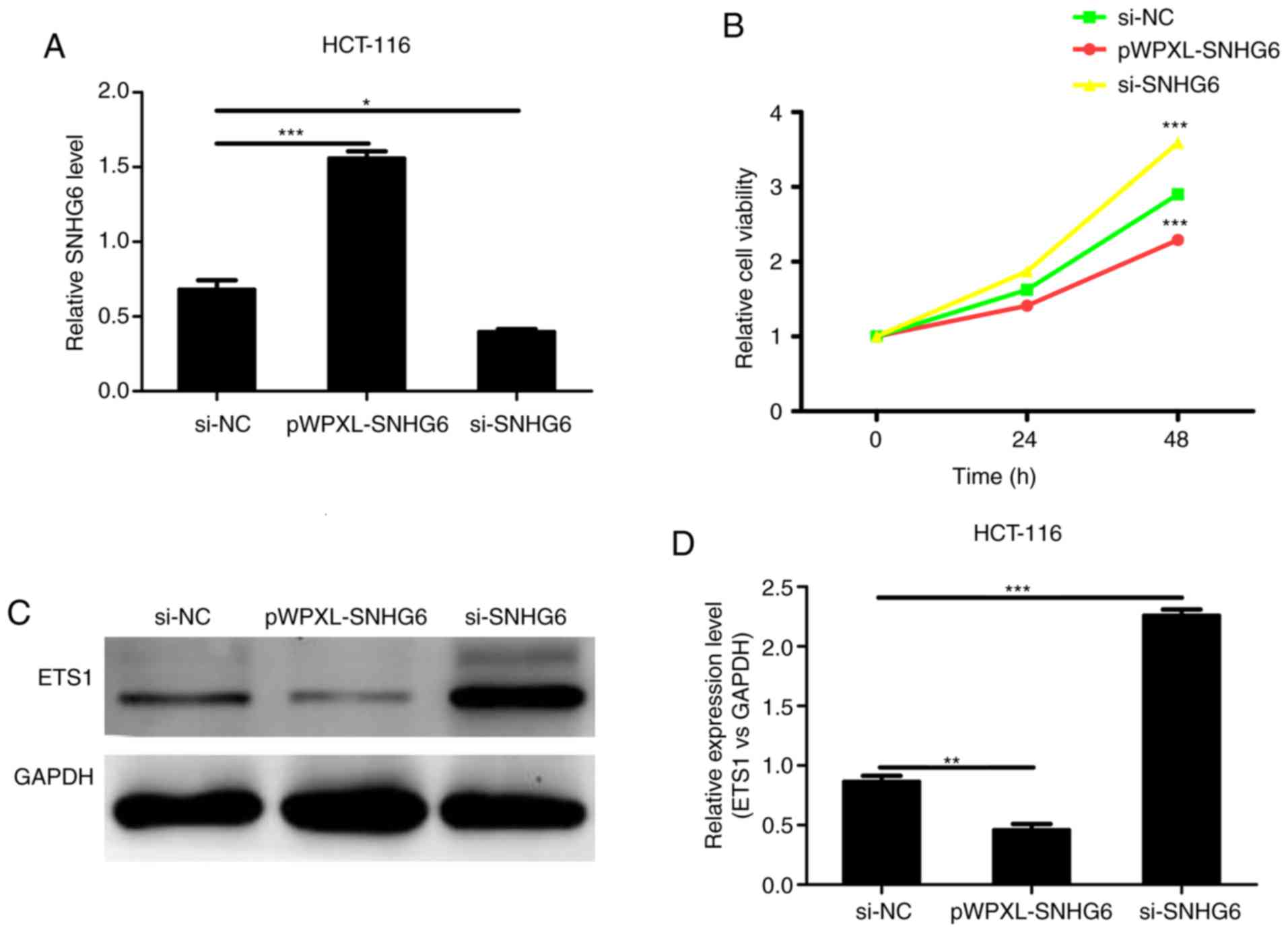
To additionally explore the role of lncRNA SNHG6 in
colon cancer cells, the cell proliferation rates were detected
in vitro following transfection with pWPXL-SNHG6 plasmids
and si-SNHG6 to overexpress or silence lncRNA SNHG6 expression,
respectively. The transfection efficiency was detected by RT-qPCR.
The RT-qPCR results demonstrated that lncRNA SNHG6 expression was
effectively upregulated following transfection with pWPXL-SNHG6;
conversely, lncRNA SNHG6 expression was markedly decreased in cells
following transfection with si-SNHG6 (Fig. 2A). Furthermore, an MTT assay was
used to detect the proliferation of colon cancer cells. As
demonstrated in Fig. 2B, the
proliferation of colon cancer cells was significantly inhibited
following overexpression of lncRNA SNHG6, whereas inhibition of
lncRNA SNHG6 significantly increased levels of cell growth. The
western blot analysis results suggested that the levels of ETS1
were downregulated when transfected with pWPXL-SNHG6 and
upregulated when transfected with si-SNHG6 (Fig. 2C). In conclusion, lncRNA SNHG6
inhibited the proliferation of colon cancer cells.
LncRNA SNHG6 inhibits proliferation
and induces apoptosis of colon cancer cells
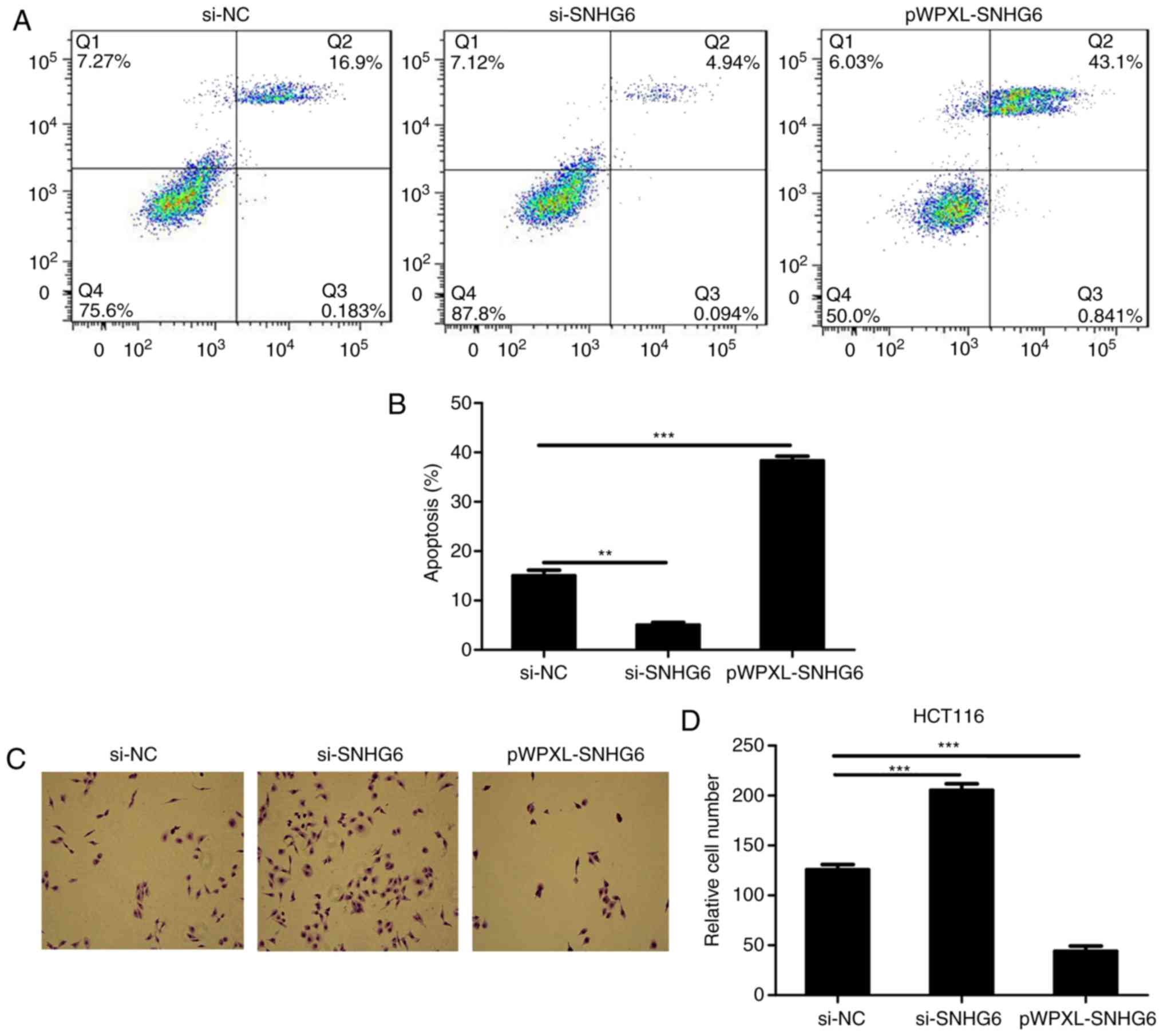
Flow cytometry was used to detect the expression of
the Annexin V and PI. As indicated in Fig. 3A, the apoptosis levels were
identified to be increased when lncRNA SNHG6 was overexpressed in
colon cells, and decreased when lncRNA SNHG6 was inhibited in colon
cells (Fig. 3A and B).
Subsequently, the Transwell assays suggested that the level of
invasion was decreased when lncRNA SNHG6 was overexpressed in
HCT116, and was upregulated following transfection with si-SNHG6 in
colon cancer cells (Fig. 3C and
D).
LncRNA SNHG6 inhibits migration of
colon cancer cells
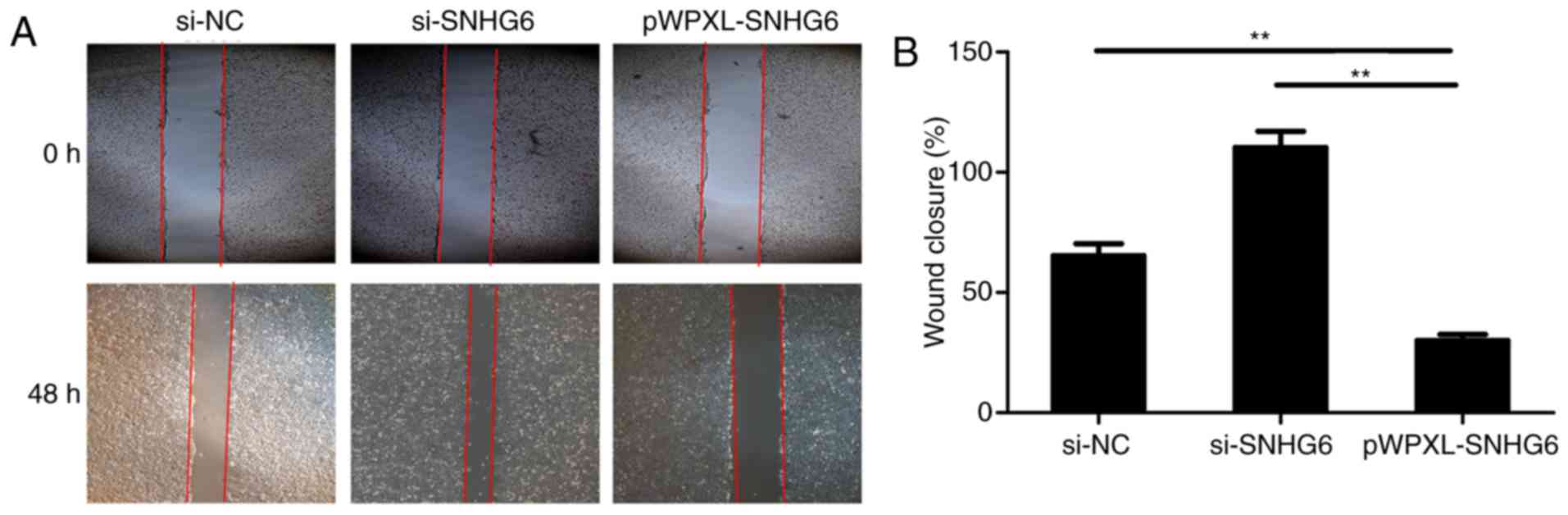
Cell scratch assays were used to detect the
migration of colon cancer cells. Overexpression of lncRNA SNHG6 in
HCT116 cells caused a decrease in cell migration levels, while
transfection with si-SNHG6 resulted in an increase in the levels of
migration in colon cancer cells (Fig.
4A and B).
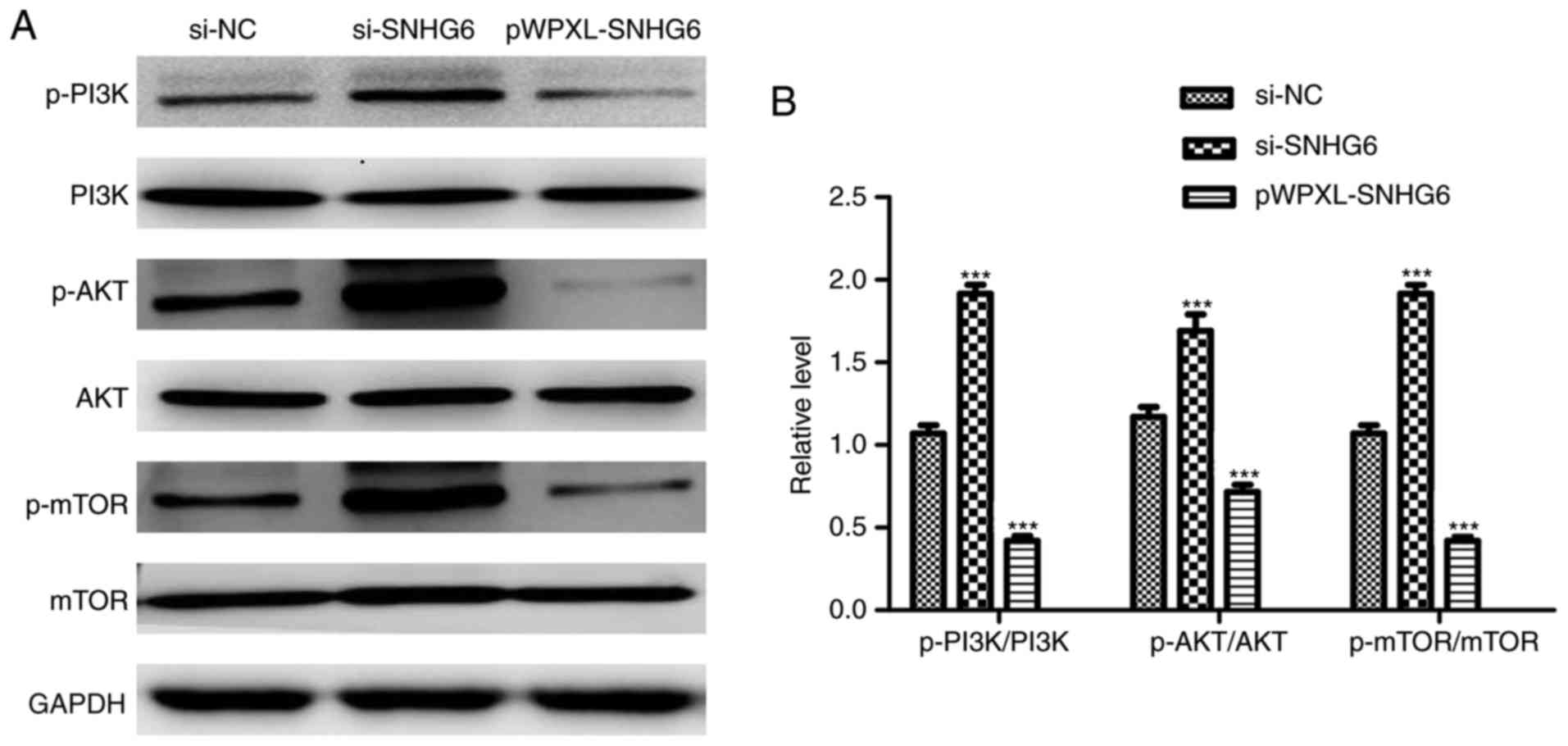
LncRNA SNHG6 regulates cell
proliferation through the PI3K/AKT/mTOR signaling pathway
Previous data have demonstrated that the
PI3K/AKT/mTOR signaling pathway promotes the development and
progression of colon cancer (18,19).
Therefore, the protein expression levels of phosphorylated-PI3K
(p-PI3K), p-AKT, p-mTOR, PI3K, AKT and mTOR were detected by
western blot analysis. The results demonstrated that the p-AKT,
p-PI3K and p-mTOR expression levels were markedly decreased
following overexpression of lncRNA SNHG6 (Fig. 5A and B), These results suggest that
lncRNA SNHG6 regulates cell proliferation through the PI3K/AKT/mTOR
signaling pathway in colon cancer.
Discussion
Colon cancer is one of the most common malignant
tumors in clinical practice, and its morbidity and mortality rates
are increasing in the majority of countries and regions throughout
the world, which poses a serious threat to human health. The
primary causes of mortality in these patients are invasion,
metastasis and chemotherapy resistance. The mechanism of metastasis
of colon cancer is complicated, and involves the abnormal
expression of multiple oncogenes and tumor suppressor genes;
previous studies have identified that abnormal lncRNA expression is
also an important factor (9–11).
Mature lncRNA may be located in the mRNA noncoding regions (3′-UTR
or introns area) of their target gene, and may result in complete
or incomplete complementary pairing, degradation or inhibition of
mRNA translation and post-transcriptional alterations in gene
expression. They may also participate in the regulation of cell
differentiation, proliferation, apoptosis, and tissue and organ
development (5,7,17).
Previous studies have confirmed that lncRNA may
regulate the expression of at least 30% of human protein-coding
genes (18), and serve a larger
role compared with protein-coding genes (19,20).
In a variety of human tumors, abnormal expression of lncRNA is
caused by gene mutation at brittle gene sites during gene transfer
activation (21). More recent
studies have identified that SNHG6 promoted growth of
nasopharyngeal tumors through targeted regulation of
lactotransferrin, and affected glioma cell aging through regulation
of cytoplasmic polyadenylation element binding protein 1 gene
expression (7,22). These data suggest that lncRNA SNHG6
is closely associated with the occurrence of tumor development.
The downregulation of lncRNA SNHG6 has been
associated with a variety of tumors: The expression of lncRNA SNHG6
in breast cancer cells was revealed to inhibit the growth of bone
metastasis and lung metastasis in situ tumors, and the
survival rate of patients with low expression of lncRNA SNHG6 was
significantly decreased compared with that of the high expression
group (22). Previous studies have
also demonstrated that lncRNA SNHG6 was significantly downregulated
in gastric cancer tissues compared with normal tissues, and
increased lncRNA SNHG6 may significantly induce cell cycle arrest
and inhibition in gastric cancer cells, and inhibit the invasion
ability of gastric cancer cells and the ability of tumor and
metastasis in vivo (9,23).
In addition, other studies have also suggested that lncRNA SNHG6
may inhibit the invasion ability of lung cancer cells (9–11).
However, the function and potential mechanism of action of lncRNA
SNHG6 in colon cancer remains to be elucidated, in particular how
SNHG6 affects cell migratory and invasive capabilities. At present,
with the development of sequencing technologies and the progression
of investigative studies, a number of lncRNAs have been identified
and investigated. It has been demonstrated that SNHG6 affected the
migration and invasion of cancer cells by altering the expression
of epithelial mesenchymal transition; this involves the
participation of multiple signaling pathways (9). The present study initially explored
the association between SNHG6 and PI3K/AKT/mTOR signaling
pathways.
A previous study hypothesized that SNHG6 represented
an unusual snoRNA gene, and there is a difference in the
pathogenesis of malignant tumors and that the knowledge base
concerning lncRNA was underdeveloped (24). In the present study, knockdown and
overexpression of SNHG6 was achieved, and qPCR used to detect the
expression level of SNHG6. Due to the limitations of the
experimental conditions in the present study including technology
and funding, the localization of lncRNA SNHG6 was not examined.
In the present study, the role of lncRNA SNHG6 in
the development of colorectal cancer was investigated; it was
identified that the expression of lncRNA SNHG6 was inhibited in
tumor cells and tissues, and that the overexpression of lncRNA
SNHG6 in cells suppressed ETS1 expression, inhibited cell
proliferation. In addition, it was confirmed that the lncRNA SNHG6
inhibited ETS1 expression by directly targeting its 3′-UTR and
activating the intrinsic invasion pathway through downregulating
the expression of PI3K/AKT/mTOR. Therefore, lncRNA SNHG6 may be a
novel therapeutic target for colon cancer. The results also
suggested that this suppression of ETS1 may a potential mechanism
of SNHG6-mediated inhibition of the viability, proliferation and
migration of colorectal cancer cells, and they provide novel
insights into the carcinogenesis of colorectal cancer. To
characterize the association between ETS1 and SNHG6 in greater
detail, correlation between sequences determined by complementarity
between sequence bases studies and luciferase tests are required.
In addition, it may be useful for the development of a treatment
approach for ETS1-activated colorectal cancer. The present study
demonstrated that lncRNA SNHG6 inhibited cell proliferation and
metastasis by targeting ETS1, and established preliminary
associations with the PI3K/AKT/mTOR signaling pathways; future
studies investigating the specific mechanisms are required.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Fund from the National Natural Science Foundation
of China (grant no. 81672427).
Availability of data and materials
The datasets generated and analyzed during the
present study are not publicly available due to further research
being performed, but are available from the corresponding author on
reasonable request.
Authors' contributions
SM and ZJ designed the study. XY, and JL performed
the experiments. RZ analyzed the data.
Ethics approval and consent to
participate
The protocol was approved by the Ethics Committee of
Cancer Hospital of China Medical University, and written informed
consent was obtained from all study participants.
Patient consent for publication
Written informed consent was obtained from all study
participants.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jones P, Cade JE, Evans CEL, Hancock N and
Greenwood DC: Does adherence to the World Cancer Research
Fund/American Institute of Cancer Research cancer prevention
guidelines reduce risk of colorectal cancer in the UK Women's
cohort study? Br J Nutr. 119:340–348. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
van der Werf A, Arthey K, Hiesmayr M, Sulz
I, Schindler K, Laviano A, Langius J and de van der Schueren M: The
determinants of reduced dietary intake in hospitalised colorectal
cancer patients. Support Care Cancer. 26:2029–2047. 2018.
View Article : Google Scholar
|
|
3
|
Han J, Hur H, Min BS, Lee KY and Kim NK:
Predictive factors for lymph node metastasis in submucosal invasive
colorectal carcinoma: A new proposal of depth of invasion for
radical surgery. World J Surg. 42:2635–2641. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Souza BU, Souza NCS, Martucci RB,
Rodrigues VD, Pinho NB, Gonzalez MC and Avesani CM: Factors
associated with sarcopenia in patients with colorectal cancer. Nutr
Cancer. 70:176–183. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Woodall M and DeLetter M: Colorectal
cancer: A collaborative approach to improve education and screening
in a rural population. Clin J Oncol Nurs. 22:69–75. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ruiz-Tovar J, Llavero C, Morales V and
Gamallo C: Effect of the application of a bundle of three measures
(intraperitoneal lavage with antibiotic solution, fascial closure
with triclosan-coated sutures and mupirocin ointment application on
the skin staples) on the surgical site infection after elective
laparoscopic colorectal cancer surgery. Surg Endosc. 32:3495–3501.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu W, Yan S, Liao X, Xiao H, Fu Z, Chen L,
Mou J, Yu H, Zhao L and Liu X: Curative versus palliative
treatments for colorectal cancer with peritoneal carcinomatosis: A
systematic review and meta-analysis. Oncotarget. 8:113202–113212.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vayrynen JP, Tuomisto A, Vayrynen SA,
Klintrup K, Karhu T, Mäkelä J, Herzig KH, Karttunen TJ and Mäkinen
MJ: Preoperative anemia in colorectal cancer: Relationships with
tumor characteristics, systemic inflammation, and survival. Sci
Rep. 8:11262018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yan K, Tian J, Shi W, Xia H and Zhu Y:
LncRNA SNHG6 is associated with poor prognosis of gastric cancer
and promotes cell proliferation and EMT through epigenetically
silencing p27 and sponging miR-101-3p. Cell Physiol Biochem.
42:999–1012. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Birgani MT, Hajjari M, Shahrisa A,
Khoshnevisan A, Shoja Z, Motahari P and Farhangi B: Long non-coding
RNA SNHG6 as a potential biomarker for hepatocellular carcinoma.
Pathol Oncol Res. 24:329–337. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cao C, Zhang T, Zhang D, Xie L, Zou X, Lei
L, Wu D and Liu L: The long non-coding RNA, SNHG6-003, functions as
a competing endogenous RNA to promote the progression of
hepatocellular carcinoma. Oncogene. 36:1112–1122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cao B, Tan S, Tang H, Chen Y and Shu P:
miR5125p suppresses proliferation, migration and invasion, and
induces apoptosis in nonsmall cell lung cancer cells by targeting
ETS1. Mol Med Rep. 19:3604–3614. 2019.PubMed/NCBI
|
|
13
|
Conrad S, Demurger F, Moradkhani K, Pichon
O, Le Caignec C, Pascal C, Thomas C, Bayart S, Perlat A, Dubourg C,
et al: 11q24.2q24.3 microdeletion in two families presenting
features of jacobsen syndrome, without intellectual disability:
Role of FLI1, ETS1, and SENCR long noncoding RNA. Am J Med Genet A.
179-993-1000. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shen C, Yin Y, Chen H, Tang S, Yin X, Zhou
Z, Zhang B and Chen Z: Neuroendocrine tumors of colon and rectum:
Validation of clinical and prognostic values of the World Health
Organization 2010 grading classifications and european
neuroendocrine tumor society staging systems. Oncotarget.
8:22123–22134. 2017.PubMed/NCBI
|
|
15
|
Expert Panel on Gastrointestinal Imaging,
; Fowler KJ, Kaur H, Cash BD, Feig BW, Gage KL, Garcia EM, Hara AK,
Herman JM, Kim DH, et al: ACR appropriateness criteria((R))
pretreatment staging of colorectal cancer. J Am Coll Radiol.
14:S234–S244. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ogawa T, Hirohashi Y, Murai A, Nishidate
T, Okita K, Wang L, Ikehara Y, Satoyoshi T, Usui A, Kubo T, et al:
ST6GALNAC1 plays important roles in enhancing cancer stem
phenotypes of colorectal cancer via the Akt pathway. Oncotarget.
8:112550–112564. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luan A, Hu MS, Leavitt T, Brett EA, Wang
KC, Longaker MT and Wan DC: Noncoding RNAs in Wound Healing: A new
and vast frontier. Adv Wound Care (New Rochelle). 7:19–27. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sallam T, Sandhu J and Tontonoz P: Long
noncoding RNA discovery in cardiovascular disease: Decoding form to
function. Circ Res. 122:155–166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen S, Zhu J, Wang F, Guan Z, Ge Y, Yang
X and Cai J: LncRNAs and their role in cancer stem cells.
Oncotarget. 8:110685–110692. 2017.PubMed/NCBI
|
|
21
|
Malhotra A, Jain M, Prakash H, Vasquez KM
and Jain A: The regulatory roles of long non-coding RNAs in the
development of chemoresistance in breast cancer. Oncotarget.
8:110671–110684. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lv P, Qiu X, Gu Y, Yang X, Xu X and Yang
Y: Long non-coding RNA SNHG6 enhances cell proliferation, migration
and invasion by regulating miR-26a-5p/MAPK6 in breast cancer.
Biomed Pharmacother. 110:294–301. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Li D, Zhao M, Huang S, Zhang Q, Lin
H, Wang W, Li K, Li Z, Huang W, et al: Long noncoding RNA SNHG6
regulates p21 expression via activation of the JNK pathway and
regulation of EZH2 in gastric cancer cells. Life Sci. 208:295–304.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Li R, Ding X, Zhang K and Qin W:
Upregulation of long non-coding RNA SNHG6 promote esophageal
squamous cell carcinoma cell malignancy and its diagnostic value.
Am J Transl Res. 11:1084–1091. 2019.PubMed/NCBI
|