Introduction
Rheumatoid arthritis (RA) is a common chronic
inflammatory joint disease characterized by synovitis, synovial
hyperplasia, pannus formation, and the destruction of bone and
cartilage (1). In China, the
incidence of RA is ~0.3% (2),
which is 0.7% lower compared with the world average level; however,
it has a high disability rate (3).
Although the causes of RA remain to be elucidated, a previous study
suggested that fibroblast-like synoviocytes (FLS), also known as
synovial fibroblasts, exert a crucial effect on initiation and
perpetuation of the immune response in patients with RA (4). FLS and macrophages are two cell types
composing the lining of normal synovial tissue. In the event of RA,
abnormal activation of macrophages can promote FLS to produce
pro-inflammatory cytokines and matrix metalloproteinase (MMPs).
These pro-inflammatory cytokines are capable of recruiting more
inflammatory cells and support continued arthropathy (5). In addition, MMPs have the ability to
degrade the extracellular matrix (ECM) resulting in damage to
cartilage and bone; for example, MMP-1 can effectively break down
collagen type II, which is the basis of articular cartilage
(6). Furthermore, tumor necrosis
factor (TNF)-α and interleukin (IL)-1, secreted in response to
lipopolysaccharide, can directly or indirectly induce the
proliferation of osteoclasts by inducing nuclear factor-κB ligand
to bind to the corresponding receptor (7,8).
Consequently, the activation of osteoclasts may lead to bone
erosion and inflammatory arthritis (9). Although a number of effective and
hypotoxic medicines have been developed, the long treatment cycle
of RA notably increases the incidence of adverse reactions.
Therefore, it is essential to understand the molecular mechanism
underlying RA and provide evidence for personalized medicine.
MicroRNAs (miRNAs/miRs) are 18–25 nt long,
endogenous non-coding RNAs, which modulate the expression of one or
more genes by regulating the degradation or translational
repression of mRNA. The molecular mechanisms underlying the
synthesis and function of miRNAs have been widely studied (10). As molecular biology has advanced,
more studies have provided evidence for a vital role of miRNAs in
the pathogenesis of RA (11,12).
In 2011, 13 differentially expressed miRNAs were identified by
comparing the differential expression of miRNAs in rheumatoid
synovial fluid monocytes with normal peripheral blood monocytes
using a miRNA microarray; miR-34a-5p was one of these
differentially expressed miRNAs (13). Previous studies have reported that
miR-34a-5p is involved in the development of RA (14,15);
however, the underlying mechanism of miR-34a-3p on FLS remains to
be elucidated. The present study constructed miR-34a-3p mimics and
inhibitor vectors to further investigate its role in RA.
Materials and methods
Patient samples and cell culture
Synovial tissue specimens of RA (7 males and 13
females; average age, 52 years old) and osteoarthritis (OA, 8 males
and 12 females; average age, 48 years old) were obtained from
patients during total knee replacement surgery or arthroscopy at
the Jining No. 1 People's Hospital from January 2016 to February
2017. In addition, arthroscopic biopsies from healthy individuals
who underwent arthroscopic surgery for knee meniscus injuries or
cruciate ligament rupture and had no history of autoimmune diseases
were recruited as a control group (11 males and 9 females; average
age, 42 years old). Written informed consent was obtained from all
patients in this study, and the study was approved by the Medical
Ethical Review Committee of Jining No. 1 People's Hospital. All RA
and OA patients fulfilled the American College of Rheumatology
criteria for the classification of the disease (16). FLS were obtained from patients with
RA and OA (RAFLS and OAFLS) by incubating synovial tissue samples
with Dulbecco's modified Eagle's medium (DMEM; cat. no. D5030;
Sigma-Aldrich; Merck KGaA) containing type II collagenase (1 mg/ml;
Sigma-Aldrich; Merck KGaA) in a humidified incubator containing 5%
CO2 at 37°C for 6 h. The same volume of 0.25% trypsin
was then added to the culture medium. Following filtration through
a 70-µm cell strainer, FLS were cultured in complete medium
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.), 100 IU/ml penicillin and 10 µg/ml streptomycin
in a humidified incubator containing 5% CO2 at 37°C.
Cell transfection
The sequence of miR-34a-3p was obtained from miRbase
(MIMAT0004557, http://www.mirbase.org/textsearch.shtml?q=miR-34a-3p).
FLS were seeded into 12-well plates (1×105 cells/well)
and transfection was conducted using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). Briefly, when cell
confluence reached 50%, miR-34a-3p mimics
(5′-CAAUCAGCAAGUAUACUGCCU-3′), miR-34a-3p inhibitor
(5′-ACAACCAGCUAAGACACUGCCA-3′) and non-sense single-strand RNA
(5′-CAGUACUUUUGUGUAGUACAA-3′; Invitrogen; Thermo Fisher Scientific,
Inc.) were diluted in DMEM (50 ng/ml), and were added to the cells
alongside 5 µl Lipofectamine® 2000. The mixture was
incubated at 37°C in a humidified atmosphere containing 5%
CO2 for 6 h, after which the medium was replaced with
complete medium. The transfection efficiencies of miR-34a-3p mimics
and inhibitor vectors were assessed after 24 h.
Cell proliferation assay
The non-transfected or transfected FLS were cultured
in 96-well plates (3×105 cells/well) under 5%
CO2 and 37°C. Cells were harvested at 0, 24 and 48 h,
respectively. Subsequently, 10% Cell Counting kit (CCK)-8 reagent
(Dojindo Molecular Technologies, Inc.) was added to the cell
culture medium and the plates were maintained at room temperature
for 4 h. The absorbance was then measured at 450 nm using a
microplate reader (Thermo Fisher Scientific, Inc.).
Flow cytometry
Cell cycle analysis was carried out as previously
described using propidium iodide (PI) staining (17). FLS transfected with miR-34a-3p
mimics, inhibitor or negative control (NC) vectors were seeded in
6-well plates (1×106 cells), and incubated for 24 h
until cell confluence reached 100%. Cells in each group were
harvested and washed with cold PBS, after which they were fixed
with 70% cold ethanol at 4°C overnight. Cells were then washed and
resuspended in PBS, and were incubated in buffer containing 50
µg/ml PI and 10 µg/ml RNase A (Beyotime Institute of Biotechnology)
for 30 min in the dark at 37°C. The fraction of cells in the
G1, S, and G2 phases of the cell cycle was
analyzed using a Coulter Epics XL flow cytometer (BD Biosciences).
The data were acquired and analyzed using FACSDiva software
(version 4.1; BD Biosciences). Integration of the area under the
curve for each of the cell cycle phases was performed with ModFit
LT software (version 3.3; BD Biosciences).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
For detection of miRNA expression levels, miRNAs
were isolated from FLS using a miRNeasy Mini kit (Qiagen GmbH), and
purified by TURBO DNA-free kit (Invitrogen; Thermo Fisher
Scientific, Inc.). Subsequently, RNA was reverse transcribed to
cDNA using a miScript II RT kit (Qiagen GmbH) as previously
described (18); the samples were
incubated at 37°C for 60 min, 95°C for 5 min and were then
maintained at 4°C. The relative expression levels of miRNAs were
analyzed using the miScript SYBR Green PCR kit (Qiagen GmbH). Each
qPCR was performed in a final volume of 20 µl containing 1X
QuantiTect SYBR Green PCR Master mix (Qiagen GmbH), 2 µl cDNA and
0.5 mM each primer. The miScript Universal primer was used as the
reverse primer for miRNA detection. In addition, for detection of
mRNA expression levels, total RNA was extracted with
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and reverse transcribed to cDNA using the Prime Script RT
reagent kit (Takara Bio, Inc.). The RT reaction conditions were as
follows: 65°C for 5 min, 30°C for 6 min and 50°C for 60 min. qPCR
was performed using the SYBR green detection kit (Takara Bio,
Inc.). The miRNA and mRNA levels were analyzed using an ABI 7500
real-time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). For detection of mRNA, the 20 µl reaction system consisted
of 10 µl 2X SYBR green master mix, 7 µl H2O, 2 µl
primers (10 µM) and 1 µl template DNA. The reaction mixtures were
denatured at 95°C for 15 min, followed by 40 cycles of annealing at
94°C for 15 sec, 55°C for 30 sec and 70°C for 30 sec. U6 was
employed as the internal reference for miRNA and β-actin was the
internal reference for mRNA. All samples were analyzed in
triplicate, and the relative expression levels were quantified
using the 2−ΔΔCq method (19). All primers are listed in Table I.
 | Table I.Primers for reverse
transcription-quantitative PCR. |
Table I.
Primers for reverse
transcription-quantitative PCR.
| Gene name | Primer
sequences |
|---|
| CDK2 | Forward:
5′-CAGTACTGCCATCCGAGAGA-3′ |
|
| Reverse:
5′-GAATGCCAGTGAGAGCAGAG-3′ |
| CDC25A | Forward:
5′-CCTCCGAGTCAACAGATTCA-3′ |
|
| Reverse:
5′-GGGTCGATGAGCTGAAAGAT-3′ |
| Cyclin D1 | Forward:
5′-GTCTTCCCGCTGGCCATGAACTAC-3′ |
|
| Reverse:
5′-GGAAGCGTGTGAGGCGGTAGTAGG-3 |
| MMP-1 | Forward:
5′-AAATGCAGGAATTCTTTGGG-3′ |
|
| Reverse:
5′-ATGGTCCACATCTGCTCTTG-3 |
| MMP-9 | Forward:
5′-TTGACAGCGACAAGAAGTGG-3′ |
|
| Reverse:
5′-GGCACAGTAGTGGCCGTAG-3 |
| TNF-α | Forward:
5′-CATCTTCTCAAAATTCGAGTGACAA-3′ |
|
| Reverse:
5′-TGGGAGTAGACAAGGTACAACCC-3′ |
| IL-6 | Forward:
5′-AGTTGCCTTCTTGGGACTGA-3′ |
|
| Reverse:
5′-TCCACGATTTCCCAGAGAAC-3 |
| MDM4 | Forward:
5′-AGGTGCGCAAGGTGAAATGT-3′ |
|
| Reverse:
5′-CCATATGCTGCTCCTGCTGAT-3′ |
| TRAF3 | Forward:
5′-ACTGCAAGAGTCAGGTTCCG-3′ |
|
| Reverse:
5′-CAAGTGTGCACTCAACTCGC-3 |
| TRAF1 | Forward:
5′-CATGCAGGAGCATGAGGCTACC-3′ |
|
| Reverse:
5′-CCACCACCCTCTGCTCCAAGC-3 |
| PTPN11 | Forward:
5′-TCAGCACAGAAATAGATGTG-3′ |
|
| Reverse:
5′-TGCTTATCAAAAGGTAGTCA-3′ |
| XIAP | Forward:
5′-GTGGTGGAAAACTGAAAAATTGG-3′ |
|
| Reverse:
5′-GAAAGTGTCGCCTGTGTTCTGA-3 |
| β-actin | Forward:
5′-CCTGGCACCCAGCACAAT-3′ |
|
| Reverse:
5′-GGGCCGGACTCGTCATAC-3 |
| U6 | Forward:
5′-CTCGCTTCGGCAGCACATA-3′ |
|
| Reverse:
5′-AACGATTCACGAATTTGCGTC-3 |
Western blot analysis
Total protein of tissues and cells were lysed in 150
µl RIPA lysis buffer containing 1% phenylmethanesulfonyl fluoride
(PMSF; Beyotime Institute of Biotechnology) according to the
manufacturer's instructions. Protein concentration was quantified
using a bicinchoninic acid protein assay (Takara Bio, Inc.), and 50
µg proteins were separated by 10% SDS-PAGE prior to being
transferred onto a polyvinylidene difluoride (PVDF) membrane (EMD
Millipore). The PVDF membrane was blocked with 5% nonfat milk in in
PBS containing 0.1% Tween-20 for 1 h at room temperature and was
then incubated with primary antibodies at 4°C overnight. The
primary antibodies were as follow: Anti-cyclin-dependent kinase 2
(CDK2; cat. no. 2546), anti-cell division cycle 25A (CDC25A; cat.
no. 3652), anti-cyclin D1 (cat. no. 2978), anti-MMP-9 (cat. no.
2270), anti-tumor necrosis factor (TNF)-α (cat. no. 3707) and
anti-interleukin (IL)-6 (cat. no. 12153; all from Cell Signaling
Technology, Inc.), β-actin (cat. no. ab8226; Abcam) and anti-MMP-1
(cat. no. ab137332; Abcam). All primary antibodies were used at a
dilution of 1:1,000. Subsequently, the membrane was incubated with
goat anti-rabbit immunoglobulin G horseradish peroxidase-linked
secondary antibodies (1:2,000; cat. no. 7074, Cell Signaling
Technology, Inc.) for 1 h at room temperature. Blots were
visualized using an enhanced chemiluminescence reagent (cat. no.
32106; Thermo Fisher Scientific, Inc.). Semi-quantification of
image density was performed using ImageJ version 1.38 software
(National Institutes of Health).
miR-34a-3p target prediction and
luciferase reporter assay
The potential targets of miR-34a-3p were predicted
using TargetScan (www.targetscan.org/vert_72) and MiRWalk 2.0
(mirwalk.umm.uni-heidelberg.de).
Subsequently, the predicted target genes were investigated using a
luciferase reporter assay. Briefly, miR-34a-3p was inserted into a
GV272 vector (Shanghai GeneChem Co., Ltd.) and fragments containing
a miR-34a-3p-binding site in the 3′-untranslated region (UTR) of
predicted genes were cloned into a GV268 plasmid vector (Shanghai
GeneChem Co., Ltd.). The QuikChange II Site-Directed Mutagenesis
kit (Agilent Technologies, Inc.) was used to mutate the binding
sites; the mutated fragments were also inserted into a GV268
plasmid vector (3′-UTR mut).
Luciferase reporter assay was performed on the 293
cell line (cat. no. CRL-3216; ATCC) using a
Dual-Luciferase® Reporter Assay system (cat. no. E1910;
Promega Corporation). Briefly, 293 cells at a density of
4×104 were transfected with 50 ng GV272-miR-204-3p, and
40 nM of either control GV268-3′-UTR or GV268-3′-UTR mut by
Lipofectamine® 2000. After 48 h, 293 cells were lysed
with 200 µl 1% sodium dodecyl sulfate (SDS) and separated by
centrifugation at 15,000 × g for 3 min at 4°C. The prepared dual
luciferase reporter mixture was added to the supernatant of lysed
cells and the relative luciferase activity normalized against
Renilla luciferase was immediately measured using a Synergy™
2 luminometer (BioTek Instruments, Inc.).
Statistical analysis
Statistical analysis was detected using GraphPad
Prism version 6.0 software (GraphPad Software, Inc.). All data are
presented as the means ± standard deviation. Differences were
analyzed using one-way analysis of variance followed by Tukey's
multiple comparison test. P<0.05 was considered to indicate a
statistically significant difference.
Results
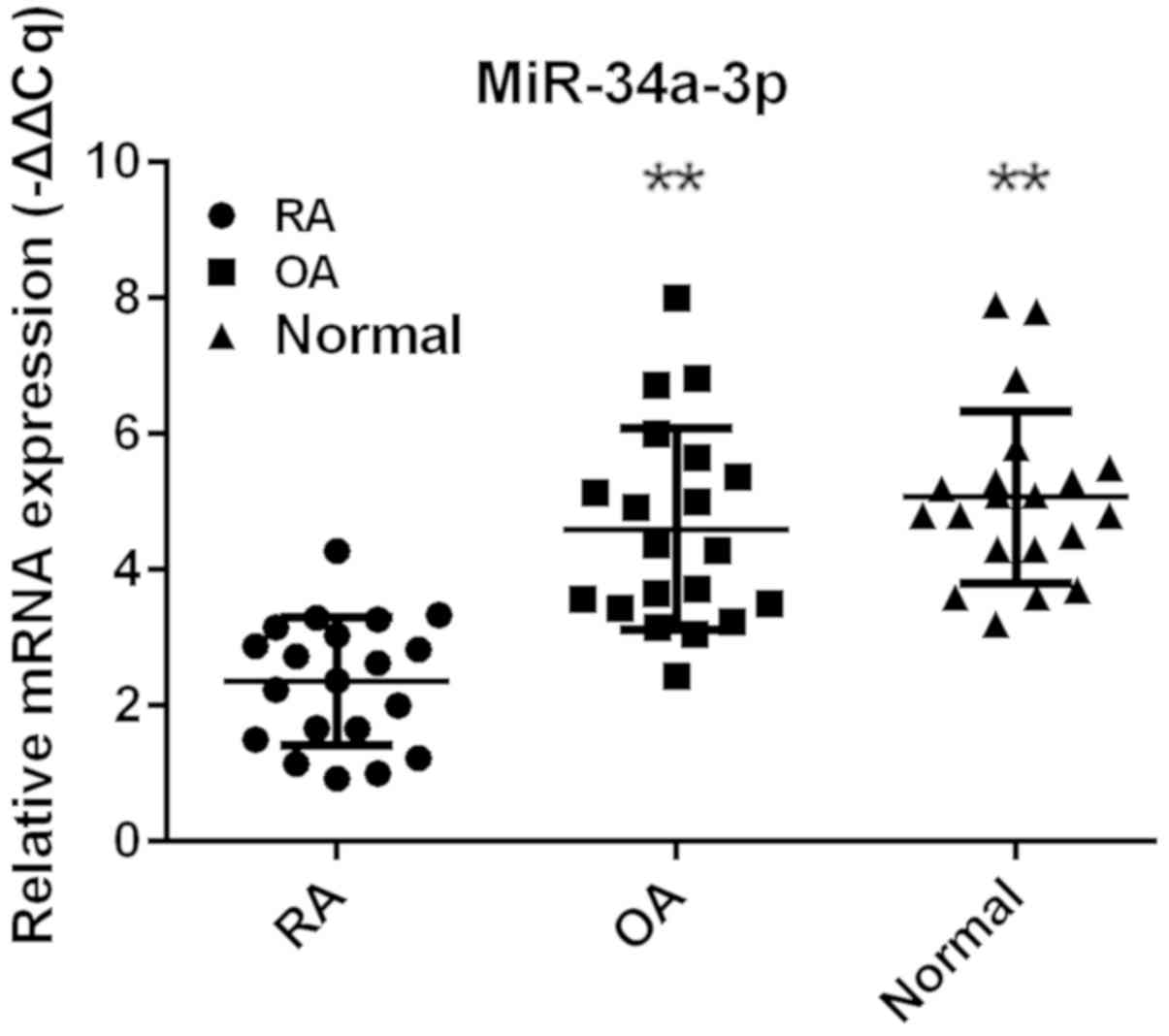
miR-34a-3p expression is decreased in
FLS from patients with RA
To identify the potential effect of miR-34a-3p on
RA, the expression levels of miR-34a-3p were compared between
different groups. The FLS purified from patients with RA and OA
were incubated under the same conditions and the results of RT-qPCR
demonstrated that the expression levels of miR-34a-3p were
generally lower in RAFLS compared with in OAFLS and normal FLS
(Fig. 1). This result indicated
the potential function of miR-34a-3p in the pathogenesis of RA.
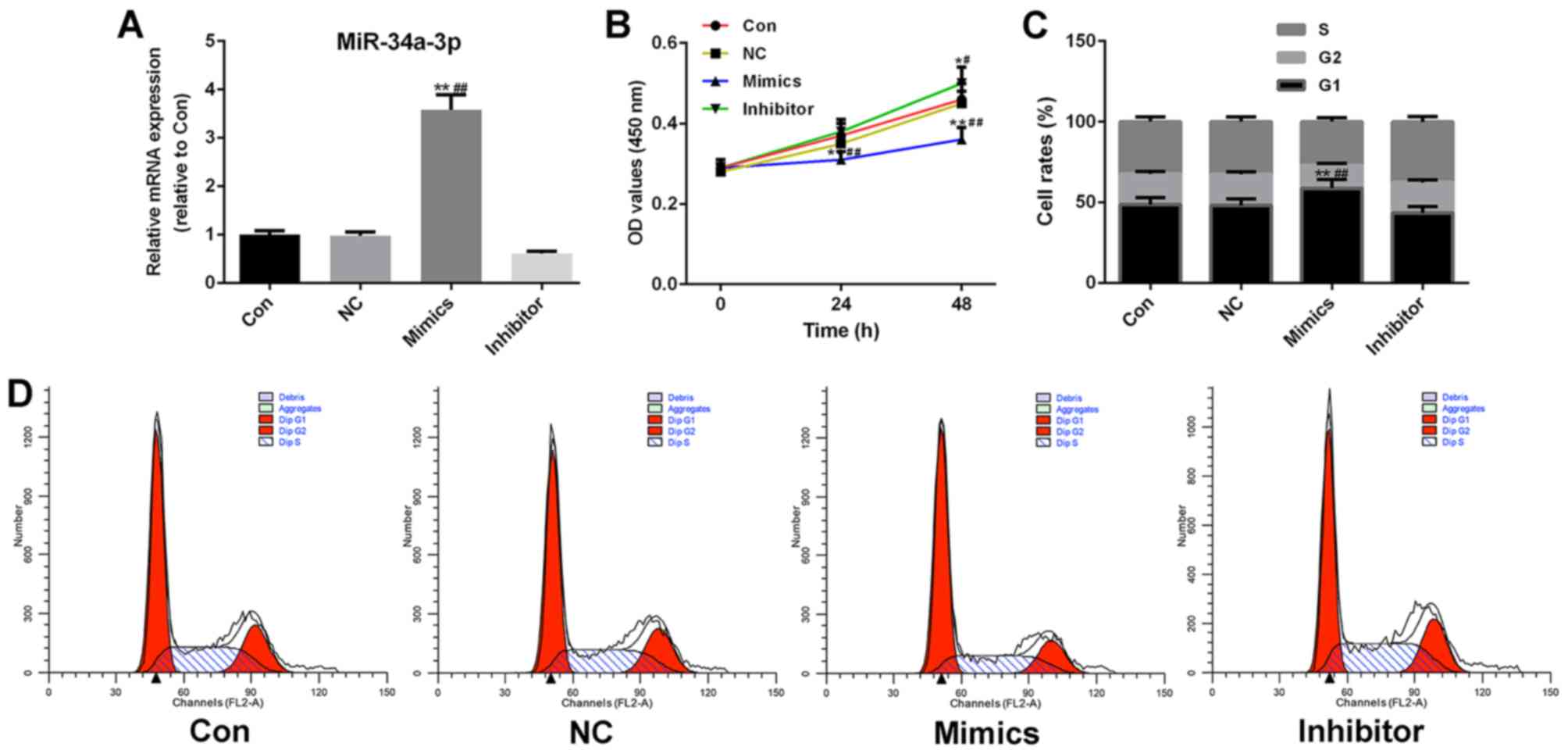
Increased expression levels of
miR-34a-3p inhibit the proliferation and cell cycle progression of
RAFLS
To investigate the potential effects of miR-34a-3p
on RAFLS, miR-34a-3p mimics and inhibitor vectors were transfected
into RAFLS (Fig. 2A). As
demonstrated in Fig. 2B,
upregulated miR-34a-3p expression had an inhibitory effect on FLS
proliferation (P<0.01). Therefore, the effects of miR-34a-3p on
the cell cycle of FLS were further investigated. The results of
flow cytometry were consistent with the CCK-8 assay; the number of
miR-34a-3p mimics-transfected FLS in G1 phase was
significantly higher compared with in the control group
(P<0.01), which suggested that high miR-34a-3p expression may
effectively inhibit the cell cycle of FLS (Fig. 2C and D). By contrast, knockdown of
miR-34a-3p promoted the cell cycle; however, this was not
significant.
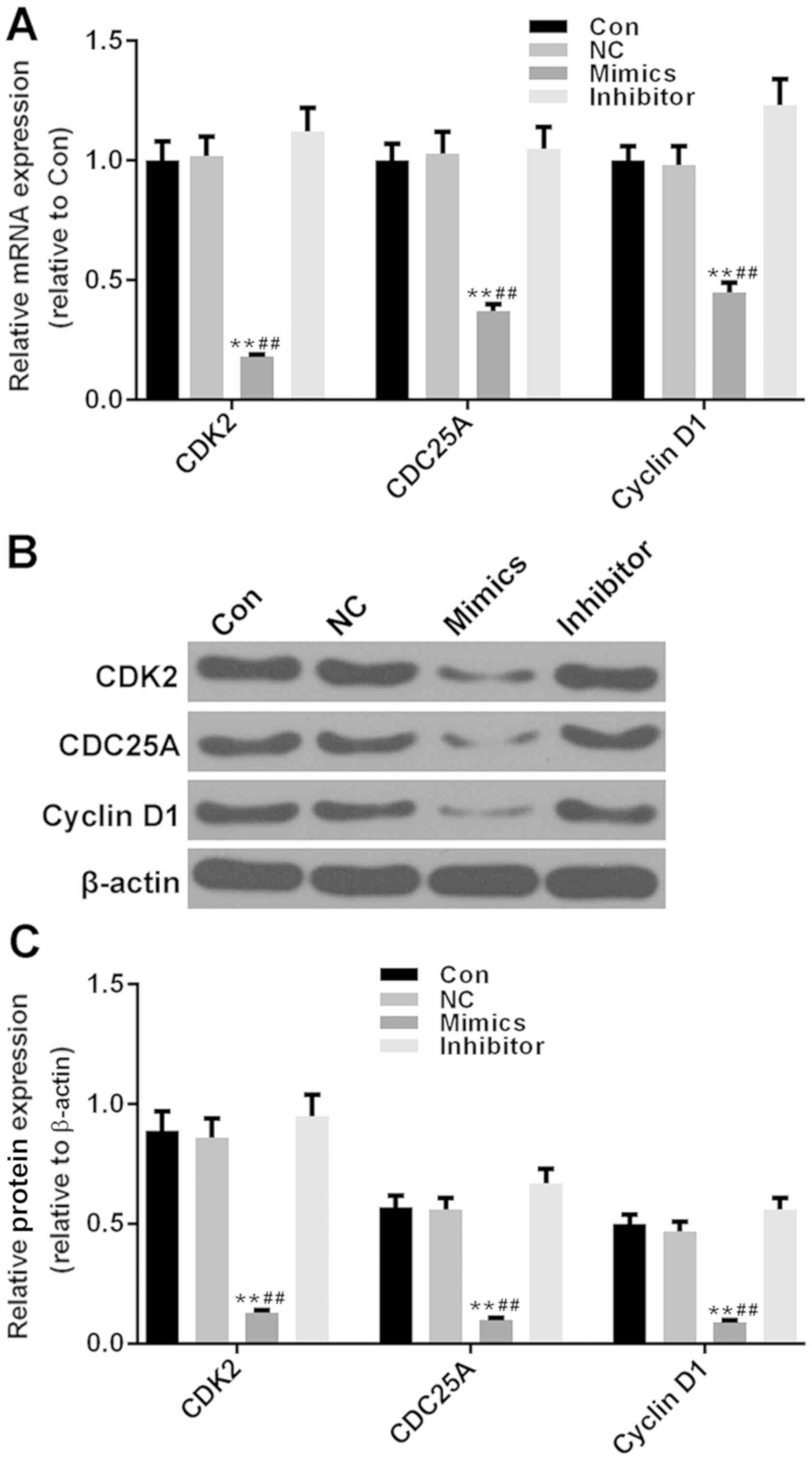
High miR-34-3p expression suppresses
the expression levels of cell cycle control genes
The cell cycle control genes serve a crucial role in
progression of the cell cycle. To further verify the inhibitory
effects of miR-34a-3p on the cell cycle of RAFLS, the expression
levels of several cell cycle control genes (CDK2, CDC25A and cyclin
D1) were measured. As demonstrated in Fig. 3A-C, the transcriptional and
post-transcriptional levels of these genes were significantly
decreased, compared with in the control and NC groups (P<0.01).
The inhibitory effects of miR-34a-3p were most marked on the mRNA
and protein expression levels of CDK2. Conversely, the miR-34a-3p
inhibitor vector appeared to exert little effects on the cell cycle
control genes. These results further verified that miR-34a-3p had
an inhibitory effect on the cell cycle progression of RAFLS.
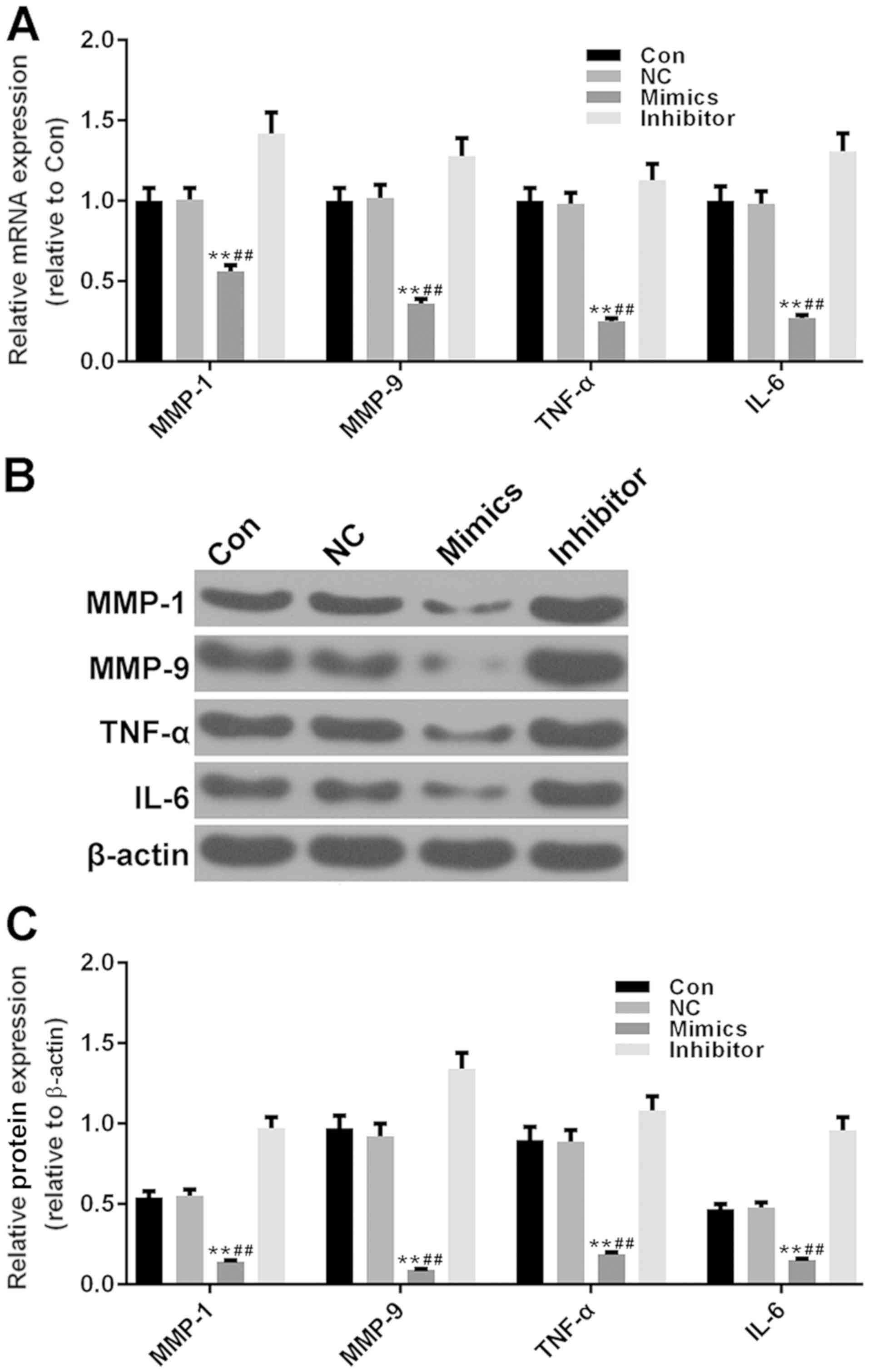
Increased miR-34a-3p expression
inhibits MMPs and pro-inflammatory cytokines in RAFLS
It has been reported that RAFLS can promote the
expression levels of MMPs and pro-inflammatory cytokines, thus
resulting in the destruction of synovial tissue (20). The present study examined the mRNA
and protein expression levels of two MMPs (MMP-1 and MMP-9) and two
pro-inflammatory cytokines (Fig.
4A-C). It was observed that the expression levels of MMP-1 and
MMP-9 were significantly decreased in the miR-34a-3p mimics group
compared with in the control group (P<0.01), which suggested
that miR-34a-3p may protect the articular cartilage through
inhibiting the expression levels of MMPs. In addition, miR-34a-3p
significantly suppressed the expression levels of TNF-α and IL-6
(P<0.01); this may result in resistance to inflammation-induced
bone destruction. Conversely, inhibition of miR-34a-3p promoted the
production of MMPs and pro-inflammatory cytokines, but not
significantly.
Affinity of miR-34a-3p to murine
double minute 4 (MDM4) is greater compared with TNF
receptor-associated factor (TRAF) 3
Two prediction algorithms (TargetScan and mirWalk)
were combined to identify the potential target genes of miR-34a-3p;
five predicted target genes were identified, MDM4, TRAF3, TRAF1,
protein tyrosine phosphatase non-receptor type 11 and X-linked
inhibitor of apoptosis (XIAP). As determined by RT-qPCR analysis,
the expression levels of four genes (MDM4, TRAF3, TRAF1 and XIAP)
were significantly decreased in the miR-34a-3p mimics group
(P<0.01), and the inhibitory effects of miR-34a-3p mimics were
the most marked on MDM4 and TRAF3 (Fig
5A). As shown in Fig. 5B, the
547–554 nt position of the MDM4 3′-UTR harbored one potential
binding site for miR-204-3p (seven bases), whereas the 5700–5706 nt
position of the TRAF3 3′-UTR included a potential binding site for
miR-204-3p (six bases). MDM4 and TRAF3 were therefore considered
potential target genes of miR-34a-3p.
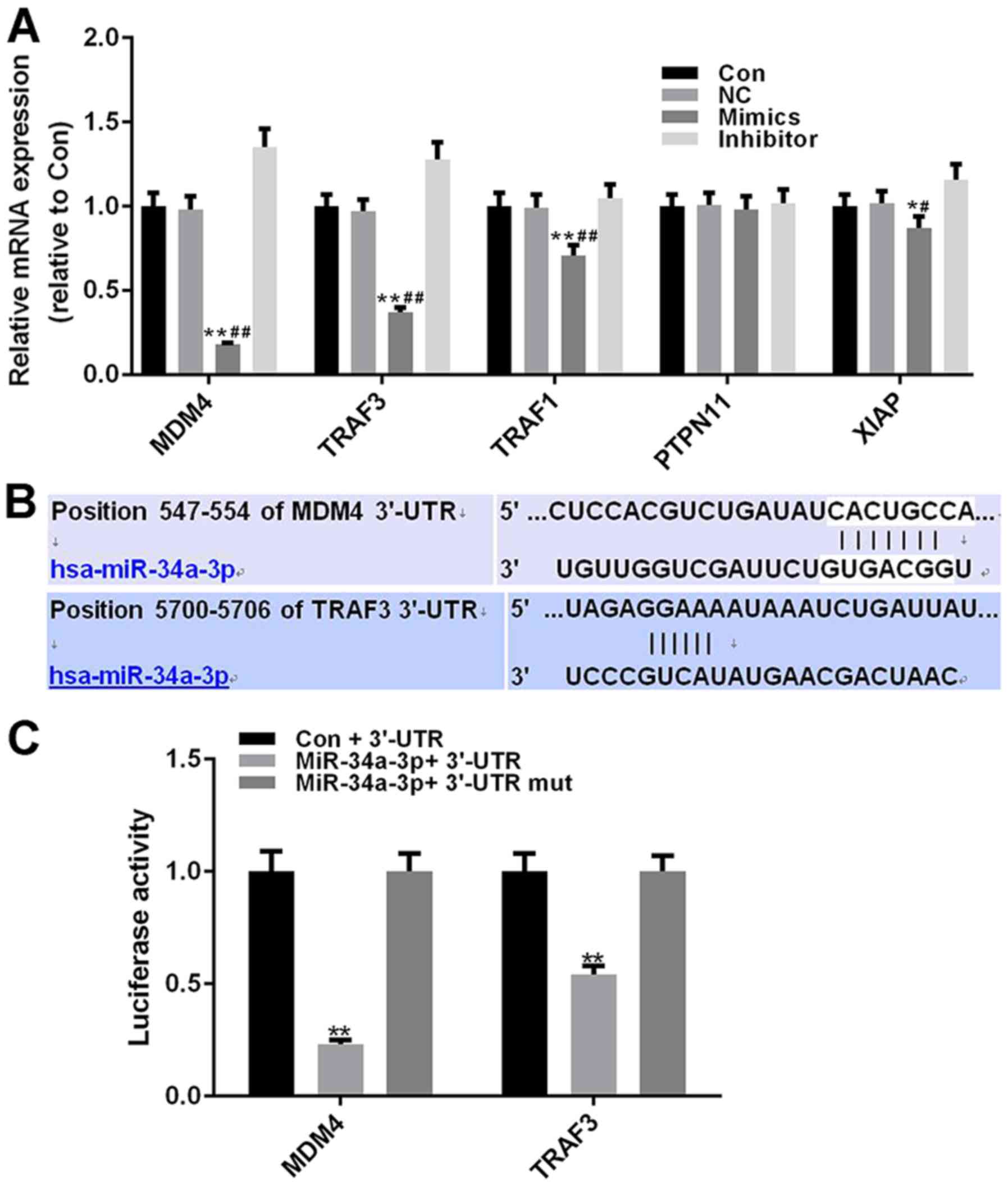 | Figure 5.Affinity of miR-34a-3p to MDM4 is
stronger compared with TRAF3. (A) mRNA expression levels of five
predicted target genes were measured in rheumatoid
arthritis-fibroblast-like synoviocytes transfected with miR-34a-3p
mimics and inhibitor vectors. β-actin was detected as an internal
control. (B) Potential miR-34a-3p binding sites in MDM4 and TRAF3.
(C) Luciferase reporter assay further verified whether miR-34a-3p
effectively targets MDM4 and TRAF3. Each value represents the mean
± standard deviation (n=3). *P<0.05, **P<0.01 vs. Con +
3′-UTR group; #P<0.05, ##P<0.01 vs. NC
group. 3′-UTR, 3′-untranslated region; Con, Control group; MDM4,
murine double minute 4; miR, microRNA; TRAF3 NC, negative control;
PTPN11, protein tyrosine phosphatase non-receptor type 11; TNF,
receptor-associated factor 3; XIAP, X-linked inhibitor of
apoptosis. |
Dual luciferase reporter plasmids were constructed
to further identify the association between miR-34a-3p and MDM4 or
TRAF3. The luciferase activity in the miR-34a-3p + MDM4 3′-UTR
group was significantly reduced compared with in the Con + MDM4
3′-UTR group (P<0.01). The luciferase activity in the miR-34a-3p
+ MDM4 3′-UTR mut group exhibited no difference compared with in
the control group. miR-34a-3p also had an effective inhibitory
effect on the luciferase activity of TRAF3 3′-UTR (P<0.01),
whereas the luciferase activity of the miR-34a-3p + TRAF 3′-UTR mut
group exhibited no difference compared with in the Con + TRAF3
3′-UTR group. Notably, the inhibitory effects of miR-34a-3p on MDM4
luciferase activity were more marked compared with on TRAF3
luciferase activity (Fig. 5C).
These findings suggested that miR-34a-3p possessed a stronger
affinity to MDM4 compared with to TRAF3.
Discussion
The present study aimed to determine the function of
the miR-34 family, which has been reported to serve an important
role in the progression of apoptotic cell death (21,22).
It has been observed that miR-34a-3p has functions opposite to
other miR-34 family members, including miR-34a-5p, miR-34b-3p/5p
and miR-34c-3p/5p (15). The onset
of OA and RA is associated with oxidative stress and inflammation;
therefore, the expression levels of miR-34a-5p were detected in
patients with RA and OA and were compared, in order to make the
results more conclusive. By comparing the expression levels of
miR-34a-3p in RAFLS with those in OAFLS, it was identified that the
miR-34a-3p expression was considerably reduced in RAFLS; therefore,
it was hypothesized that miR-34a-3p may have an ameliorating effect
on RA. To further determine the potential role of miR-34a-3p,
overexpression and knockdown of miR-34a-3p were artificially
induced, and the proliferation and activation of RAFLS were
detected.
Pannus formation is an important characteristic of
RA, which is characterized by angiogenic factor-mediated
angiogenesis and abnormal hyperplasia of the synovium (23). The pannus, which is composed mostly
of FLS, is formed alongside a marked infiltration of lymphocytes
and macrophages (24). The
increased number of FLS is a key participant in the progression of
RA; FLS can attach to articular cartilage and secrete several types
of pro-inflammatory cytokines and MMPs to invade the ECM and
cartilage, thus inducing severe joint damage and dysfunction
(25,26). In the present study, enhanced
miR-34a-3p expression significantly inhibited FLS proliferation,
which may greatly decrease the production of pro-inflammatory
cytokines and MMPs, partly mitigating inflammation and bone
destruction in patients with RA. In addition, the CDK family serves
a vital role in regulating cell cycle progression; CDK2 can induce
the G1/S transition and promote DNA replication
(27). CDC25A and cyclin D1
contribute to activating CDK2, and cyclin D1 can also induce
phosphorylation of the retinoblastoma tumor suppressor protein
family, activating E2F transcription factors and ultimately driving
G1/S progression (28,29).
The results of the present study indicated that increased
miR-34a-3p expression may suppress the expression levels of CDK2,
CDC25A and cyclin D1, in order to elicit cell cycle arrest of FLS;
these results may explain why the number of cells in G1
phase was much higher in the miR-34a-3p mimics group. MMPs produced
by FLS enhance the migration and invasion of FLS, and previous
studies have demonstrated that knockdown of SUMO-conjugating enzyme
UBC9 or sphingosine kinase 1 notably mitigates the disease
progression in RA by blocking the expression of MMPs (30,31).
Combined with these studies and the findings of the present study,
miR-34a-3p may effectively inhibit cell proliferation, and the
migratory and invasive capacities of FLS, and may thus protect
articular cartilage from MMPs.
RA is a chronic inflammatory disease, and the
functional mechanisms of several inflammatory cytokines have been
studied in depth. Previous studies have demonstrated that the
expression levels of TNF-α and IL-6 are closely associated with
synovitis and joint destruction (32–34).
TNF-α stimulation can lead to fibrosis and the inflammatory
response of FLS, resulting in production of IL-6, vascular cell
adhesion molecule-1 and vascular endothelial growth factor, which
participate in the pathophysiology of RA (35,36).
In the current study, the expression levels of TNF-α and IL-6 are
markedly decreased with increased expression levels of miR-34a-3p,
which may suggest that miR-34a-3p expression had the ability to
radically decrease inflammation in RA. A previous study
demonstrated that coenzyme Q10, an endogenous antioxidant, can
effectively downregulate inflammatory cytokines and oxidative
stress by inhibiting TNF-α and IL-6 expression levels (37), and tocilizumab is considered a
promising agent in RA treatment as it can specifically target IL-6
(38). These data indicate a
potential treatment effect of miR-34a-3p on RA.
In the present study, it was suggested that MDM4 may
be a potential target gene of miR-34a-3p, according to prediction
software (TargetScan and MiRWalk). In addition, the predictions
were verified using a luciferase assay, which was helpful in
further investigating the molecular mechanism underlying the
effects of miR-34a-3p on the pathophysiology of RA. A previous
study demonstrated a close association between the expression of
MDM4 in FLS and the hyperplasia phenotype of RA synovial tissues
(39). Therefore, it was
hypothesized that the treatment effect of miR-34a-3p on RA may be
dependent on miR-34a-3p effectively silencing MDM4 expression;
however, further study is required to confirm this. Niederer et
al (15) reported that the
mechanism by which miR-34a affects RA may be that miR-34a inhibits
apoptosis of RA synovial fibroblasts by upregulating the expression
of its direct target XIAP. Another study also identified that
miR-34a-deficient mice are resistant to collagen-induced arthritis,
and that miR-34a is an epigenetic regulator of dendritic cell
function that may contribute to RA, suggesting that RA regression
can be aided using a miR-34a inhibitor (40). A previous study also identified
that inhibition of miR-34a can improve arthritis in mice by
decreasing the percentage of T cells, cytokine expression and bone
loss (15). The two possible
mature miRNAs of miR-125a, miR-125a-3p and miR-125a-5p, have
inverse effects on invasion and migration of lung cancer cells
(41). The 3p arm presents more
individually regulated targets based on poorly sequence
conservation (42). Therefore, the
present study hypothesized that miR-34a-3p may exert a protective
effect that may different from that of miR-34a-5p in RA. The
development of RA is associated with inflammation and immunity, and
miR-34a can participate in RA through different pathways.
Therefore, further research is required on the mechanism of action
of miR-34a in RA. Notably, miR-34a-3p serves a crucial role in the
pathogenesis of RA. The present study demonstrated that it may
inhibit FLS proliferation and suppress the production of
pro-inflammatory cytokines and MMPs, greatly ameliorating articular
cartilage destruction. In conclusion, miR-34a-3p may be considered
a novel therapeutic target for RA; however, its underlying
mechanism requires further research.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
CFH and DW made substantial contributions to the
conception and design of the present study. LHZ performed data
acquisition, analysis and interpretation. CFH and DW drafted the
article and critically revised it for important intellectual
content. All authors approved the final version to be published.
All authors agreed to be accountable for all aspects of the work in
ensuring that questions associated with the accuracy or integrity
of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Medical
Ethical Review Committee of Jining No. 1 People's Hospital. All
procedures performed using human samples were in accordance with
the ethical standards of the institutional and/or national research
committee and with the 1964 Helsinki declaration and its later
amendments or comparable ethical standards. Written informed
consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Smolen JS, Aletaha D and McInnes IB:
Rheumatoid arthritis. Lancet. 388:2023–2038. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li ZG, Liu Y, Xu HJ, Chen ZW, Bao CD, Gu
JR, Zhao DB, An Y, Hwang LJ, Wang L, et al: Efficacy and safety of
tofacitinib in chinese patients with rheumatoid arthritis. Chin Med
J (Engl). 131:2683–2692. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gibofsky A: Overview of epidemiology,
pathophysiology, and diagnosis of rheumatoid arthritis. Am J Manag
Care. 18:S295–S302. 2012.PubMed/NCBI
|
|
4
|
Hawtree S, Muthana M and Wilson AG: The
role of histone deacetylases in rheumatoid arthritis
fibroblast-like synoviocytes. Biochem Soc Trans. 41:783–788. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Filer A, Ward LSC, Kemble S, Davies CS,
Munir H, Rogers R, Raza K, Buckley CD, Nash GB and McGettrick HM:
Identification of a transitional fibroblast function in very early
rheumatoid arthritis. Ann Rheum Dis. 76:2105–2112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hirohata S and Sakakibara J:
Angioneogenesis in rheumatoid arthritis. Lancet. 354:423–424. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Danks L, Komatsu N, Guerrini MM, Sawa S,
Armaka M, Kollias G, Nakashima T and Takayanagi H: RANKL expressed
on synovial fibroblasts is primarily responsible for bone erosions
during joint inflammation. Ann Rheum Dis. 75:1187–1195. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim HR, Kim KW, Kim BM, Jung HG, Cho ML
and Lee SH: Reciprocal activation of CD4+ T cells and synovial
fibroblasts by stromal cell-derived factor 1 promotes RANKL
expression and osteoclastogenesis in rheumatoid arthritis.
Arthritis Rheumatol. 66:538–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jung YK, Kang YM and Han S: Osteoclasts in
the inflammatory arthritis: Implications for pathologic osteolysis.
Immune Netw. 19:e22019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiao C and Rajewsky K: MicroRNA control in
the immune system: Basic principles. Cell. 136:26–36. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ruedel A, Dietrich P, Schubert T,
Hofmeister S, Hellerbrand C and Bosserhoff AK: Expression and
function of microRNA-188-5p in activated rheumatoid arthritis
synovial fibroblasts. Int J Clin Exp Pathol. 8:6607–6616.
2015.PubMed/NCBI
|
|
12
|
Tavasolian F, Abdollahi E, Rezaei R,
Momtazi-Borojeni AA, Henrotin Y and Sahebkar A: Altered expression
of microRNAs in rheumatoid arthritis. J Cell Biochem. 119:478–487.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ji JD, Kim TH, Lee B, Na KS, Choi SJ, Lee
YH and Song GG: Integrated analysis of microRNA and mRNA expression
profiles in rheumatoid arthritis synovial monocytes. J Rheum Dis.
18:253–263. 2011. View Article : Google Scholar
|
|
14
|
Dang Q, Yang F, Lei H, Liu X, Yan M, Huang
H, Fan X and Li Y: Inhibition of microRNA-34a ameliorates murine
collagen-induced arthritis. Exp Ther Med. 14:1633–1639. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Niederer F, Trenkmann M, Ospelt C,
Karouzakis E, Neidhart M, Stanczyk J, Kolling C, Gay RE, Detmar M,
Gay S, et al: Down-regulation of microRNA-34a* in rheumatoid
arthritis synovial fibroblasts promotes apoptosis resistance.
Arthritis Rheum. 64:1771–1779. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arnett FC, Edworthy SM, Bloch DA, McShane
DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS,
et al: The American Rheumatism Association 1987 revised criteria
for the classification of rheumatoid arthritis. Arthritis Rheum.
31:315–324. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bailon-Moscoso N, González-Arévalo G,
Velásquez-Rojas G, Malagon O, Vidari G, Zentella-Dehesa A,
Ratovitski EA and Ostrosky-Wegman P: Phytometabolite
dehydroleucodine induces cell cycle arrest, apoptosis, and DNA
damage in human astrocytoma cells through p73/p53 regulation. PLoS
One. 10:e01365272015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H
and Yu H: Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p
expression in colon cancer. Am J Transl Res. 6:391–401.
2014.PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bustamante MF, Garcia-Carbonell R,
Whisenant KD and Guma M: Fibroblast-like synoviocyte metabolism in
the pathogenesis of rheumatoid arthritis. Arthritis Res Ther.
19:1102017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sotillo E, Laver T, Mellert H, Schelter
JM, Cleary MA, McMahon S and Thomas-Tikhonenko A: Myc
overexpression brings out unexpected antiapoptotic effects of
miR-34a. Oncogene. 30:2587–2594. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mudduluru G, Ceppi P, Kumarswamy R,
Scagliotti GV, Papotti M and Allgayer H: Regulation of Axl receptor
tyrosine kinase expression by miR-34a and miR-199a/b in solid
cancer. Oncogene. 30:2888–2899. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang W, Chen L, Jiang Y and Shen Y:
miR-26a-5p regulates synovial fibroblast invasion in patients with
rheumatoid arthritis by targeting Smad 1. Med Sci Monit.
24:5178–5184. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Audo R, Deckert V, Daien CI, Che H,
Elhmioui J, Lemaire S, Pais de Barros JP, Desrumaux C, Combe B,
Hahne M, et al: PhosphoLipid transfer protein (PLTP) exerts a
direct pro-inflammatory effect on rheumatoid arthritis (RA)
fibroblasts-like-synoviocytes (FLS) independently of its lipid
transfer activity. PLoS One. 13:e01938152018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ruedel A, Dietrich P, Schubert T,
Hofmeister S, Hellerbrand C and Bosserhoff AK: Expression and
function of microRNA-188-5p in activated rheumatoid arthritis
synovial fibroblasts. Int J Clin Exp Pathol. 8:4953–4962.
2015.PubMed/NCBI
|
|
26
|
Jia W, Wu W, Yang D, Xiao C, Su Z, Huang
Z, Li Z, Qin M, Huang M, Liu S, et al: Histone demethylase JMJD3
regulates fibroblast-like synoviocyte-mediated proliferation and
joint destruction in rheumatoid arthritis. FASEB J. 32:4031–4042.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bačević K, Lossaint G, Achour TN, Georget
V, Fisher D and Dulić V: Cdk2 strengthens the intra-S checkpoint
and counteracts cell cycle exit induced by DNA damage. Sci Rep.
7:134292017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sherr CJ, Beach D and Shapiro GI:
Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov.
6:353–367. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou H, Shen T, Luo Y, Liu L, Chen W, Xu
B, Han X, Pang J, Rivera CA and Huang S: The antitumor activity of
the fungicide ciclopirox. Int J Cancer. 127:2467–2477. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li F, Li X, Kou L, Li Y, Meng F and Ma F:
SUMO-conjugating enzyme UBC9 promotes proliferation and migration
of fibroblast-like synoviocytes in rheumatoid arthritis.
Inflammation. 37:1134–1141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yuan H, Yang P, Zhou D, Gao W, Qiu Z, Fang
F, Ding S and Xiao W: Knockdown of sphingosine kinase 1 inhibits
the migration and invasion of human rheumatoid arthritis
fibroblast-like synoviocytes by down-regulating the PI3K/AKT
activation and MMP-2/9 production in vitro. Mol Biol Rep.
41:5157–5165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mo WX, Yin SS, Chen H, Zhou C, Zhou JX,
Zhao LD, Fei YY, Yang HX, Guo JB, Mao YJ, et al: Chemotaxis of Vδ2
T cells to the joints contributes to the pathogenesis of rheumatoid
arthritis. Ann Rheum Dis. 76:2075–2084. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wei ST, Sun YH, Zong SH and Xiang YB:
Serum levels of IL-6 and TNF-α may correlate with activity and
severity of rheumatoid arthritis. Med Sci Monit. 21:4030–4038.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Guo Q, Wang Y, Xu D, Nossent J, Pavlos NJ
and Xu J: Rheumatoid arthritis: Pathological mechanisms and modern
pharmacologic therapies. Bone Res. 6:152018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nishina N, Kaneko Y, Kameda H, Kuwana M
and Takeuchi T: Reduction of plasma IL-6 but not TNF-α by
methotrexate in patients with early rheumatoid arthritis: A
potential biomarker for radiographic progression. Clin Rheumatol.
32:1661–1666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Siegfried G, Basak A, Cromlish JA,
Benjannet S, Marcinkiewicz J, Chrétien M, Seidah NG and Khatib AM:
The secretory proprotein convertases furin, PC5, and PC7 activate
VEGF-C to induce tumorigenesis. J Clin Invest. 111:1723–1732. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Abdollahzad H, Aghdashi MA, Asghari
Jafarabadi M and Alipour B: Effects of coenzyme Q10 supplementation
on inflammatory cytokines (TNF-α, IL-6) and oxidative stress in
rheumatoid arthritis patients: A randomized controlled trial. Arch
Med Res. 46:527–533. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim GW, Lee NR, Pi RH, Lim YS, Lee YM, Lee
JM, Jeong HS and Chung SH: IL-6 inhibitors for treatment of
rheumatoid arthritis: Past, present, and future. Arch Pharm Res.
38:575–584. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu N, Wang Y, Li D, Chen G, Sun R, Zhu R,
Sun S, Liu H, Yang G and Dong T: MDM4 overexpression contributes to
synoviocyte proliferation in patients with rheumatoid arthritis.
Biochem Biophys Res Commun. 401:417–421. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kurowska-Stolarska M, Alivernini S,
Melchor EG, Elmesmari A, Tolusso B, Tange C, Petricca L, Gilchrist
DS, Di Sante G, Keijzer C, et al: MicroRNA-34a dependent regulation
of AXL controls the activation of dendritic cells in inflammatory
arthritis. Nat Commun. 8:158772017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jiang L, Huang Q, Zhang S, Zhang Q, Chang
J, Qiu X and Wang E: Hsa-miR-125a-3p and hsa-miR-125a-5p are
downregulated in non-small cell lung cancer and have inverse
effects on invasion and migration of lung cancer cells. BMC Cancer.
10:3182010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Córdova-Rivas S, Fraire-Soto I,
Mercado-Casas Torres A, Servín-González LS, Granados-López AJ,
López-Hernández Y, Reyes-Estrada CA, Gutiérrez-Hernández R,
Castañeda-Delgado JE, Ramírez-Hernández L, et al: 5p and 3p strands
of miR-34 family members have differential effects in cell
proliferation, migration, and invasion in cervical cancer cells.
Int J Mol Sci. 20:2019. View Article : Google Scholar
|