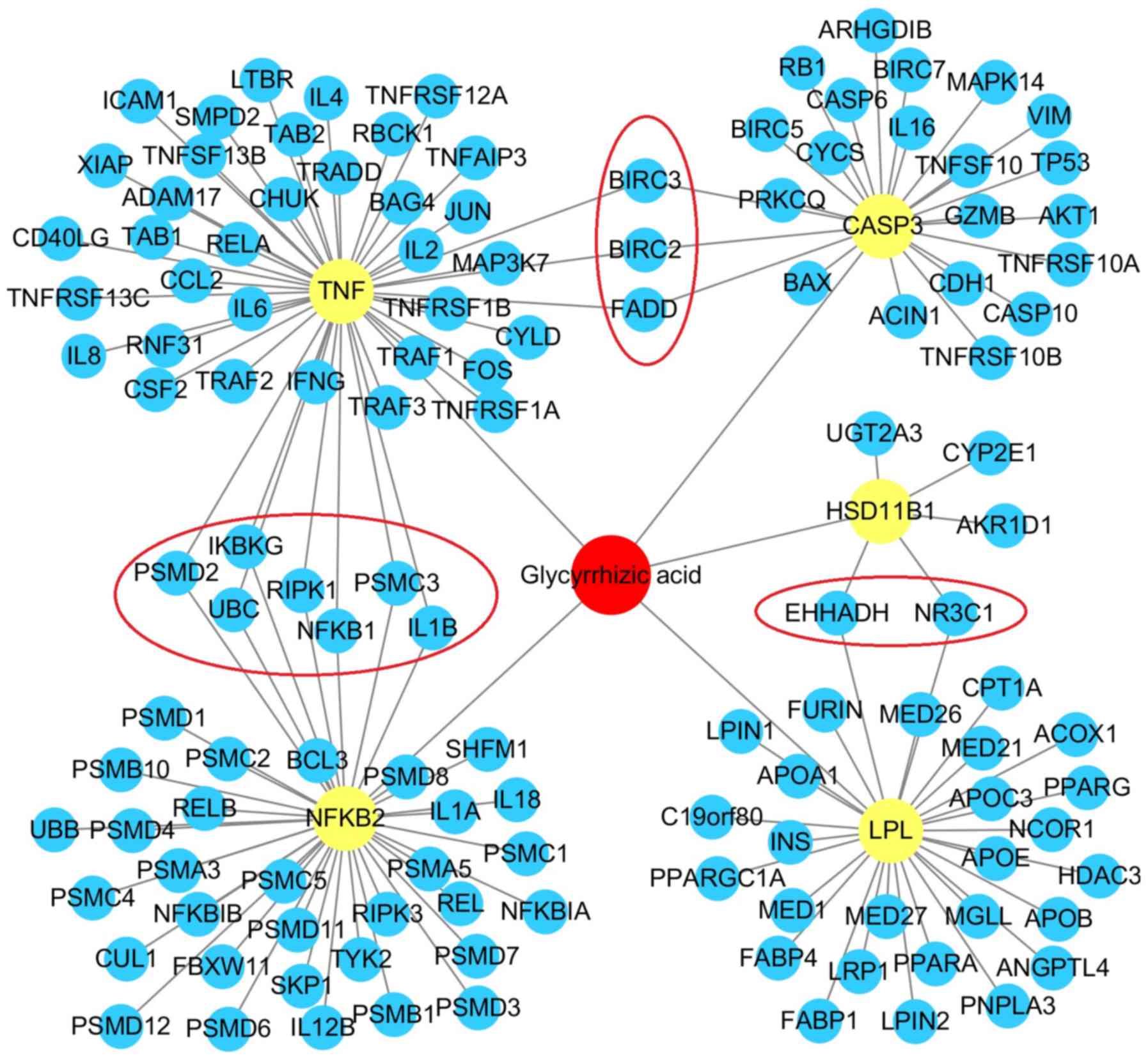
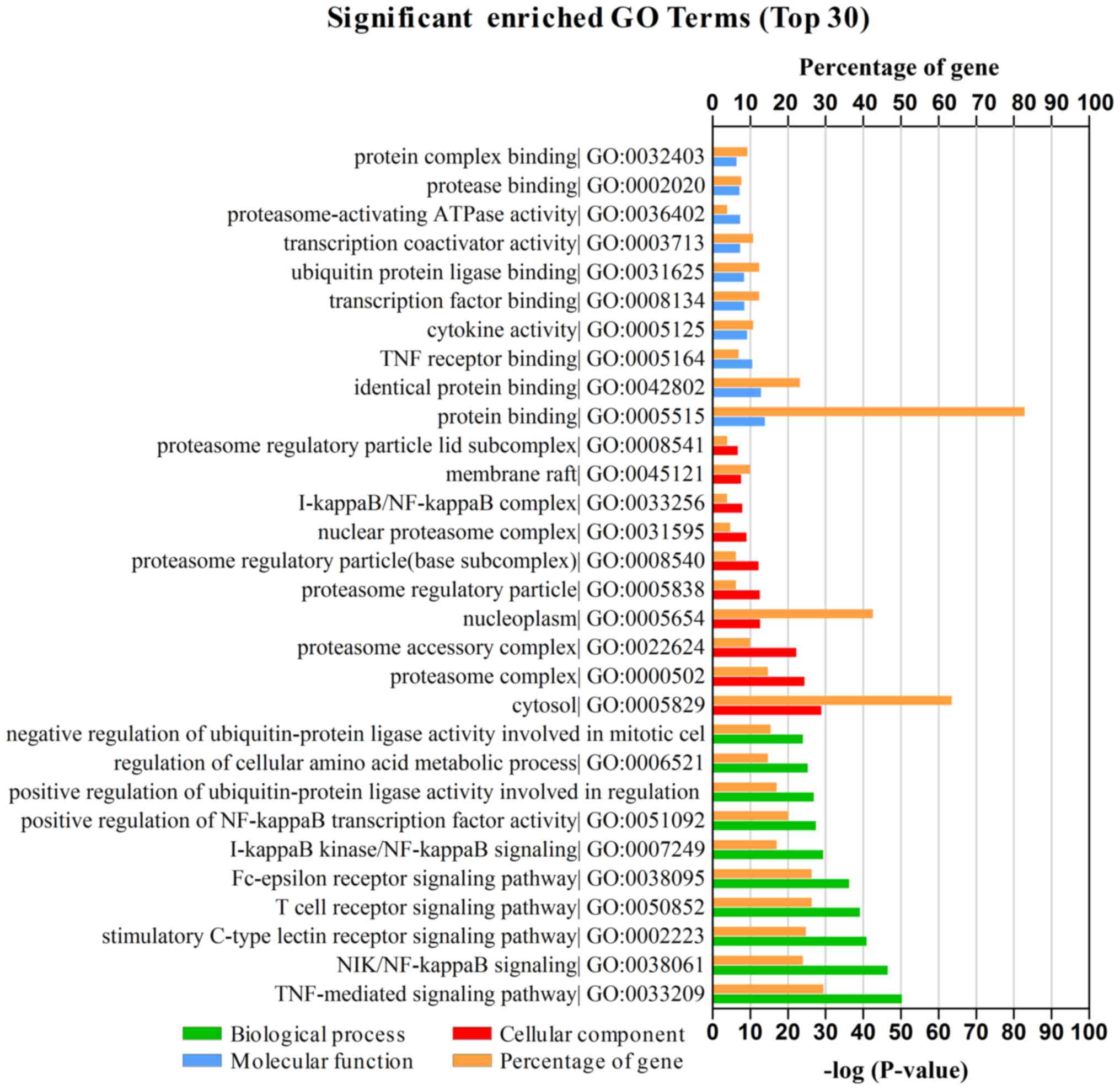
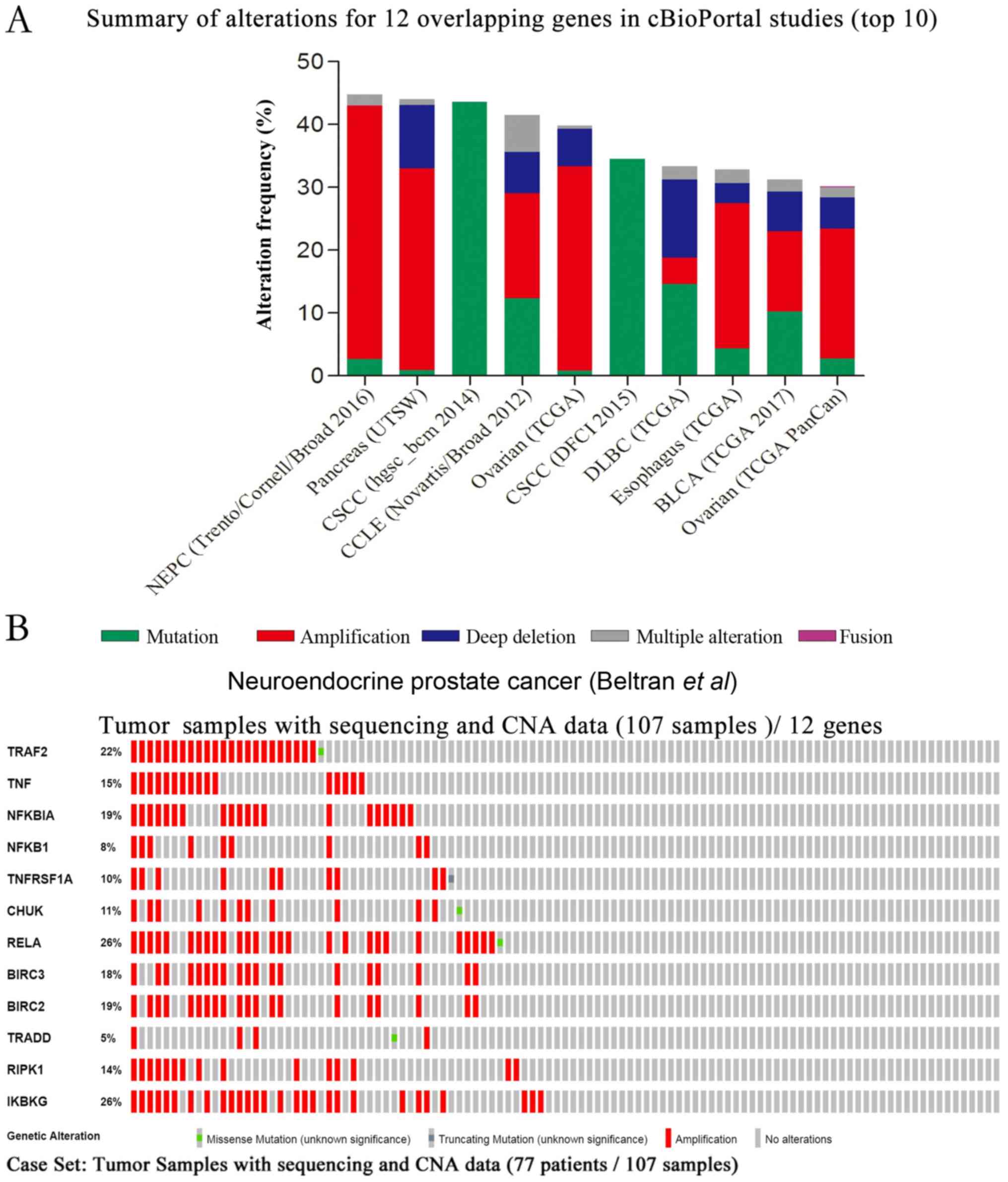
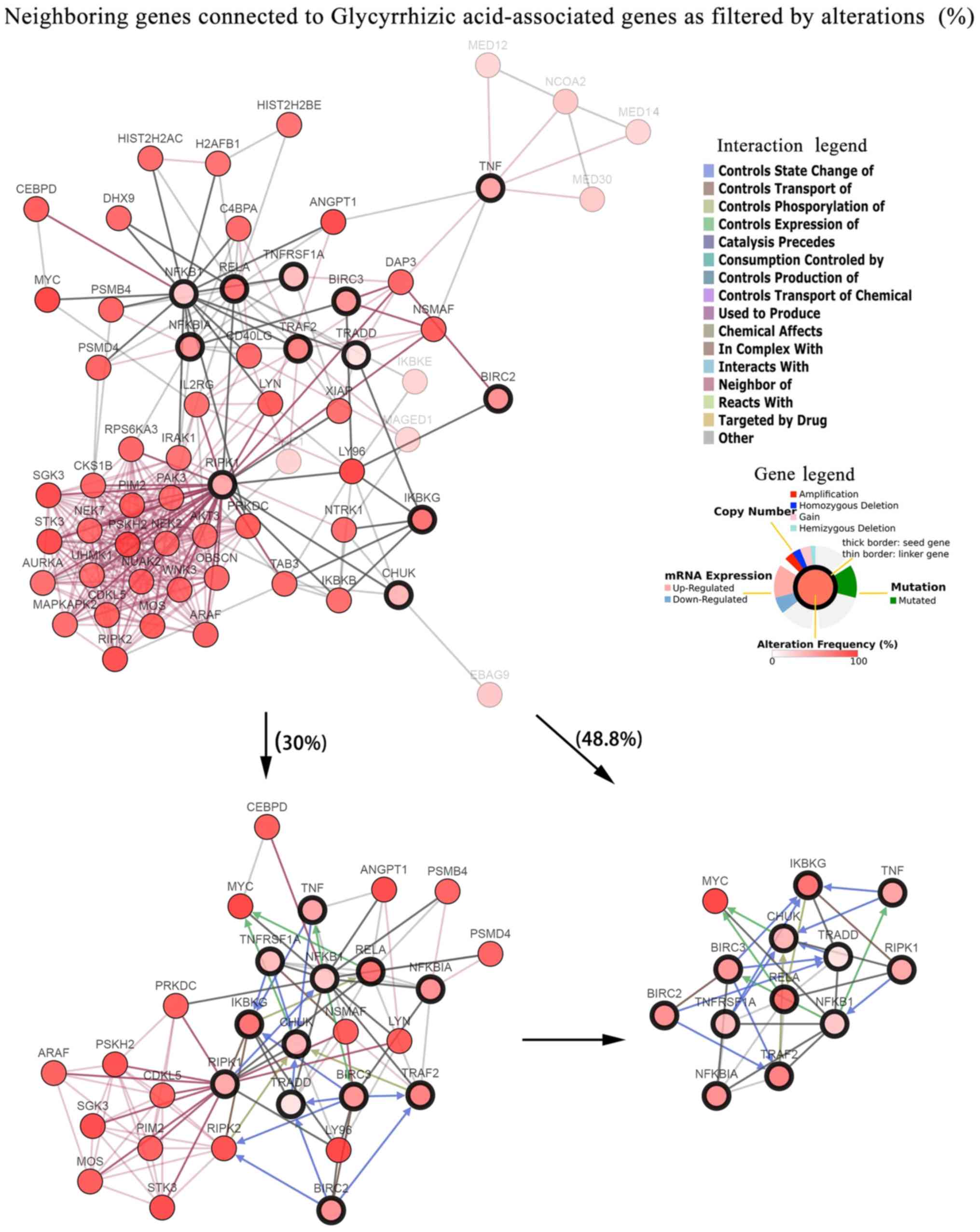
|
1
|
Saif MW, Li J, Lamb L, Kaley K, Elligers
K, Jiang Z, Bussom S, Liu SH and Cheng YC: First-in-human phase II
trial of the botanical formulation PHY906 with capecitabine as
second-line therapy in patients with advanced pancreatic cancer.
Cancer Chemother Pharmacol. 73:373–380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ming LJ and Yin AC: Therapeutic effects of
glycyrrhizic acid. Nat Prod Commun. 8:415–418. 2013.PubMed/NCBI
|
|
3
|
Farrukh MR, Nissar UA, Kaiser PJ, Afnan Q,
Sharma PR, Bhushan S and Tasduq SA: Glycyrrhizic acid (GA) inhibits
reactive oxygen Species mediated photodamage by blocking ER stress
and MAPK pathway in UV-B irradiated human skin fibroblasts. J
Photochem Photobiol B. 148:351–357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fouladi S, Masjedi M, Ghasemi R, G Hakemi
M and Eskandari N: The in vitro impact of glycyrrhizic acid on
CD4+ T lymphocytes through OX40 receptor in the patients
with allergic rhinitis. Inflammation. 41:1690–1701. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He SQ, Gao M, Fu YF and Zhang YN:
Glycyrrhizic acid inhibits leukemia cell growth and migration via
blocking AKT/mTOR/STAT3 signaling. Int J Clin Exp Pathol.
8:5175–5181. 2015.PubMed/NCBI
|
|
6
|
Hou S, Zhang T, Li Y, Guo F and Jin X:
Glycyrrhizic acid prevents diabetic nephropathy by activating
AMPK/SIRT1/PGC-1α signaling in db/db mice. J Diabetes Res.
2017:28659122017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kiratipaiboon C, Tengamnuay P and
Chanvorachote P: Glycyrrhizic acid attenuates stem cell-like
phenotypes of human dermal papilla cells. Phytomedicine.
22:1269–1278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li C, Peng S, Liu X, Han C, Wang X, Jin T,
Liu S, Wang W, Xie X, He X, et al: Glycyrrhizin, a direct HMGB1
antagonist, ameliorates inflammatory infiltration in a model of
autoimmune thyroiditis via inhibition of TLR2-HMGB1 signaling.
Thyroid. 27:722–731. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin SC, Chu PY, Liao WT, Wu MY, Tsui KH,
Lin LT, Huang CH, Chen LL and Li CJ: Glycyrrhizic acid induces
human MDA-MB-231 breast cancer cell death and autophagy via the
ROS-mitochondrial pathway. Oncol Rep. 39:703–710. 2018.PubMed/NCBI
|
|
10
|
Refahi S, Pourissa M, Zirak MR and Hadadi
G: Modulation expression of tumor necrosis factor α in the
radiation-induced lung injury by glycyrrhizic acid. J Med Phys.
40:95–101. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang LY, Cheng KC, Li Y, Niu CS, Cheng JT
and Niu HS: Glycyrrhizic acid increases glucagon like peptide-1
secretion via TGR5 activation in type 1-like diabetic rats. Biomed
Pharmacother. 95:599–604. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang YM and Du GQ: Glycyrrhizic acid
prevents enteritis through reduction of NF-κB p65 and p38MAPK
expression in rat. Mol Med Rep. 13:3639–3646. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang J, Shi Y, Chen H, Wang X, Chen Y and
Yang B: Glycyrrhizic acid attenuates myocardial injury: Involvement
of RIP140/NF-κB Pathway. Biomed Pharmacother. 95:62–67. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang H, Zhang R, Chen J, Shi M, Li W and
Zhang X: High mobility group Box1 inhibitor glycyrrhizic acid
attenuates kidney injury in streptozotocin-induced diabetic rats.
Kidney Blood Press Res. 42:894–904. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Relling MV and Evans WE: Pharmacogenomics
in the clinic. Nature. 526:343–350. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mooney SD: Progress towards the
integration of pharmacogenomics in practice. Hum Genet.
134:459–465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Campedel L, Kossaï M, Blanc-Durand P,
Rouprêt M, Seisen T, Compérat E, Spano JP and Malouf G:
Neuroendocrine prostate cancer: Natural history, molecular
features, therapeutic management and future directions. Bull
Cancer. 104:789–799. 2017.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Su G, Morris JH, Demchak B and Bader GD:
Biological network exploration with Cytoscape 3. Curr Protoc
Bioinformatics. 47:8.13.1–24. 2014. View Article : Google Scholar
|
|
19
|
Guo Y, Bao Y, Ma M and Yang W:
Identification of key candidate genes and pathways in colorectal
cancer by integrated bioinformatical analysis. Int J Mol Sci.
18(pii): E7222017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Canault B, Bourg S, Vayer P and Bonnet P:
Comprehensive network map of ADME-Tox databases. Mol Inform.
36:2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Coelho-Santos V, Leitão RA, Cardoso FL,
Palmela I, Rito M, Barbosa M, Brito MA, Fontes-Ribeiro CA and Silva
AP: The TNF-α/NF-κB signaling pathway has a key role in
methamphetamine-induced blood-brain barrier dysfunction. J Cereb
Blood Flow Metab. 35:1260–1271. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kang R, Xie Y, Zhang Q, Hou W, Jiang Q,
Zhu S, Liu J, Zeng D, Wang H, Bartlett DL, et al: Intracellular
HMGB1 as a novel tumor suppressor of pancreatic cancer. Cell Res.
27:916–932. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Beltran H, Prandi D, Mosquera JM, Benelli
M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV,
Varambally S, et al: Divergent clonal evolution of
castration-resistant neuroendocrine prostate cancer. Nat Med.
22:298–305. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Asl MN and Hosseinzadeh H: Review of
pharmacological effects of Glycyrrhiza sp. and its bioactive
compounds. Phytother Res. 22:709–724. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brumatti G, Salmanidis M and Ekert PG:
Crossing paths: Interactions between the cell death machinery and
growth factor survival signals. Cell Mol Life Sci. 67:1619–1630.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hayden MS and Ghosh S: Regulation of NF-κB
by TNF family cytokines. Semin Immunol. 26:253–266. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yaw HP, Ton SH, Chin HF, Karim MK,
Fernando HA and Kadir KA: Modulation of lipid metabolism in
glycyrrhizic acid-treated rats fed on a high-calorie diet and
exposed to short or long-term stress. Int J Physiol Pathophysiol
Pharmacol. 7:61–75. 2015.PubMed/NCBI
|
|
29
|
Chen S, Jiang S, Zheng W, Tu B, Liu S,
Ruan H and Fan C: RelA/p65 inhibition prevents tendon adhesion by
modulating inflammation, cell proliferation, and apoptosis. Cell
Death Dis. 8:e27102017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miot C, Imai K, Imai C, Mancini AJ, Kucuk
ZY, Kawai T, Nishikomori R, Ito E, Pellier I, Dupuis Girod S, et
al: Hematopoietic stem cell transplantation in 29 patients
hemizygous for hypomorphic IKBKG/NEMO mutations. Blood.
130:1456–1467. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wei B, Liang J, Hu J, Mi Y, Ruan J, Zhang
J, Wang Z, Hu Q, Jiang H and Ding Q: TRAF2 is a valuable prognostic
biomarker in patients with prostate cancer. Med Sci Monit.
23:4192–4204. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wei B, Ruan J, Mi Y, Hu J, Zhang J, Wang
Z, Hu Q, Jiang H and Ding Q: Knockdown of TNF receptor-associated
factor 2 (TRAF2) modulates in vitro growth of TRAIL-treated
prostate cancer cells. Biomed Pharmacother. 93:462–469. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jin H, Dong YY, Zhang H, Cui Y, Xie K and
Lou G: shRNA depletion of cIAP1 sensitizes human ovarian cancer
cells to anticancer agent-induced apoptosis. Oncol Res. 22:167–176.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seidelin JB: Regulation of antiapoptotic
and cytoprotective pathways in colonic epithelial cells in
ulcerative colitis. Scand J Gastroenterol. 50 (Suppl 1):S1–S29.
2015. View Article : Google Scholar
|
|
35
|
Hostetler BJ, Uchakina ON, Ban H and
McKallip RJ: Treatment of hematological malignancies with
glycyrrhizic acid. Anticancer Res. 37:997–1004. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim KJ, Choi JS, Kim KW and Jeong JW: The
anti-angiogenic activities of glycyrrhizic acid in tumor
progression. Phytother Res. 27:841–846. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Doss GP, Agoramoorthy G and Chakraborty C:
TNF/TNFR: Drug target for autoimmune diseases and immune-mediated
inflammatory diseases. Front Biosci (Landmark Ed). 19:1028–1040.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Berman-Booty LD and Knudsen KE: Models of
neuroendocrine prostate cancer. Endocr Relat Cancer. 22:R33–R49.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guo R, Tu Y, Xie S, Liu XS, Song Y, Wang
S, Chen X and Lu L: A role for receptor-interacting protein
kinase-1 in neutrophil extracellular trap formation in patients
with systemic lupus erythematosus: A preliminary study. Cell
Physiol Biochem. 45:2317–2328. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ando K, Sakoda M, Ueno S, Hiwatashi K,
Iino S, Minami K, Kawasaki Y, Hashiguchi M, Tanoue K, Mataki Y, et
al: Clinical implication of the relationship between high mobility
group Box-1 and tumor differentiation in hepatocellular carcinoma.
Anticancer Res. 38:3411–3418. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y, Li L, Qi C, Hua S, Fei X, Gong F
and Fang M: Glycyrrhizin alleviates Con A-induced hepatitis by
differentially regulating the production of IL-17 and IL-25. Biomed
Pharmacother. 110:692–699. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dujmovic I, Mangano K, Pekmezovic T,
Quattrocchi C, Mesaros S, Stojsavljevic N, Nicoletti F and Drulovic
J: The analysis of IL-1 beta and its naturally occurring inhibitors
in multiple sclerosis: The elevation of IL-1 receptor antagonist
and IL-1 receptor type II after steroid therapy. J Neuroimmunol.
207:101–106. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shin MS, Kang Y, Wahl ER, Park HJ, Lazova
R, Leng L, Mamula M, Krishnaswamy S, Bucala R and Kang I:
Macrophage migration inhibitory factor regulates U1 small nuclear
RNP immune complex-mediated activation of the NLRP3 inflammasome.
Arthritis Rheumatol. 71:109–120. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ehmann F, Caneva L, Prasad K, Paulmichl M,
Maliepaard M, Llerena A, Ingelman-Sundberg M and Papaluca-Amati M:
Pharmacogenomic information in drug labels: European medicines
agency perspective. Pharmacogenomics J. 15:201–210. 2015.
View Article : Google Scholar : PubMed/NCBI
|