Introduction
Glycyrrhizic acid (GA) is a triterpene glycoside
isolated from the root of the licorice plant. In Japan and China,
GA has been developed as an anti-inflammatory hepatoprotective drug
in cases of chronic hepatitis (1).
GA has also been determined to exhibit a wide range of
pharmacological and biological activities, including anti-ulcer,
anti-allergic, antioxidant, anti-diabetic and anti-tumor effects
(2). In recent years, an
increasing number of studies have focused on determining the
underlying mechanisms and targets of GA (3–14).
However, the lack of systematic dissection of drug-target
mechanisms between GA-associated primary and secondary targets is a
great obstacle to the research and development of susceptibility
indicators for clinical applications. To address this obstacle, the
following should be determined: i) Whether significant interactions
exist between recently reported targets, including high mobility
group box 1 (HMGB1), and well-documented targets mediated by GA,
including caspase-3 (CASP3), tumor necrosis factor (TNF) and
nuclear factor κB subunit 2 (NFKB2) (1,8,14);
ii) whether there are key hub elements among targets that can
improve the understanding of interrelated biological actions
mediated by GA, including inflammatory tumors or apoptosis
regulation in immune-associated diseases; and iii) whether the
systematic assessment of previously published articles can provide
predictive leads to uncover molecular events mediated by GA.
In recent years, the genetic alteration of ~20 genes
has impacted ~80 approved drugs and, as such, has affected their
applicability in a clinical setting (15). Pharmacogenomics (PG) has been
developed to determine the associations between genetic variants
and drug responses and to create an evidenced-based strategy for
improving the administration of drugs (16). To remove analytical obstacles, the
PG-based approach may be suitable for mining existing drug-target
data obtained from numerous multicenter genomics studies. PG may
also be effective for focusing research scope and direction as well
as determining drug-target interactions. The current study queried
multiple online sources (Drugbank, PubChem, PharmaGKB, STRING,
DAVID and cBioPortal) to optimize candidate target genes and to
construct a functional activity network associated with GA. The
results revealed that the main target pathway mediated by GA is
closely associated with the TNF axis. Furthermore, an overlapping
gene set extracted from protein-protein interaction (PPI) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analyses may be
the key element to the pathogenesis of neuroendocrine prostate
cancer, which is a rare malignancy with a poor prognosis,
associated with inflammatory induction, and mostly occurs as an
adaptive response following intensive androgen deprivation
treatment (17). Thus, the present
study provided an insight into GA-mediated anti-cancer activity and
may have implications for the treatment of inflammatory malignant
neuroendocrine prostate cancer.
Materials and methods
Identification of drug targets and
visualization of interactions
GA drug-target information was searched using
multiple online sources, including Drugbank (https://www.drugbank.ca/), PharmaGKB (https://www.pharmgkb.org/) and PubChem (https://pubchem.ncbi.nlm.nih.gov/).
Identification of the direct protein targets of GA was performed
using DrugBank. A total of five primary protein targets associated
with GA were identified. Interaction networks for GA-associated
primary protein targets were generated from the STRING database
(https://string-db.org/) and integrated into seven
active interaction sources (textmining, experiments, databases,
co-expression, neighborhood, gene fusion and co-occurrence). The
data were then used to output a total of 138 functional partners.
The visualized interaction network of integrated GA and GA target
data was produced using Cytoscape (version 3.61; http://cytoscape.org/), an open-source software
package for visualizing complex networks. Bioinformatics analysis
was performed as previously described (18).
Gene Ontology (GO) and KEGG
analysis
Functional and pathway enrichment for the target
gene set of GA were analyzed using DAVID (version 6.8; http://david.ncifcrf.gov/), a tool for gene functional
annotation, visualization and integrated discovery (19). A total of 129 GA-associated target
genes were specifically queried and integrated into the GO and KEGG
analysis tools in DAVID. The top 30 GO terms with P<0.05 are
presented. The significance of the identified genes from the
drug-target analysis was verified by replacing significant
candidates with other random genes. Validated genes were then
systematically sorted based on KEGG analysis. The top 10 pathways
with P<0.05 were selected. Statistical analysis was subsequently
performed using DAVID, and data were analyzed using the Fisher's
exact test.
Analysis of GA-associated tumor
genetic alterations using cBioPortal
To determine the connectivity of GA-associated
target genes within different cancer genomic studies, GA-associated
data were integrated for further analysis using cBioPortal
(http://www.cbioportal.org/), an
open-source platform that provides visualization, analysis and
downloadable large-scale cancer genomic datasets (20). The visualization of genetic
alterations across tumor samples was produced using OncoPrint tool.
Interaction networks of the overlapping genes from the enrichment
analysis were generated via multiple visualization analyses. The
underlying data could then be connected to clinical profiles to
promote novel discoveries in drug-associated biological functions.
The overlapping genes of pathway enrichment analysis was performed
with Venn analysis (Venny software; version 2.1; http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Results
Characterization of GA bioactivity and
visualization of the GA-associated target network via
Cytoscape
Drugbank and PubChem are free bioinformatics and
cheminformatics resources that allow for the mining of detailed
drug data with comprehensive drug-target information (21). The current study queried Drugbank
using GA as the input and subsequently checked against PubChem.
This resulted in output DB13751, categorizing GA as an
anti-inflammatory agent used in the alimentary tract and for liver
therapy, grouping GA as an approved experimental drug. Five primary
protein targets of GA [corticosteroid 11-β-dehydrogenase isozyme 1
(HSD11B1), TNF, CASP3, NFKB2 and lipoprotein lipase (LPL)] were
revealed (Table I). To further
expand the analyses, the secondary protein targets of GA were
detected via PPI analysis using STRING. A total of 138 functional
partners were identified and determined to be associated with
GA-associated primary protein targets (Table SI).
 | Table I.Identification of primary protein
targets of glycyrrhizic acid using DrugBank. |
Table I.
Identification of primary protein
targets of glycyrrhizic acid using DrugBank.
| Searched drug | Targets |
|---|
|
|
|---|
| # | DB_ID | Gene_Symbol | Uniprot_ID | Entrez_ID | Actions |
|---|
| 1 | DB13751 | HSD11B1 | P28845 | 3290 | Antagonist |
| 2 | DB13751 | TNF | P01375 | 7124 | Antagonist |
| 3 | DB13751 | CASP3 | P42574 | 836 | Antagonist |
| 4 | DB13751 | NFKB2 | Q00653 | 4791 | Translocation
inhibitor |
| 5 | DB13751 | LPL | P06858 | 4023 | Inducer |
A drug-target interaction network was constructed by
integrating data into Cytoscape (version 3.61) (18). The primary and secondary PPIs of
GA, including HSD11B1 (5 PPIs), TNF (44 PPIs), CASP3 (23 PPIs),
NFKB2 (38 PPIs) and LPL (28 PPIs), are presented in Fig. 1. Additionally, when compared with
HSD11B1 and LPL, three primary protein targets (TNF, NFKB2 and
CASP3) possessed up to 10 crossed secondary protein targets
[baculoviral IAP repeat-containing protein 3 (BIRC3), BIRC2,
FAS-associated death domain protein (FADD), 26S proteasome
non-ATPase regulatory subunit 2 (PSMD2), NF-κB essential modulator
(IKBKG), ubiquitin C (UBC), receptor-interacting
serine/threonine-protein kinase 1 (RIPK1), NF-κB subunit 1 (NFKB1),
26S proteasome regulatory subunit 6A (PSMC3) and interleukin 1β
(IL1B)]. This indicated that the main target pathway mediated by GA
is closely associated with the TNF axis.
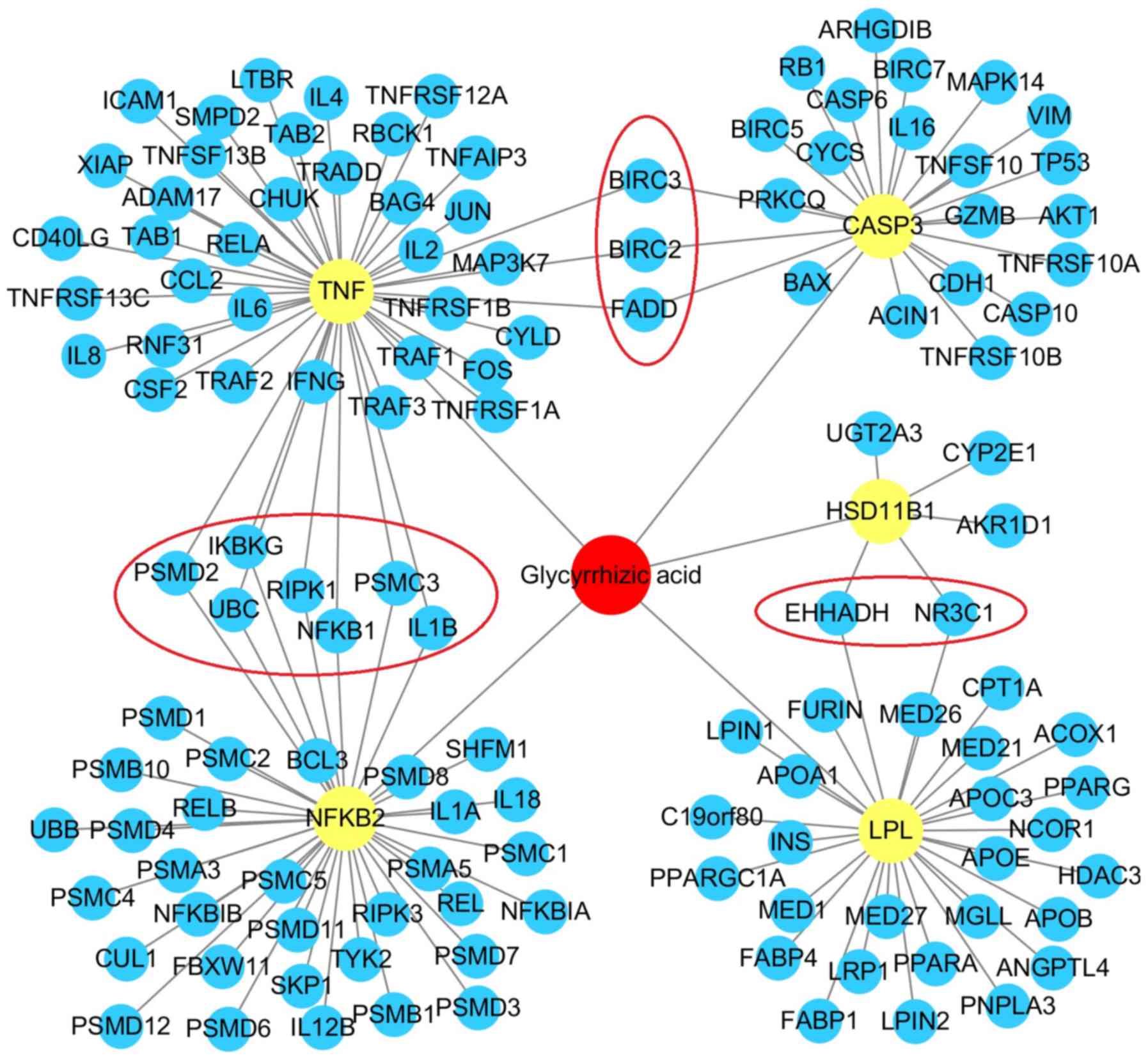 | Figure 1.Drug-target interaction network of
GA-associated protein targets. GA is presented in red, primary
protein targets including HSD11B1, TNF, CASP3, NFKB2 and LPL are
presented in yellow, and secondary protein targets that interact
with GA-associated primary protein targets are presented in blue.
Crossed secondary protein targets (BIRC3, BIRC3, FADD, PSMD2,
IKBKG, UBC, RIPK1, NFKB1, PSMC3, IL1B, EHHADH are NR3C1) are
presented as red circles. The network exhibited a total of 138
protein-protein interactions. GA, glycyrrhizic acid. |
GO analysis and signaling pathway
enrichments connected to GA-associated gene sets
To assess the functional attributes of a
GA-associated gene set (129 genes from Homo sapiens), GO and
pathway enrichment analyses were performed using DAVID with
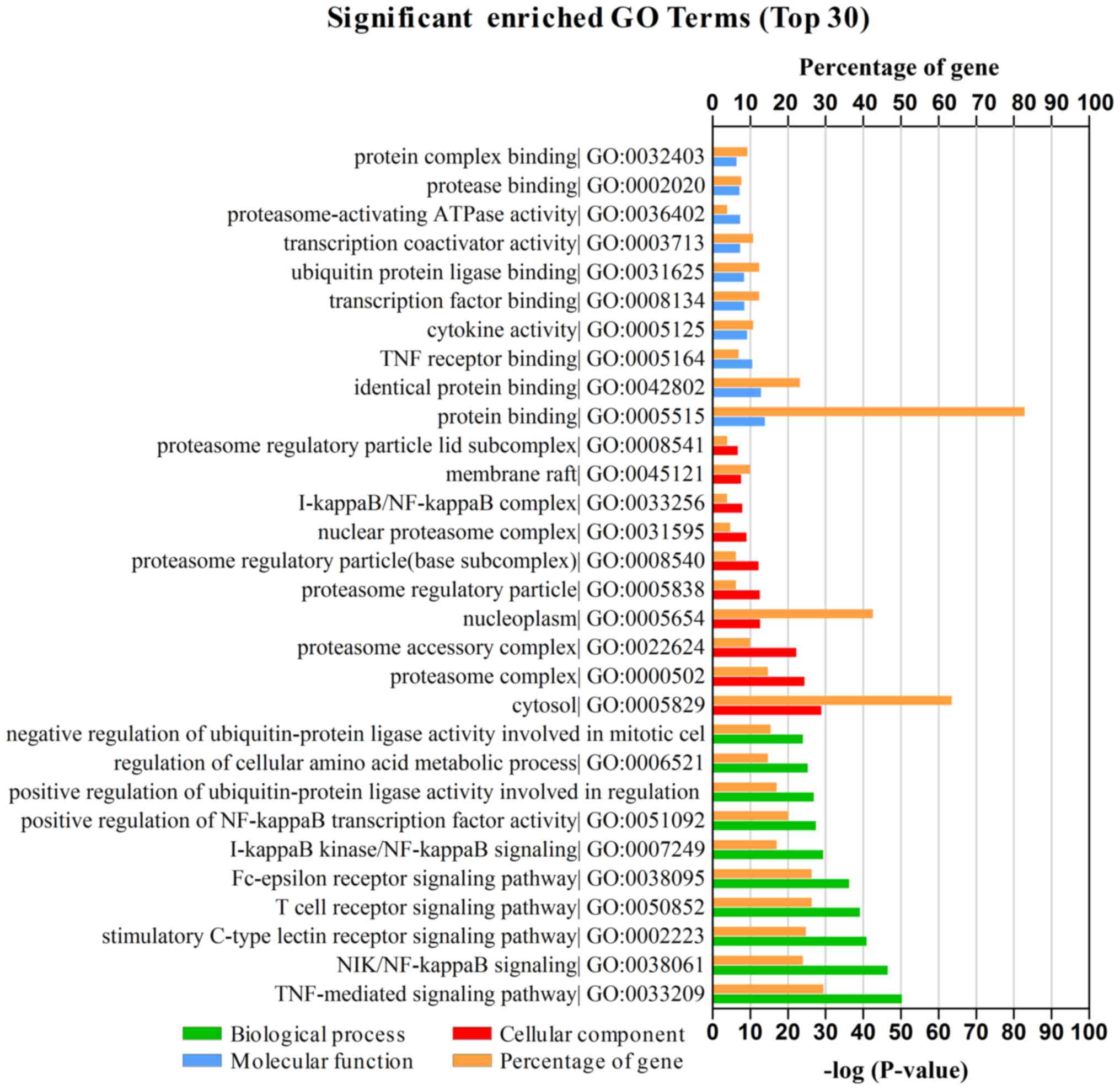
P<0.05 as the cut-off criterion (19). As presented in Fig. 2, GA-associated genes were
classified into three functional groups: Biological processes (BP),
cellular components (CC) and molecular functions (MF). In the BP
group, genes were mainly enriched in ‘TNF-mediated signaling
pathway’ and ‘NIK/NF-κB signaling’. In the CC group, genes were
largely enriched in ‘cytosol’, ‘proteasome complex’ and
‘nucleoplasm’. In the MF group, genes were primarily enriched in
‘protein binding’. The GO results therefore indicated that most
GA-mediated genes were significantly enriched in ‘TNF-mediated
signaling pathway’, ‘protein binding’, ‘cytosol’ and
‘nucleoplasm’.
The top 10 KEGG pathways connected to GA-associated
genes are presented in Table II
and were as follows: ‘TNF signaling pathway’ (34 genes),
‘Epstein-Barr virus infection’ (41 genes), ‘NF-κB signaling
pathway’ (28 genes), ‘apoptosis’ (24 genes), ‘proteasome’ (20
genes), ‘herpes simplex infection’ (28 genes), ‘NOD-like receptor
signaling pathway’ (18 genes), ‘toxoplasmosis’ (23 genes),
‘osteoclast differentiation’ (23 genes) and ‘measles’ (22 genes).
In this functional enrichment analysis, the GA-associated gene set
was indicated to be closely associated with TNF and TNF-related
signaling pathways, which possess potential biological functions,
including: i) The regulation of gene transcription associated with
survival, proliferation and inflammation via the TNF/NF-κB
signaling pathway; and ii) the control of cell apoptosis via the
TNF/caspase family signaling pathway. On account of a direct
association with the TNF axis, as determined via the interactive
network analysis of GA-associated protein targets, the current
study focused the subsequent analysis on the TNF and TNF-associated
signaling pathways (Fig. 1). Thus,
based on the results of the GO and KEGG pathway analysis, in
combination with the determination of GA-associated protein
targets, it was revealed that the main biological and functional
association exists between GA and the key hub elements of TNF and
the TNF-associated pathway (crossed targets including BIRC2, BIRC3,
RIPK1, IKBKG and NFKB1; Fig. 1),
which are closely associated with certain physiological or
pathological regulatory mechanisms including apoptosis,
anti-apoptosis, cell survival, cell proliferation, cell
differentiation and inflammation (22).
 | Table II.List of significant enriched
signaling pathways of the glycyrrhizic acid-associated genes. |
Table II.
List of significant enriched
signaling pathways of the glycyrrhizic acid-associated genes.
| Pathway name | #Gene | P-value | Entrez_ID |
|---|
| TNF signaling
pathway | 34 |
1.97×10−33 | 7185 1437 7186 6347
7124 4792 4790 6885 207 7132 9530 2353 836 7133 602 3553 1147 7187
3383 3569 5970 8772 330 10454 23118 329 8717 843 3725 1432 8737
8517 11035 7128 |
| Epstein-Barr virus
infection | 41 |
2.99×10−33 | 7185 7186 4793 7431
7979 4792 4790 4791 6885 207 5707 3458 5708 5709 5710 9861 5713
1147 5714 7187 3383 5970 5971 7157 5925 10454 23118 8717 7297 5705
5704 5718 5702 5717 3725 8737 1432 5701 5700 8517 7128 |
| NF-κB signaling
pathway | 28 |
2.78×10−27 | 7185 7186 7124 331
4792 4790 4791 6885 7132 3553 1147 7187 3383 4055 5970 5971 115650
330 10454 23118 329 8717 5588 10673 959 8737 8517 7128 |
| Apoptosis | 24 |
2.24×10−25 | 7186 7124 331 5970
54205 7157 4792 4790 8772 330 329 8717 8797 207 843 7132 839 836
8743 8795 581 8737 8517 1147 |
| Proteasome | 20 |
1.31×10−22 | 5699 7979 5705 5718
5689 5704 5717 5686 5702 5701 5684 3458 5707 5700 5708 5709 5710
9861 5713 5714 |
| Herpes simplex
infection | 28 |
4.51×10−18 | 7185 7186 7124 6347
4793 4792 4790 6885 7132 2353 836 3458 3553 8454 1147 7187 3569
5970 54205 7157 8772 6500 10454 23118 7297 3725 8517 3593 |
| NOD-like receptor
signaling pathway | 18 |
2.19×10−17 | 3569 7124 6347 5970
3606 4793 4792 4790 330 10454 329 23118 6885 1432 8517 3553 7128
1147 |
| Toxoplasmosis | 23 |
4.57×10−17 | 7124 331 4793 5970
54205 79444 4792 4790 330 10454 329 23118 6885 207 7297 7132 836
959 1432 3458 8517 3593 1147 |
| Osteoclast
differentiation | 23 |
4.67×10−16 | 7186 7124 5970 5468
5971 4792 4790 4791 10454 23118 6885 7297 207 7132 1540 2353 3725
1432 8517 3458 3553 1147 3552 |
| Measles | 22 |
9.04×10−15 | 3565 3569 4793 5970
7157 4792 4790 23118 6885 8797 207 7297 5588 8743 8795 3458 3553
3593 7128 3552 1147 3558 |
Mining genetic alterations of
GA-associated genes in cBioPortal cancer studies
Although DrugBank categorizes GA as an
anti-inflammatory agent used for hepatic protection or peptic ulcer
treatment, the results of the enrichment analysis indicated that it
may also be associated with apoptosis, survival and proliferation.
Furthermore, a previous study revealed that GA demonstrates marked
efficacy in the treatment of inflammation-induced migration or
invasion in pancreatic cancer (23). To further confirm if susceptible
cancers are associated with GA-associated genes, cBioPortal was
utilized to analyze the genetic alterations of GA-associated genes
in different types of human cancer. Since the TNF axis is a main
target module of GA (Fig. 1), and
since the NF-κB and apoptosis pathways are associated with the TNF
signaling pathway, an overlapping gene set was extracted [TNF
receptor associated factor 2 (TRAF2), TNF, NFKB inhibitor-α
(NFKBIA), NFKB1, TNF receptor superfamily member 1A (TNFRSF1A),
component of inhibitor of nuclear factor-κB kinase complex (CHUK),
RELA proto-oncogene, NF-κB subunit (RELA), BIRC3, BIRC2, TNFRSF1A
associated via death domain (TRADD), RIPK1 and IKBKG] from the
genes enriched in TNF, NF-κB and apoptosis signaling pathways using
Venn analysis (Fig. S1). A total
of 12 overlapping genes were utilized to cross-check their genomic
changes and clinical profiles in cBioPortal studies. After querying
the overlapping gene set of all listed cancer studies, genomic
alterations (top 10 cancer studies) were determined to range
between 30.09 and 44.8% (Fig. 3A).
As the study by Beltran et al (24) demonstrated the most prominent
genetic alterations, their queried genes were further explored in
the current study. A visual summary of the documented genetic
changes was created using OncoPrint and is presented in Fig. 3B. The results revealed that 48% of
sequenced samples possessed an alteration in at least one gene, and
that frequency changes ranged between 5 and 26%, which were
associated with the majority of gene amplifications. The query
contained no genetic pairs with mutually exclusive alterations, and
66 gene pairs with concurrent alterations were determined, in which
27 gene pairs were significant (P<0.05; data not shown).
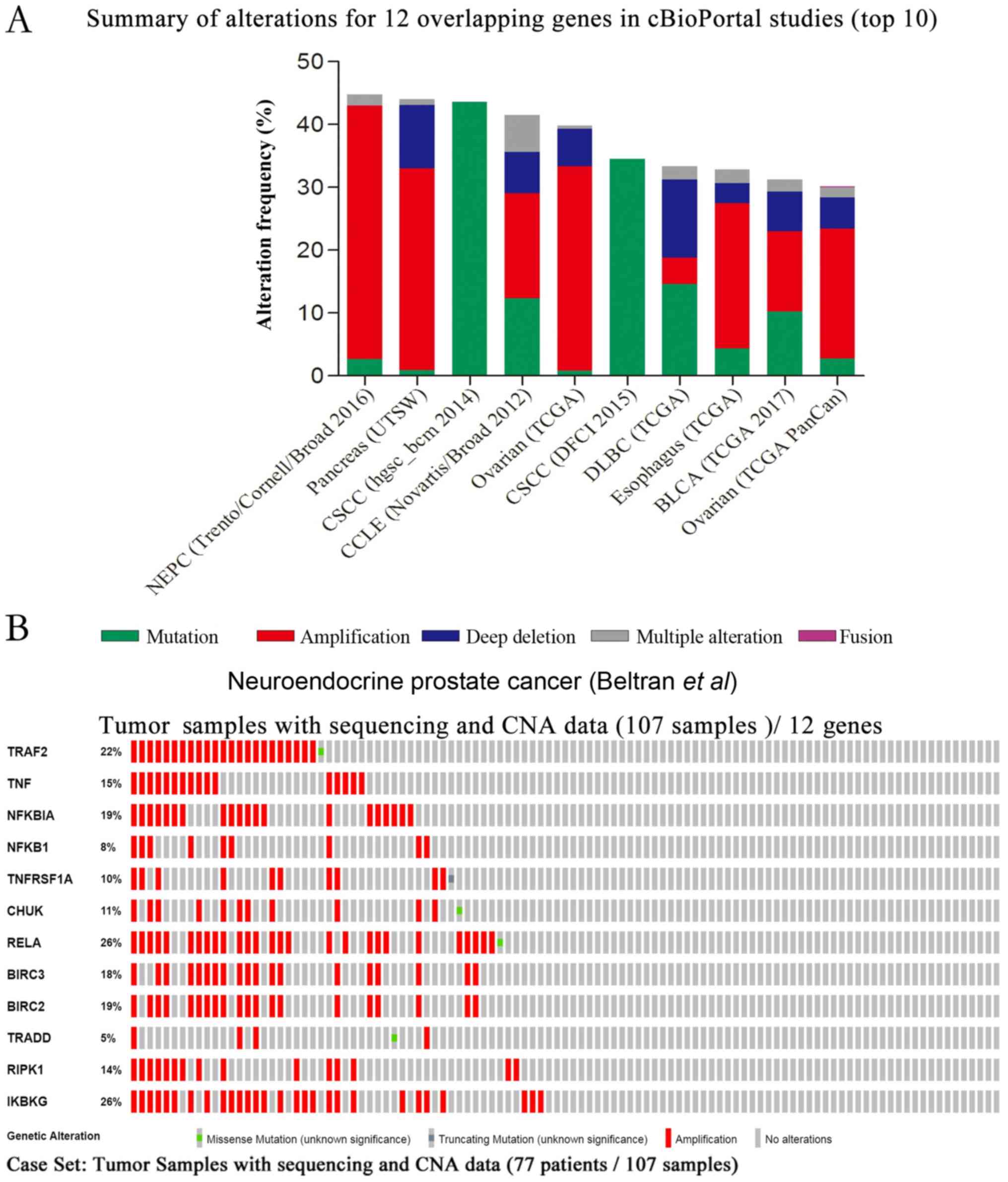 | Figure 3.Summary of the genetic changes of 12
GA-associated overlapping genes, obtained from cBioPortal cancer
genomic studies. (A) Overview of the top 10 genomic changes of 12
overlapping genes (TRAF2, TNF, NFKBIA, NFKB1, TNFRSF1A, CHUK, RELA,
BIRC3, BIRC2, TRADD, RIPK1 and IKBKG) in cBioPortal genomic
datasets available from all 225 listed studies. (B) OncoPrint
profile of the genetic alterations of 12 overlapping genes in
samples of neuroendocrine prostate cancer (Beltran et al)
(24). The results revealed
alterations in 51 (48%) of 107 sequenced samples. CNA, copy-number
alteration; GA, glycyrrhizic acid. |
Furthermore, cBioPortal provided interaction
analyses for overlapping genes and built networks to display genes
that were altered in neuroendocrine prostate cancer (Fig. 4). The current study constructed a
network that comprised all neighbors of the overlapping genes from
the pathway enrichment analysis. The network was then further
modified by highlighting the neighbors of five key hub genes
obtained from the PPI analysis. Five hub genes were also identified
in crossed GA-associated secondary protein targets, including
NFKB1, BIRC3, BIRC2, RIPK1 and IKBKG. To reduce the network
complexity, only neighbors with higher alterations and queried
genes were presented by filtering the frequency of genetic
alterations. A gene network including the 12 overlapping genes and
avian myelocytomatosis viral oncogene homolog (MYC; Entrez ID,
4609) was identified by filtering neighbor alterations (≥48.8%).
Comparatively, with the filter condition of neighbor alterations
≥30%, a cluster of 29 genes were observed. The complexity of
interactions between the 12 GA-associated overlapping genes and
their most relevant altered gene pairs in samples from the studies
by Beltran et al (24) were
presented on refined and unrefined networks.
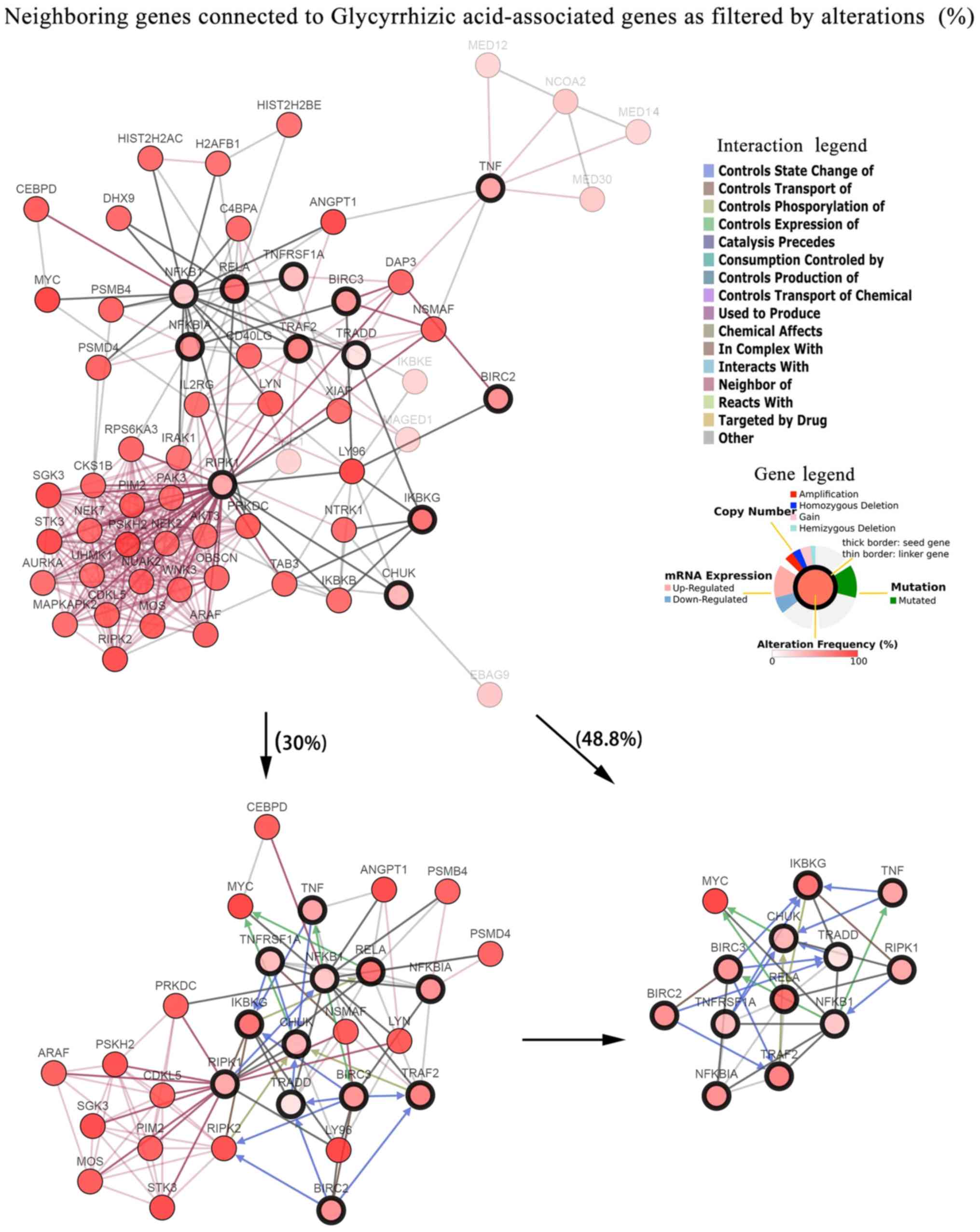 | Figure 4.Visualized gene networks of
GA-associated genes in neuroendocrine prostate cancer. Mined genes
(TRAF2, TNF, NFKBIA, NFKB1, TNFRSF1A, CHUK, RELA, BIRC3, BIRC2,
TRADD, RIPK1 and IKBKG; marked with a thick black border) were
applied as seeds to capture their interactions, which were
sequenced as altered genes in the current study. Neighbors are
presented by adjusting filters to 30% and maximal genetic
alterations. |
Discussion
Licorice root is a traditional medicine that has
been utilized to treat viral infections, peptic ulcers, lipidemia
and hyperglycemia (2,25). A considerable number of components
have been extracted from licorice, including flavonoids and
triterpenes, but GA is currently considered to be the main
bioactive herbal compound (25). A
wide range of pharmacological properties and multiple targets have
been identified for GA via independent clinical and experimental
studies. However, it is not fully known how GA serves a role in the
regulation of certain biological processes. Therefore, a systematic
method to bridge GA to its targets is required to connect these
observed biological activities. The current study performed a
PG-based analysis to elucidate the network interactions of GA and
to assess its potential clinical application. PG-based analysis is
comprised of four steps: i) GA is used as a seed to search for its
primary direct targets by querying multiple drug bioinformatics and
cheminformatics databases; ii) an assessment of the secondary
indirect targets connected to GA-associated primary targets is
performed by integrating data into a PPI network analysis tool;
iii) pristine interaction networks associated with GA are filtered
via GO and pathway enrichment analyses; and iv) the existence of
genetic changes are validated for the refined target gene from
large-scale samples obtained via cBioPortal cancer genomic studies.
Different from experimental techniques that identify GA-associated
targets, PG-based analysis is performed only via a rationalized
hypothesis that integrates multiple online sources. PG-based
analysis can generate effective results that may lead to the
optimization of experiments to assess the complex mechanisms used
by drugs.
The current study identified 5 GA-associated primary
protein targets (HSD11B1, TNF, CASP3, NFKB2 and LPL), 138
functional partners of the primary targets, 30 enriched GO terms
and 10 enriched pathways connected to GA-associated target genes. A
target network center surrounding GA was then constructed based on
the aforementioned results. Two obvious modules were determined in
this network. Module one contained three primary targets (TNF,
CASP3 and NFKB2) and 10 crossed secondary protein targets (BIRC3,
BIRC2, FADD, PSMD2, IKBKG, UBC, RIPK1, NFKB1, PSMC3 and IL1B) that
were closely associated with TNF axis-mediated regulation of
apoptosis, immunity and inflammation. TNF-α may therefore activate
pro-apoptotic and anti-apoptotic pathways. TNF-α can induce
apoptosis by activating caspase-family proteins (caspase-3,
caspase-8 and caspase-10). However, it may also inhibit apoptosis
via NF-κB, which induces the expression of anti-apoptotic genes
including Bcl-2 (22,26). In addition, cytokines of the TNF
family may induce gene transcription to regulate inflammation, cell
survival, cell differentiation and cell proliferation, primarily
via the activation of the NF-κB pathway (27). Module two contained two primary
targets (HSD11B1 and LPL) and two crossed secondary protein targets
(EHHADH and NR3C1) that were closely associated with the regulation
of lipometabolism (28).
Functional characteristic analysis of 129 GA-associated target
genes was performed via KEGG. It was revealed that 34 genes were
classified as validated targets of GA and were significantly
enriched in the TNF signaling pathway. BP analysis of GA-associated
target genes revealed that 29.5% of genes were enriched in
‘TNF-mediated signaling pathway’, which was the most statistically
significant response. Module 1, associated with the TNF axis, was
then considered to be the core analysis subset, and it was
speculated that bioactivities may be initiated via the interaction
between GA and its targets at the origin of various molecular
events.
The TNF pathway is strongly associated with the
physiological or pathological regulation of apoptosis, cell
survival, cell proliferation and inflammation (22). In recent years, the GA-mediated
regulation of inflammation has been studied, particularly in
hepatoprotective processes. However the GA-mediated regulation of
apoptosis, cell proliferation and cell survival in certain
diseases, such as cancer, remains unclear. Connectivity between the
TNF axis and GA-mediated biological functions in cancer were
assessed by determining the genetic changes of 12 overlapping genes
(TRAF2, TNF, NFKBIA, NFKB1, TNFRSF1A, CHUK, RELA, BIRC3, BIRC2,
TRADD, RIPK1 and IKBKG) revealed by TNF and TNF-associated
signaling pathways (TNF/NF-κB signaling pathway and TNF/caspase
family apoptosis signaling pathway). In the current study, the
connectivity between GA-associated targets and cancer was revealed
via queries made using cBioPortal. In the case of neuroendocrine
prostate cancer, frequency changes of the gene set ranged between 5
and 26%, and the abrupt genetic alterations centered on expression
amplification. RELA (with a 26% change) is a proto-oncogene that
usually forms a complex with NFKB, which subsequently moves to the
nucleus and activates the transcription of specific genes (29). IKBKG (26%), an NF-κB essential
modulator, encodes the subunit of the inhibitor of NF-κB kinase
complex, which activates NF-κB and induces the activation of genes
relating to survival, inflammation and other pathways. Similarly to
RELA, hypomorphic mutations of IKBKG may result in immunodeficiency
(30). TRAF2 (22%), a mediator of
TNF receptor anti-apoptotic signaling, is indispensable for the
TNF-α-mediated activation of MAPK8/JNK and NF-κB (31). TRADD, a TNF receptor-associated
apoptotic signal transducer, interacts with TRAF2, resulting in the
direct inhibition of caspase activation by recruiting
inhibitor-of-apoptosis proteins (IAPs) (32). In addition, other IAPs inhibit
apoptosis. For example, BIRC2 (19%) and BIRC3 (18%) usually form a
complex by binding to TRAF1/2 (33,34).
Previous studies have indicated that the anti-cancer activities of
GA are closely associated with anti-proliferative or
anti-angiogenic signals linked to the TNF axis (5,9,35,36).
Therefore, the results of the PG-based analysis indicated that
GA-associated core targets may therapeutically benefit
neuroendocrine prostate cancer.
It has been established that TNF binds to TNFR1/2 to
regulate distinct downstream pathways under certain conditions. The
binding of TNF to TNFR1 activates MAPK/NF-κB/caspase signaling
pathways to mediate the regulation of inflammatory cytokines,
vascular effects or apoptosis (37). In addition, binding may activate
the PI3K-AKT signaling pathway to mediate the regulation of cell
survival (37). Three primary
targets (TNF, CASP3 and NFKB2) of the TNF axis exhibit GA-mediated
inhibitory activity. TNFR1 of the TNF/NF-κB signaling pathway and
TNFR2 of the PI3K-AKT signaling pathway are considered to be the
most relevant targets. However, the PI3K-AKT signaling pathway was
not included in the top 10 pathways of KEGG. The GA-mediated
downregulation of this pathway has been previously demonstrated as
an important mechanism involved in the anti-cancer effects of GA in
the growth and migration of leukemia cells (5). Therefore, the current study may only
provide a reference for future biological studies. Despite this,
significant genetic alterations of TRAF2 (22%), BIRC2 19%) and
BIRC3 (18%) were identified in the current study and may therefore
serve primary regulatory roles.
The PG-based analysis of the present study was
further utilized to mine potential interactions connected to
GA-associated overlapping targets in neuroendocrine prostate cancer
by constructing a visual network using cBioPortal. It is possible
to identify key targets for the treatment of neuroendocrine
prostate cancer. Although neuroendocrine prostate cancer is
extremely rare, with a prevalence of <1%, it is a lethal and
aggressive subtype of prostate cancer, which includes the
resistance mechanism of non-androgen receptors (38). The loss or inactivation of RB
transcriptional corepressor 1 and p53, or the overexpression of the
MYC family (particularly N-My and C-Myc) serve important roles in
the development of aggressive prostate cancer and metastasis
(38). The current study revealed
that the MYC target gene (44.8%) exhibited the highest alteration
by filtering the interactive network connected to overlapping
targets associated with GA. Furthermore, RIPK1 (14%) was deemed to
serve an important role as it connected 12 of the 29 genes with
>30% genetic alterations in neuroendocrine prostate cancer. The
majority of targets that interacted with RIPK1 were considered to
be proto-oncogenes, including LYN proto-oncogene, Src family
tyrosine kinase (33.6%), A-Raf proto-oncogene, serine/threonine
kinase (30.8%), Pim-2 proto-oncogene, serine/threonine kinase
(31.8%) and MOS proto-oncogene, serine/threonine kinase (33.6%). In
response to tissue damage and pathogen recognition, the encoded
protein of RIPK1 serves an important role in the regulation of
inflammation and cell death (39).
This may provide an increased understanding of underlying malignant
disease mechanisms and may lead to the development of GA-associated
chemoprevention in neuroendocrine prostate cancer.
However, certain previously demonstrated effects of
GA were not identified among candidate genes in the present study.
For example, high mobility group box 1 (HMGB1) is a nuclear
DNA-binding protein that regulates transcription in several
cellular processes including inflammation, cell differentiation and
tumor cell migration (40). In
particular, the impact of GA on HMGB1 is of great biological
relevance to the pharmacological effects of GA in
immunoinflammatory diseases. GA has been demonstrated to inhibit
HMGB1 expression, resulting in the upregulation of heme oxygenase 1
expression (41). By inhibiting
the interaction of HMGB1 with its receptor toll like receptor 2, GA
may have the potential to inhibit the activation of the innate
immune system, including the production of IL-1 and TNF-α, which
are implicated in the pathogenesis of immunoinflammatory and
autoimmune diseases such as multiple sclerosis and rheumatoid
arthritis (42). Although
biological evidence indicates that the GA-mediated inhibition of
HMGB1 is a principal pharmacological effect, the PG analysis of the
current study did not determine HMGB1 as target of GA. The reason
why certain demonstrated targets in vitro, such as HMGB1 and
the inflammasome, were not shown in the present analysis may that
the interaction is indirect, and thus these are not primary
targets. Alternatively, it may be that the databases were not
up-to-date with respect to predicting the secondary interactions.
For this reason, it is hypothesized that other genes not included
in the analysis of the current study remain worthy of assessment
in vitro to determine all the pharmacological effects of GA;
in particular, the effects of GA on the pleiotropic cytokine
macrophage migration inhibitory factor, which is implicated in the
pathogenesis of autoimmune diseases and cancer, and is an activator
of the inflammasome (43). There
remain many obstacles to validating associations based on PG
analysis and their translation into clinical practice. Although
~15% of approved drugs include PG information on the label, only a
fraction of these are deemed actionable (44).
Based on the PG analysis strategy implemented in the
current study, a network of 138 functional interactions was
constructed to identify GA-associated primary and secondary
targets. The main biological effects mediated by the drug-target
network of GA were enriched via the regulation of the TNF axis, as
determined using integrated analyses. The impacts of TNF axis core
elements were further evaluated in a specific type of cancer via
the cBioPortal altered genes network. A simple and flexible
interface was also constructed to test hypotheses regarding
significant genetic alterations and subsequent phenotype
expression, and to develop the potential use of GA in
neuroendocrine prostate cancer. Furthermore, the analytical method
used in the current study may help elucidate the side effects of
GA. However, further experimentation is required to determine which
effects are dominant in the treatment of neuroendocrine prostate
cancer, therapeutic or side effects.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Miss Caina Gao
(Affiliated Children's Hospital of Zhejiang University), Miss Jie
Bao and Dr Hang Mu (both (Yangtze Delta Region Institute of
Tsinghua University) for assisting in the preparation of the
manuscript.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SW, LL and LS conceived and designed the current
study. SW and LL performed data analysis. SW wrote the manuscript.
SW and LL revised the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Saif MW, Li J, Lamb L, Kaley K, Elligers
K, Jiang Z, Bussom S, Liu SH and Cheng YC: First-in-human phase II
trial of the botanical formulation PHY906 with capecitabine as
second-line therapy in patients with advanced pancreatic cancer.
Cancer Chemother Pharmacol. 73:373–380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ming LJ and Yin AC: Therapeutic effects of
glycyrrhizic acid. Nat Prod Commun. 8:415–418. 2013.PubMed/NCBI
|
|
3
|
Farrukh MR, Nissar UA, Kaiser PJ, Afnan Q,
Sharma PR, Bhushan S and Tasduq SA: Glycyrrhizic acid (GA) inhibits
reactive oxygen Species mediated photodamage by blocking ER stress
and MAPK pathway in UV-B irradiated human skin fibroblasts. J
Photochem Photobiol B. 148:351–357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fouladi S, Masjedi M, Ghasemi R, G Hakemi
M and Eskandari N: The in vitro impact of glycyrrhizic acid on
CD4+ T lymphocytes through OX40 receptor in the patients
with allergic rhinitis. Inflammation. 41:1690–1701. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He SQ, Gao M, Fu YF and Zhang YN:
Glycyrrhizic acid inhibits leukemia cell growth and migration via
blocking AKT/mTOR/STAT3 signaling. Int J Clin Exp Pathol.
8:5175–5181. 2015.PubMed/NCBI
|
|
6
|
Hou S, Zhang T, Li Y, Guo F and Jin X:
Glycyrrhizic acid prevents diabetic nephropathy by activating
AMPK/SIRT1/PGC-1α signaling in db/db mice. J Diabetes Res.
2017:28659122017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kiratipaiboon C, Tengamnuay P and
Chanvorachote P: Glycyrrhizic acid attenuates stem cell-like
phenotypes of human dermal papilla cells. Phytomedicine.
22:1269–1278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li C, Peng S, Liu X, Han C, Wang X, Jin T,
Liu S, Wang W, Xie X, He X, et al: Glycyrrhizin, a direct HMGB1
antagonist, ameliorates inflammatory infiltration in a model of
autoimmune thyroiditis via inhibition of TLR2-HMGB1 signaling.
Thyroid. 27:722–731. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin SC, Chu PY, Liao WT, Wu MY, Tsui KH,
Lin LT, Huang CH, Chen LL and Li CJ: Glycyrrhizic acid induces
human MDA-MB-231 breast cancer cell death and autophagy via the
ROS-mitochondrial pathway. Oncol Rep. 39:703–710. 2018.PubMed/NCBI
|
|
10
|
Refahi S, Pourissa M, Zirak MR and Hadadi
G: Modulation expression of tumor necrosis factor α in the
radiation-induced lung injury by glycyrrhizic acid. J Med Phys.
40:95–101. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang LY, Cheng KC, Li Y, Niu CS, Cheng JT
and Niu HS: Glycyrrhizic acid increases glucagon like peptide-1
secretion via TGR5 activation in type 1-like diabetic rats. Biomed
Pharmacother. 95:599–604. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang YM and Du GQ: Glycyrrhizic acid
prevents enteritis through reduction of NF-κB p65 and p38MAPK
expression in rat. Mol Med Rep. 13:3639–3646. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang J, Shi Y, Chen H, Wang X, Chen Y and
Yang B: Glycyrrhizic acid attenuates myocardial injury: Involvement
of RIP140/NF-κB Pathway. Biomed Pharmacother. 95:62–67. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang H, Zhang R, Chen J, Shi M, Li W and
Zhang X: High mobility group Box1 inhibitor glycyrrhizic acid
attenuates kidney injury in streptozotocin-induced diabetic rats.
Kidney Blood Press Res. 42:894–904. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Relling MV and Evans WE: Pharmacogenomics
in the clinic. Nature. 526:343–350. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mooney SD: Progress towards the
integration of pharmacogenomics in practice. Hum Genet.
134:459–465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Campedel L, Kossaï M, Blanc-Durand P,
Rouprêt M, Seisen T, Compérat E, Spano JP and Malouf G:
Neuroendocrine prostate cancer: Natural history, molecular
features, therapeutic management and future directions. Bull
Cancer. 104:789–799. 2017.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Su G, Morris JH, Demchak B and Bader GD:
Biological network exploration with Cytoscape 3. Curr Protoc
Bioinformatics. 47:8.13.1–24. 2014. View Article : Google Scholar
|
|
19
|
Guo Y, Bao Y, Ma M and Yang W:
Identification of key candidate genes and pathways in colorectal
cancer by integrated bioinformatical analysis. Int J Mol Sci.
18(pii): E7222017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Canault B, Bourg S, Vayer P and Bonnet P:
Comprehensive network map of ADME-Tox databases. Mol Inform.
36:2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Coelho-Santos V, Leitão RA, Cardoso FL,
Palmela I, Rito M, Barbosa M, Brito MA, Fontes-Ribeiro CA and Silva
AP: The TNF-α/NF-κB signaling pathway has a key role in
methamphetamine-induced blood-brain barrier dysfunction. J Cereb
Blood Flow Metab. 35:1260–1271. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kang R, Xie Y, Zhang Q, Hou W, Jiang Q,
Zhu S, Liu J, Zeng D, Wang H, Bartlett DL, et al: Intracellular
HMGB1 as a novel tumor suppressor of pancreatic cancer. Cell Res.
27:916–932. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Beltran H, Prandi D, Mosquera JM, Benelli
M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV,
Varambally S, et al: Divergent clonal evolution of
castration-resistant neuroendocrine prostate cancer. Nat Med.
22:298–305. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Asl MN and Hosseinzadeh H: Review of
pharmacological effects of Glycyrrhiza sp. and its bioactive
compounds. Phytother Res. 22:709–724. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brumatti G, Salmanidis M and Ekert PG:
Crossing paths: Interactions between the cell death machinery and
growth factor survival signals. Cell Mol Life Sci. 67:1619–1630.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hayden MS and Ghosh S: Regulation of NF-κB
by TNF family cytokines. Semin Immunol. 26:253–266. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yaw HP, Ton SH, Chin HF, Karim MK,
Fernando HA and Kadir KA: Modulation of lipid metabolism in
glycyrrhizic acid-treated rats fed on a high-calorie diet and
exposed to short or long-term stress. Int J Physiol Pathophysiol
Pharmacol. 7:61–75. 2015.PubMed/NCBI
|
|
29
|
Chen S, Jiang S, Zheng W, Tu B, Liu S,
Ruan H and Fan C: RelA/p65 inhibition prevents tendon adhesion by
modulating inflammation, cell proliferation, and apoptosis. Cell
Death Dis. 8:e27102017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miot C, Imai K, Imai C, Mancini AJ, Kucuk
ZY, Kawai T, Nishikomori R, Ito E, Pellier I, Dupuis Girod S, et
al: Hematopoietic stem cell transplantation in 29 patients
hemizygous for hypomorphic IKBKG/NEMO mutations. Blood.
130:1456–1467. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wei B, Liang J, Hu J, Mi Y, Ruan J, Zhang
J, Wang Z, Hu Q, Jiang H and Ding Q: TRAF2 is a valuable prognostic
biomarker in patients with prostate cancer. Med Sci Monit.
23:4192–4204. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wei B, Ruan J, Mi Y, Hu J, Zhang J, Wang
Z, Hu Q, Jiang H and Ding Q: Knockdown of TNF receptor-associated
factor 2 (TRAF2) modulates in vitro growth of TRAIL-treated
prostate cancer cells. Biomed Pharmacother. 93:462–469. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jin H, Dong YY, Zhang H, Cui Y, Xie K and
Lou G: shRNA depletion of cIAP1 sensitizes human ovarian cancer
cells to anticancer agent-induced apoptosis. Oncol Res. 22:167–176.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seidelin JB: Regulation of antiapoptotic
and cytoprotective pathways in colonic epithelial cells in
ulcerative colitis. Scand J Gastroenterol. 50 (Suppl 1):S1–S29.
2015. View Article : Google Scholar
|
|
35
|
Hostetler BJ, Uchakina ON, Ban H and
McKallip RJ: Treatment of hematological malignancies with
glycyrrhizic acid. Anticancer Res. 37:997–1004. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim KJ, Choi JS, Kim KW and Jeong JW: The
anti-angiogenic activities of glycyrrhizic acid in tumor
progression. Phytother Res. 27:841–846. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Doss GP, Agoramoorthy G and Chakraborty C:
TNF/TNFR: Drug target for autoimmune diseases and immune-mediated
inflammatory diseases. Front Biosci (Landmark Ed). 19:1028–1040.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Berman-Booty LD and Knudsen KE: Models of
neuroendocrine prostate cancer. Endocr Relat Cancer. 22:R33–R49.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guo R, Tu Y, Xie S, Liu XS, Song Y, Wang
S, Chen X and Lu L: A role for receptor-interacting protein
kinase-1 in neutrophil extracellular trap formation in patients
with systemic lupus erythematosus: A preliminary study. Cell
Physiol Biochem. 45:2317–2328. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ando K, Sakoda M, Ueno S, Hiwatashi K,
Iino S, Minami K, Kawasaki Y, Hashiguchi M, Tanoue K, Mataki Y, et
al: Clinical implication of the relationship between high mobility
group Box-1 and tumor differentiation in hepatocellular carcinoma.
Anticancer Res. 38:3411–3418. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y, Li L, Qi C, Hua S, Fei X, Gong F
and Fang M: Glycyrrhizin alleviates Con A-induced hepatitis by
differentially regulating the production of IL-17 and IL-25. Biomed
Pharmacother. 110:692–699. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dujmovic I, Mangano K, Pekmezovic T,
Quattrocchi C, Mesaros S, Stojsavljevic N, Nicoletti F and Drulovic
J: The analysis of IL-1 beta and its naturally occurring inhibitors
in multiple sclerosis: The elevation of IL-1 receptor antagonist
and IL-1 receptor type II after steroid therapy. J Neuroimmunol.
207:101–106. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shin MS, Kang Y, Wahl ER, Park HJ, Lazova
R, Leng L, Mamula M, Krishnaswamy S, Bucala R and Kang I:
Macrophage migration inhibitory factor regulates U1 small nuclear
RNP immune complex-mediated activation of the NLRP3 inflammasome.
Arthritis Rheumatol. 71:109–120. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ehmann F, Caneva L, Prasad K, Paulmichl M,
Maliepaard M, Llerena A, Ingelman-Sundberg M and Papaluca-Amati M:
Pharmacogenomic information in drug labels: European medicines
agency perspective. Pharmacogenomics J. 15:201–210. 2015.
View Article : Google Scholar : PubMed/NCBI
|