Introduction
Esophageal cancer (EC) is one of the most common
types of cancer in the world, particularly in China (1). Two major subtypes of esophageal
cancer are esophageal squamous cell carcinoma (ESCC), and
adenocarcinoma. The International Agency for Research on Cancer
reported that ESCC was most common in South-Eastern and Central
Asia (79% of the total global EC cases) and China had about half of
all ESCC cases on worldwide in 2012 (2). Despite advances in diagnosis and
treatments, ESCC continues to have poor prognosis due to
progressive metastasis (3). The
molecular mechanisms of ESCC invasion leading to metastasis remain
poorly understood. Therefore, understanding the molecular
mechanisms of progressive metastasis will help to ascertain
promising therapeutic targets that may benefit the development of
alternative and novel approaches in ESCC therapy.
MicroRNAs (miRNAs/miRs) constitute a class of
endogenous small non-coding RNAs that regulate post-transcriptional
suppression or translational repression of their target mRNAs
(4,5). Accumulating evidence indicates the
essential roles of miRNAs in the initiation and progression of
several types of cancer in humans (6,7). A
number of miRNA expression profiling studies have demonstrated that
miRNA expression signatures are associated with ESCC development
and progression (8,9). miRNA profiling of primary esophageal
cancer tissue and paired paracancerous tissue of patients with ESCC
demonstrated an association between four miRNAs and patient
outcomes (10). One of these
miRNAs was miR-212. High miR-212 expression was associated with
short-term survival and poor clinical outcome in patients with ESCC
(11), indicating that this miRNA
may contribute to the development and metastasis of ESCC. However,
the biological functions of miR-212 in ESCC progression remain
unknown.
In the present study, the aim was to explore the
roles of miR-212 in ESCC cell proliferation, migration and
invasion, and the potential underlying mechanisms. Using the in
vitro cell model of lentivirus-mediated gain-of-function, it
was demonstrated that miR-212 had no significant effect on ESCC
cell proliferation; rather, miR-212 facilitated migration and
invasion of ESCC cells by promoting the epithelial-mesenchymal
transition (EMT) process and increasing the protein levels of the
extracellular matrix (ECM)-degrading enzymes matrix
metalloproteinase (MMP)2, MMP9 and urokinase-type plasminogen
activator (uPa).
The miR-212 level was previously associated with
poor outcome in patients with ESCC (11), thus the potential of therapeutic
drugs that inhibit the miR-212-induced ESCC cell migration was
investigated. A number of drugs, including LY294002 (PI3K
inhibitor) (12), rapamycin (mTOR
inhibitor) (13),
5-(Tetradecyloxy)-2-furoic acid (TOFA; an acetyl-CoA carboxylase 1
inhibitor) (14), metformin
(15), propranolol (16) and berberine (17), were chosen based on: i) The
classical signaling molecules that are associated with cancer
development, such as the PI3K-Akt/mTOR-p70S6K and AMP-activated
protein kinase (AMPK) pathways (12–14);
ii) drugs, such as metformin and propranolol, that have
demonstrated anti-tumor functions in several types of cancer; and
iii) drugs, such as berberine, that demonstrated an inhibitory
function against ESCC cell proliferation and migration (17). It was found that only berberine, an
alkaloid extracted from Ranunculaceae coptis rhizome, and
none of the other tested drugs, displayed an inhibitory function
against miR-212 overexpression-induced ESCC cell migration.
Materials and methods
Reagents and antibodies
All-in-One™ miRNA first-strand cDNA synthesis kit,
All-in-One™ miRNA qPCR detection kit and all quantitative (q)PCR
primers were purchased from GeneCopoeia, Inc. Primerscript™ RT
reagent kit with gDNA eraser and SYBR premix Ex Taq II were
obtained from Takara Biotechnology Co., Ltd.
3-(4,5-Dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide (MTT)
was purchased from Beyotime Institute of Biotechnology. The cell
culture media, including RPMI-1640 medium and DMEM, were obtained
from Corning Inc. Fetal bovine serum (FBS) was purchased from Clark
Bioscience. All primary antibodies, including those against
E-cadherin (3195), β-catenin (8480), vimentin (5741), Twist1
(46702) and GAPDH (5174) were purchased from Cell Signaling
Technology, Inc. The secondary antibody used was horseradish
peroxidase-labeled goat anti-rabbit IgG (H+L; A0208) obtained from
Beyotime Institute of Biotechnology. Berberine hydrochloride was
purchased from Shanghai YuanYe Biotechnology Co., Ltd. Metformin
and rapamycin were purchased from Selleck Chemicals. Propranolol
was obtained from Sigma-Aldrich (Merck KGaA), LY294002 was ordered
from Beyotime Institute of Biotechnology, and TOFA was purchased
from Santa Cruz Biotechnology, Inc.
Cell culture and lentivirus
infection
Three human ESCC cell lines KYSE-450, TE-1 and
Eca109 were obtained from Cobioer Biosciences, Co., Ltd. KYSE-450
and TE-1 cells were cultured in RPMI-1640 medium, containing 10%
FBS, and Eca109 was cultured in DMEM supplemented with 10% FBS at
37°C in 5% CO2. All culture media contained 100 µg/ml
penicillin and 100 µg/ml streptomycin (Beijing Solarbio Science
& Technology Co., Ltd.).
Viral particles of lentiviral vectors LV10-(U6/RFP
& Puro) expressing a scrambled control (Con,
5′-TTCTCCGAACGTGTCACGT-3′) and mature hsa-miR-212 (miR-212,
5′-ACCUUGGCUCUAGACUGCUUACU-3′) were purchased from Shanghai
GenePharma Co., Ltd. The viral titers of miR-212- and the
Con-expressing vectors were at a concentration of 1×108
transduction (T)U/ml. Cells (2×105) were seeded into a
3.5-cm dish for viral infection. The cells were infected with 0,
10, 20 and 40 µl lentivirus to determine the multiplicity of
infection (MOI). The MOI was 0, 5, 10 and 20 when the cells were
infected with 0, 10, 20 and 40 µl lentivirus, respectively. Based
on the cell proliferation and migration for each MOI, 20 µl
lentivirus or MOI 10 was chosen throughout the present study. Cells
were cultured in 6-well plate in medium with 5% FBS for
approximately 24 h, and subsequently infected with 20 µl Con and
miR-212 vector-expressing viral particles, in the presence of 5
µg/ml polybrene. At 6 h after infection, the supernatant was
removed. After 24 h transfection, puromycin (5 µg/ml) was added to
the culture medium every 3 days for a selection of successful
transfected cells to become a population of stable cells expressing
miR-212 or scrambled control. Unless otherwise specified, all the
cells used in the subsequent experiments were those stably
expressing miR-212 after lentivirus transfection.
Cell proliferation assay
An MTT assay was performed to measure
miR-212-overexpressed cell proliferation. Briefly, KYSE-450
(4×103 cells/well), TE-1 (5×103 cells/well)
and Eca109 (4×103 cells/well) cells were plated into
96-well culture plates and incubated overnight at 37°C in 5%
CO2. MTT (10 µl) was added to each well at a final
concentration of 5 mg/ml at 0, 24, 48, and 72 h. After 4 h
incubation at 37°C, the blue MTT formazan crystals were dissolved
in 100 µl/well DMSO. The absorbance at 490 nm was measured via the
Multiskan spectrum microplate reader (Thermo Fisher Scientific,
Inc.). All experiments were repeated three times.
Cell migration and invasion
assays
Cell migration and invasion were assayed using a
6.5-mm diameter Transwell chamber with 8-µm micropores (Corning
Inc.). For invasion assays, the upper chamber was pre-coated with
50 µl Matrigel (1 mg/ml) at 37°C for 8 h. Then, the Matrigel was
removed from the chamber. For migration and invasion assays, 200 µl
of the cell suspension (1×105 cells/ml in serum-free
medium) was placed in the upper chamber without serum, and 600 µl
of 10% FBS-containing medium was added to the bottom chamber. For
the drug treatment experiments, the drug at final concentration
indicated below was added to the cell suspension. After incubation
for 36 h, cells were fixed with 4% paraformaldehyde for 15 min at
room temperature and then stained with 0.1% crystal violet for
another 15 min. Non-migrated or non-invaded cells on the upper
surface of the chambers were cleaned with a cotton swab, and
migrated cells or invaded cells in five different fields per
chamber were counted using a phase contrast microscope under ×100
magnification (Nikon TS100; Nikon Corporation).
Western blot analysis
Cells were washed with cold PBS and lysed with RIPA
lysis buffer (Wuhan Boster Biological Technology, Ltd.). The
protein concentration was determined by the BCA method. Protein (30
µg) was subjected to SDS-PAGE on a 10% gel and transferred onto a
nitrocellulose membrane (GE Healthcare). Following blocking with 5%
non-fat milk for 1 h at room temperature, the membrane was
incubated at 4°C overnight with primary antibodies against each of
the following proteins: E-cadherin, β-catenin, vimentin, Twist1 and
GAPDH (all 1:1,000 dilution). Following washing, the membrane was
incubated with corresponding horseradish peroxidase-conjugated
secondary antibody (1:1,000 dilution) for 1 h at room temperature.
The membrane was washed and immersed in SuperSignal West Pico
Chemiluminescent Substrate detection kit (Thermo Fisher Scientific,
Inc.). Bound secondary antibody was detected by the Amersham™
Imager 600 system (GE Healthcare Bio-Sciences). Image J software
v1.8.0.112 (National Institutes of Health) was used to quantify all
protein band densities. The level of the tested proteins is
represented by determining the ratio of the integrated density of
tested protein relative to that of the GAPDH loading control.
RNA extraction and reverse
transcription (RT)-qPCR
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). To detect miR-212
expression, cDNA was synthesized from 1 µg total RNA by using
All-in-One™ miRNA first-strand cDNA synthesis kit (GeneCopoeia,
Inc.) according to the manufacturer's instructions. All-in-One™
miRNA qPCR kit (GeneCopoeia, Inc.) was used to amplify miR-212. The
qPCR reaction was carried out for 40 cycles with the following
temperature profile: Initial denaturation at 95°C for 10 min,
annealing at 62.5°C for 20 sec and extension at 72°C for 10 sec. U6
was used as the internal control housekeeping gene. The primers of
has-miR-212 and U6 were purchased from GeneCopoeia, Inc.
To detect the mRNA levels of MMP2, MMP9 and uPA,
cDNA was synthesized from 1 µg total RNA using PrimerScript™ RT
reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd.)
following the manufacturer's instructions. The resulting cDNA was
analyzed by RT-qPCR using the SYBR Premix Ex Taq II (Takara
Biotechnology Co., Ltd.). The qPCR reaction was run by 40 cycles of
a two-step thermocycling program: Denaturation at 95°C for 30 sec
and extension at 95°C for 5 sec. GAPDH was used as the internal
control. The primer sequences for qPCRs are listed in Table I.
 | Table I.Sequences of the primers used in the
reverse transcription-quantitative PCR experiments. |
Table I.
Sequences of the primers used in the
reverse transcription-quantitative PCR experiments.
| Gene (Acc.
no.) | Primer
sequence |
|---|
| MMP2
(NM_004530.5) | F:
5′-TTGACGGTAAGGACGGACTC-3′ |
|
| R:
5′-TCTCAAAGTTGTAGGTGGTGGA-3′ |
| MMP9
(NM_004994.2) | F:
5′-GACAGCGACAAGAAGTGGGG-3′ |
|
| R:
5′-TCAGGGCGAGGACCATAGAG-3′ |
| uPA
(NM_002658.4) | F:
5′-CACACTGCTTCATTGATTACCCA-3′ |
|
| R:
5′-AAGGCAATGTCGTTGTGGTG-3′ |
| GAPDH
(NM_002046) | F:
5′-GCAGGGGGGAGCCAAAAGGGT-3′ |
|
| R:
5′-TGGGTGGCAGTGATGGCATGG-3′ |
All qPCR reactions were run on a QuantStudio™ Dx
Real-Time PCR Instrument (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The relative levels of miR-212 and indicated
genes were normalized to the corresponding internal control and
quantified based on the 2−ΔΔCq method (18).
Statistical analysis
All data are expressed as the mean ± standard
deviation and were analyzed using ANOVA. Levene's test was used to
assess the equality of variances of each group. Tukey's test was
used to analyze the equal variances while Games-Howell's test was
used to assess the unequal variances in multiple comparisons.
P<0.05 was considered to indicate a statistically significant
difference. All analyses were performed with SPSS version 22.0 (IBM
Corp).
Results
miR-212 has no effect on ESCC cell
proliferation
miR-212-overexpressing ESCC cell lines were
generated by lentiviral infection to explore the function and the
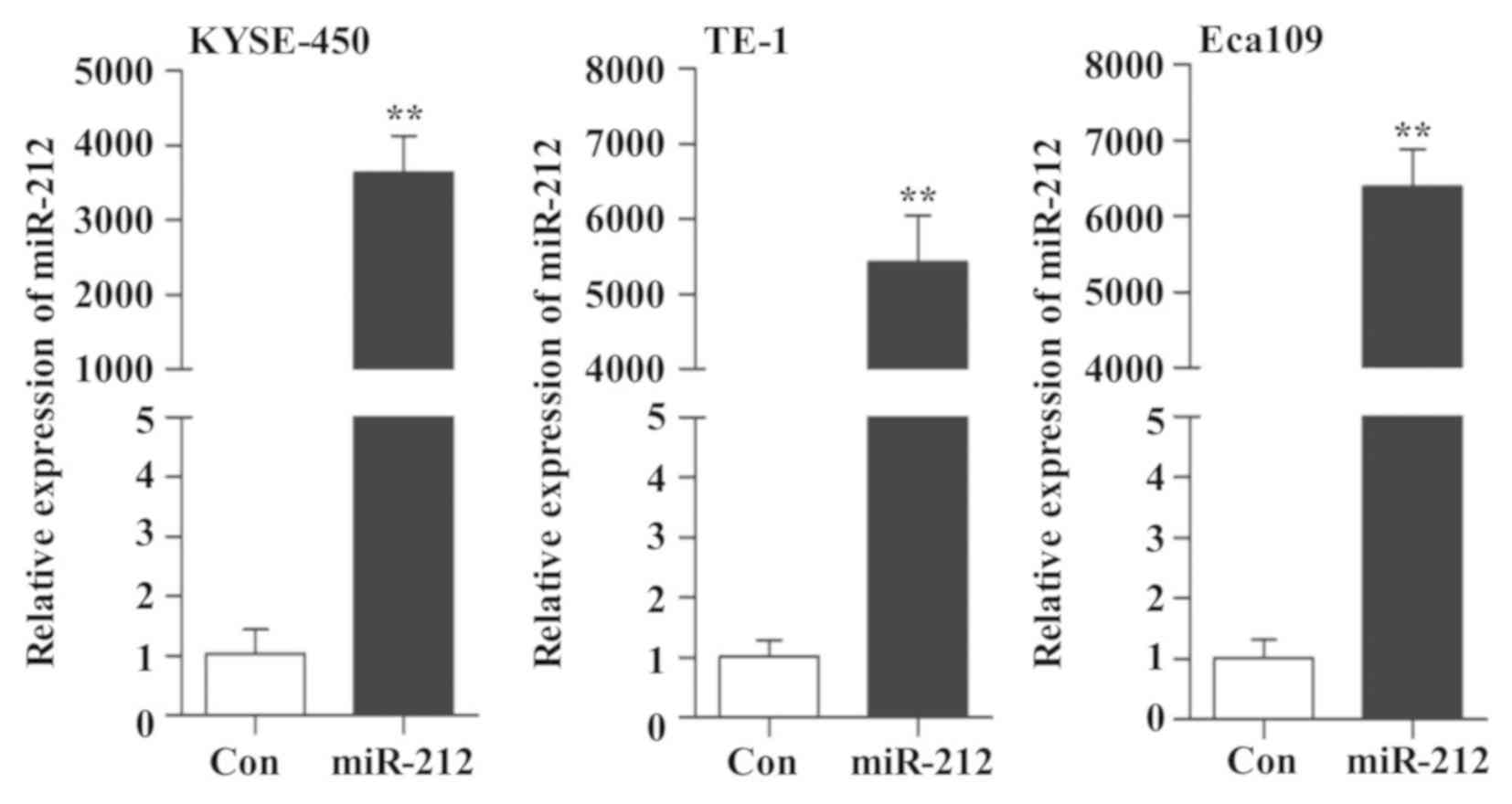
possible mechanism of miR-212 in ESCC cells. As indicated in
Fig. 1, overexpression of miR-212
was confirmed by RT-qPCR. Therefore, this in vitro cell
model was used to study the role of miR-212 in subsequent
experiments.
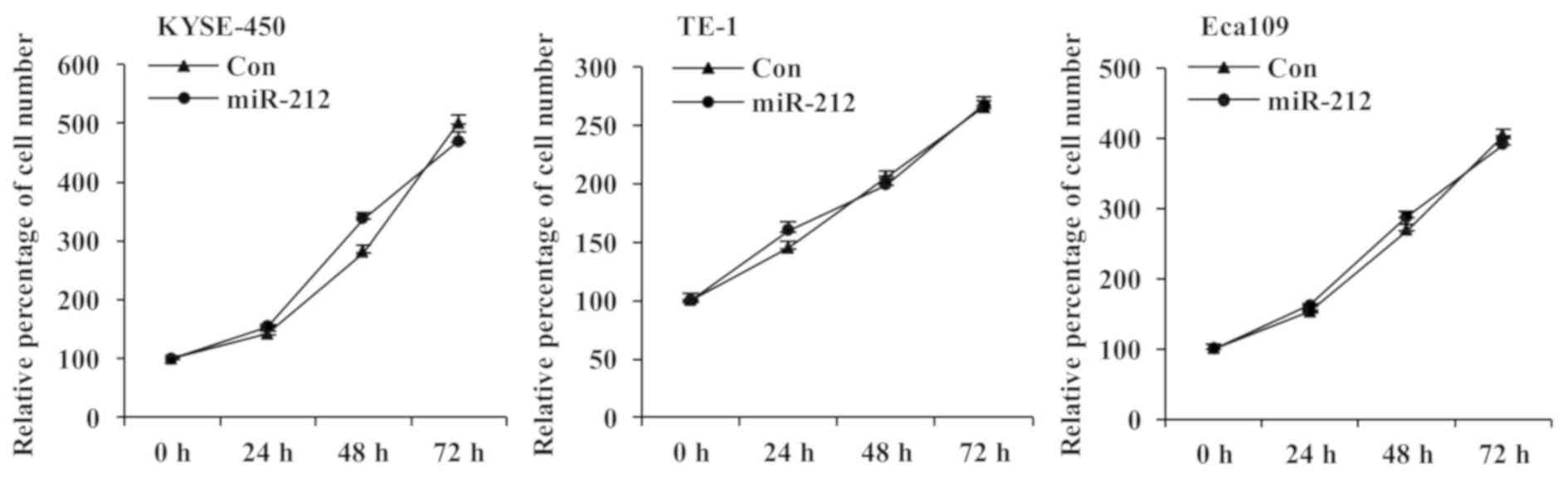
The effect of overexpressing miR-212 on the
proliferation of ESCC cells was investigated by MTT assay. As shown
in Fig. 2, the cell proliferation
of all three miR-212-overexpressing ESCC cell lines was similar to
that of the corresponding control cell lines at all tested time
points. These results indicate that miR-212 has no significant
effect on ESCC cell proliferation.
miR-212 promotes ESCC cell migration
and invasion
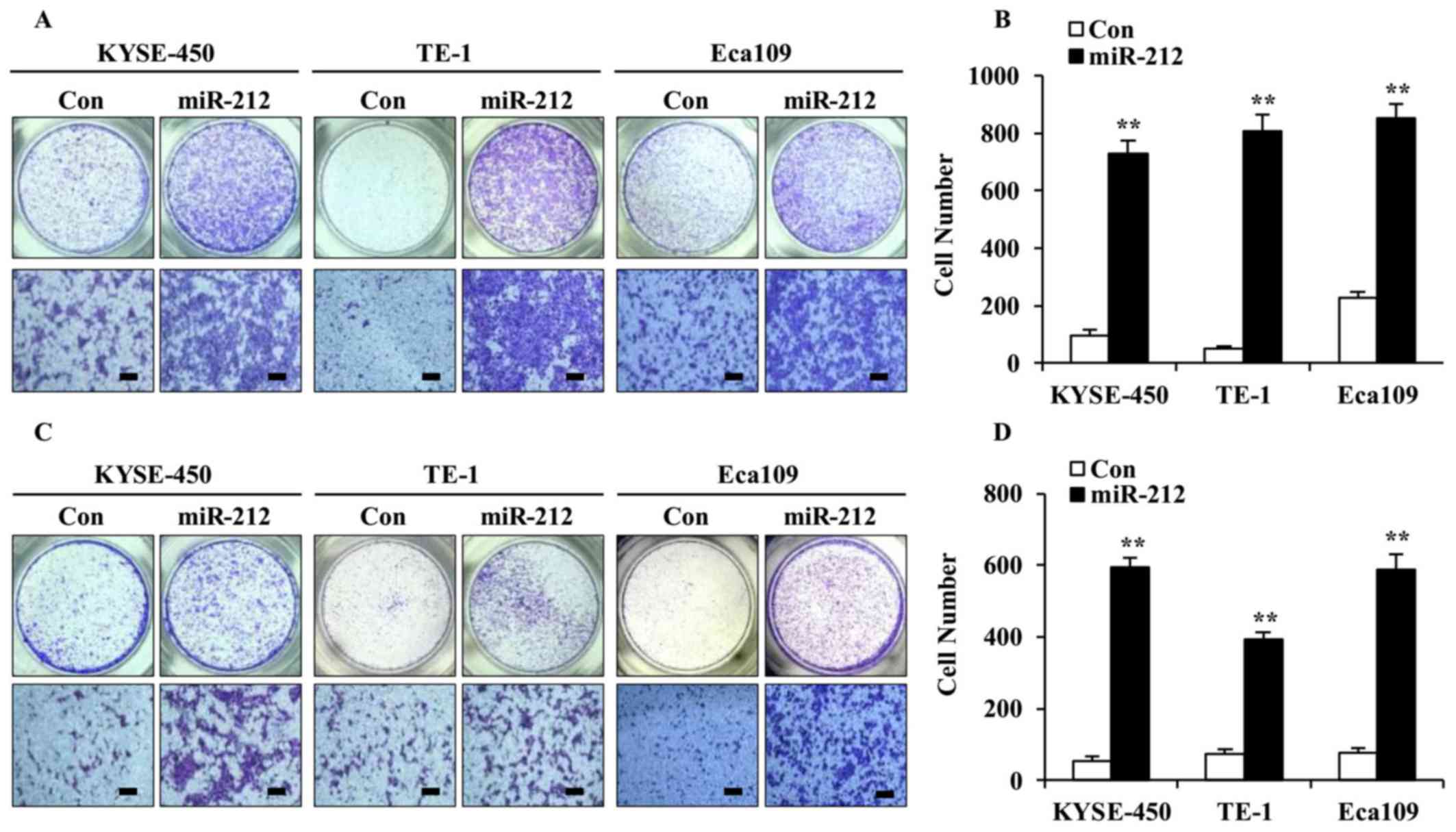
As high expression of miR-212 was associated with
poor outcome in patients with ESCC (11), the effect of miR-212 on ESCC cell
migration and invasion was examined using Transwell assays. As
shown in Fig. 3, overexpression of
miR-212 significantly increased cell migration (Fig. 3A and B) as well as invasion
(Fig. 3C and D), compared with the
corresponding controls. These results suggest that miR-212 plays a
facilitative role in ESCC cell migration and invasion.
miR-212 overexpression promotes EMT
and enhances the level of ECM-degrading enzymes in ESCC cells
Cancer cells often undergo EMT before gaining their
metastatic potential (19). To
elucidate further mechanistic insights into the role of miR-212 in
promoting ESCC cell migration and invasion, western blot analyses
were performed to examine the expression of EMT-associated markers.
As shown in Fig. 4A and C,
overexpression of miR-212 led to the decrease of E-cadherin
(epithelial marker) and the increase of β-catenin, vimentin, and
transcription factor Twist1 (mesenchymal markers). This indicates
that miR-212 may promote EMT in ESCC cells.
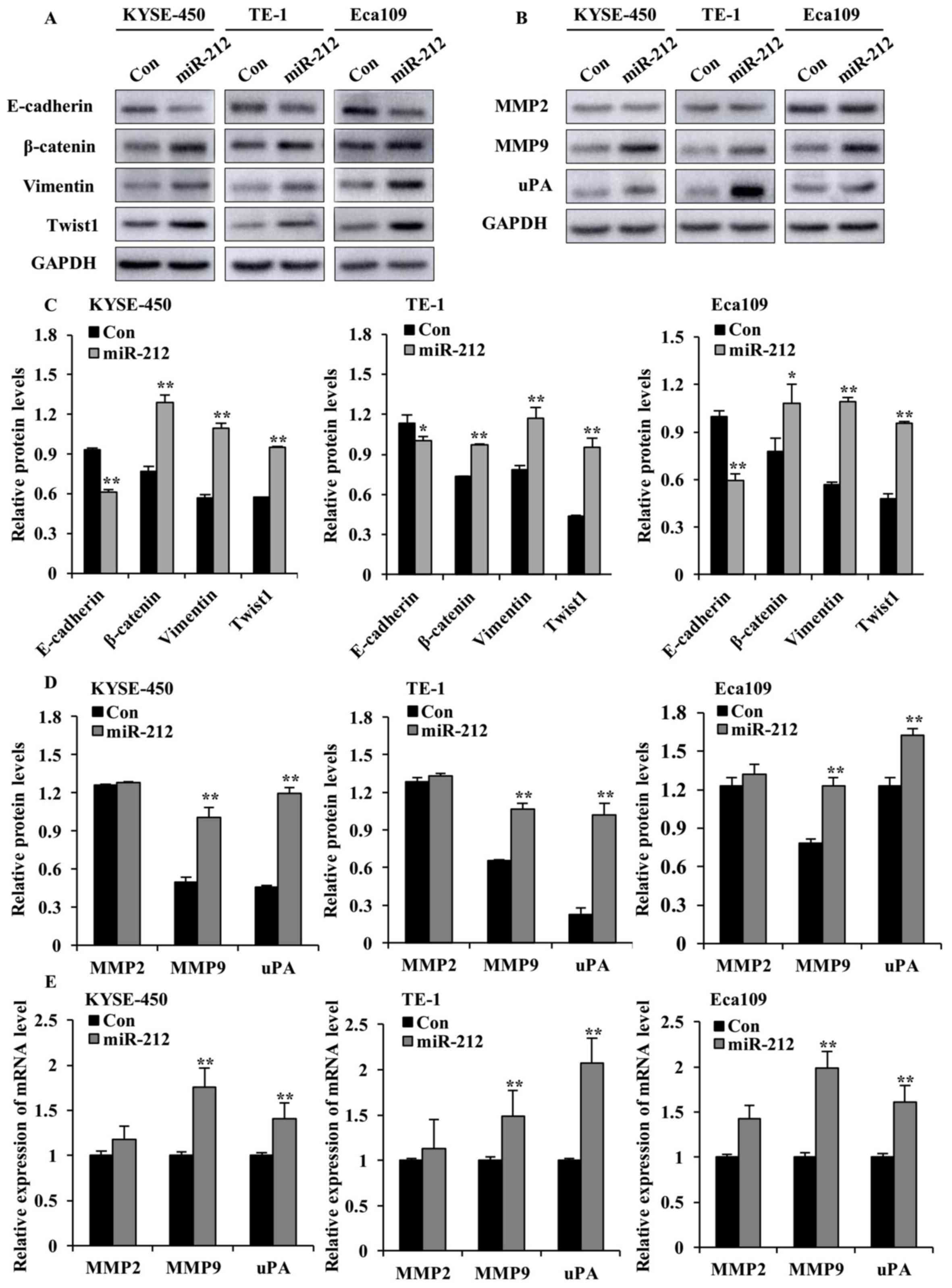 | Figure 4.miR-212 overexpression enhances the
levels of EMT markers, MMP2, MMP9 and uPA in ESCC cells. (A)
Western blot analysis of the expression of E-cadherin, β-catenin,
vimentin and Twist1, and (B) MMP2, MMP9 and uPA in
miR-212-overexpressed (miR-212) and Con-transfected ESCC cells. (C)
Quantification plots of the protein levels detected in (A). (D)
Quantification plots of the protein levels detected in (B). The
y-axis represents the ratio of the integrated density of tested
protein relative to GAPDH loading control. (E) Reverse
transcription-quantitative PCR results of mRNA levels of MMP2, MMP9
and uPA in miR-212-overexpressed and Con ESCC cells. The y-axis
represents the relative mRNA level normalized to the level in the
respective Con group. The relative mRNA level is the ratio of
indicated mRNA level to the GAPDH level in each group. Data are
expressed as mean ± SD. *P<0.05 and **P<0.01 vs. the
respective Con cells. ESCC, esophageal squamous cell carcinoma;
miR, microRNA; Con, scrambled control; MMP, matrix
metalloproteinase; uPA, urokinase-type plasminogen activator. |
Increased proteinase activity in the ECM surrounding
malignant tumor cells has been implicated in metastatic processes
(20). Hence, the protein
expression of uPA, MMP2 and MMP9, which are key ECM proteinases and
markers of cancer cell invasion (21), was examined. As shown in Fig. 4B, D and E, overexpression of
miR-212 significantly increased the protein and mRNA levels of MMP9
and uPA in the three ESCC cell lines, compared with the
corresponding controls. Interestingly, miR-212 did not cause any
significant alteration of MMP2 protein and mRNA levels.
Collectively, these findings indicate that miR-212-induced ESCC
cell migration and invasion may be mediated by promoting EMT and
increasing the level of ECM proteinases.
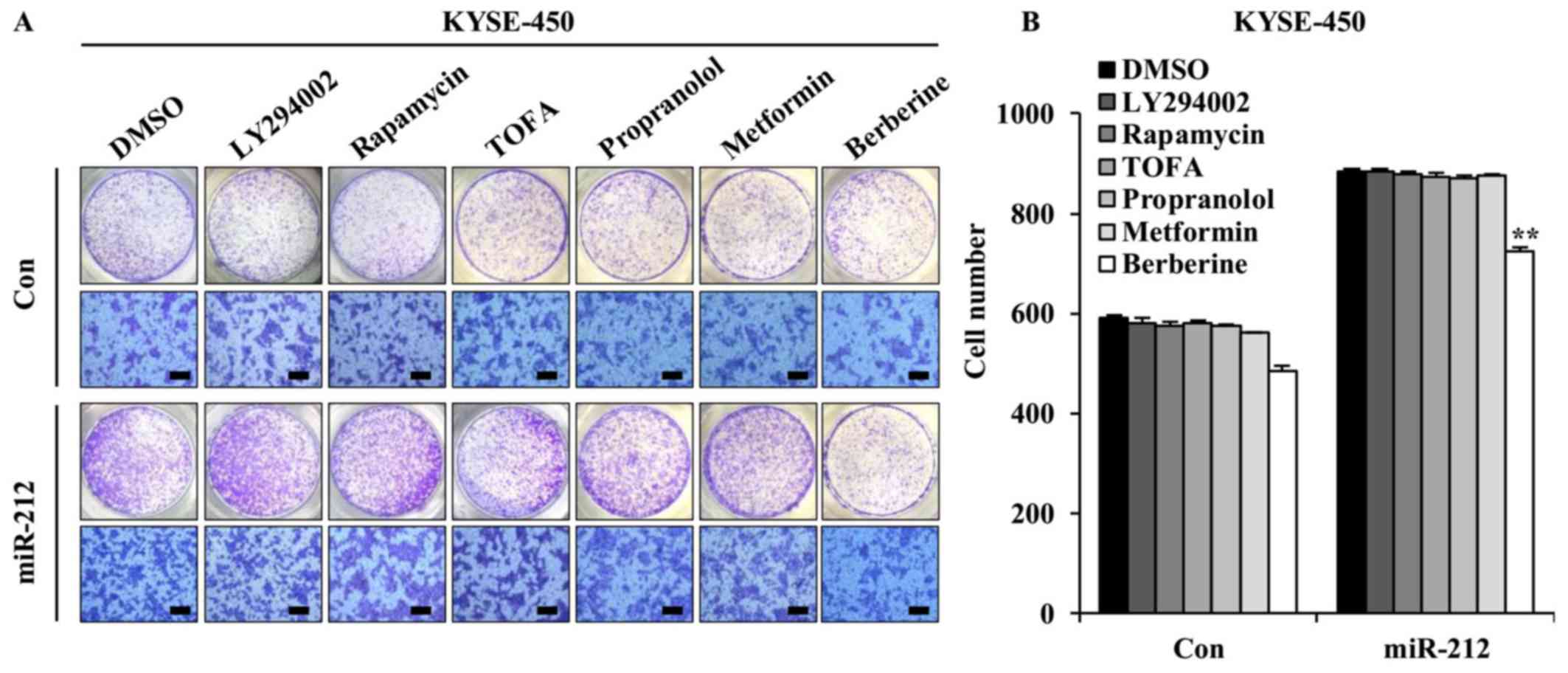
Berberine inhibits miR-212-induced
migration of ESCC cells
As miR-212 was demonstrated to promote ESCC cell
migration and invasion, the potential of therapeutic drugs against
metastasis of ESCC with high miR-212 expression was explored. The
effect of LY294002 (PI3K inhibitor), rapamycin (mTOR inhibitor),
TOFA (acetyl-CoA carboxylase 1 inhibitor inhibitor), metformin,
propranolol and berberine were tested. A Transwell migration assay
was performed to detect the effect of these drugs on
miR-212-induced ESCC cell migration. The concentrations of these
drugs here used were based on preliminary results obtained from
miR-212-overexpressed KYSE-450 cells and vector control KYSE-450
cells (data not shown). Compared with the vehicle-treated control
cells, with the exception of berberine, none of these drugs caused
changes in miR-212-induced KYSE-450 cell migration (Fig. 5), indicating that high motility of
ESCC cells initiated by miR-212 may be triggered by activation of
multiple signaling cascades.
It was previously shown that berberine, a
traditional Chinese medicine, displayed antitumor activity against
ESCC cells by regulating a number of signaling pathways (17). Accumulating evidence indicates that
berberine prevents cell motility by inhibition of EMT in a number
of cancer cell types including esophageal cancer cells (22,23).
Based on the results shown in Fig.
5, the inhibitory function of berberine on miR-212-induced cell
migration was further investigated by focusing on
miR-212-overexpressed KYSE-450 cells. The Transwell migration was
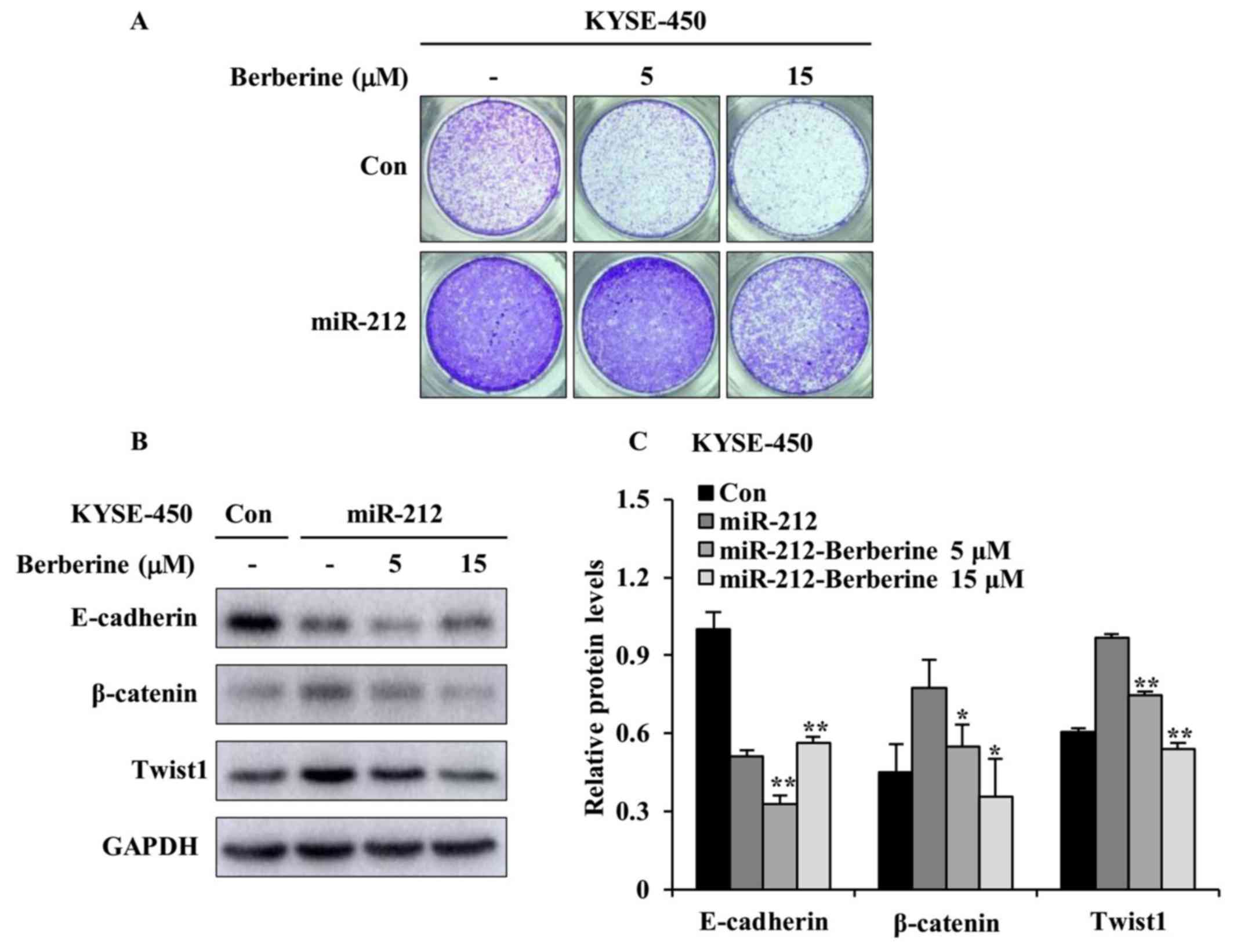
conducted with or without berberine treatment. As shown in Fig. 6A, the intact images of the migrated
cell on the lower surface of the chambers revealed that berberine
had an inhibitory effect on miR-212-induced cell migration when
compared with that of DMSO treatment in miR-212-overexpressed
KYSE-450 cells. Such inhibitory effect on cell migration was more
evident with berberine at 15 µM than 5 µM. In an effort to study
whether blocking EMT is a potential mechanism by which berberine
plays the inhibitory effect on miR-212-induced cell migration,
western blotting was conducted to examine the protein levels of
EMT-related signal markers. As shown in Fig. 6B and C, berberine treatment clearly
augmented the level of E-cadherin and attenuated the levels of
β-catenin and Twist1 when comparing with DMSO treatment in
miR-212-overexpressed KYSE-450 cells. Taken together, these results
imply that berberine prevents miR-212-induced ESCC cell migration
by inhibiting EMT process.
Discussion
Accumulating evidence has indicated that miRNAs
participate in the progression of several types of cancer by
targeting multiple genes involved in cancer progression and
metastasis, and the aberrant expression of miRNAs contributes to
the pathogenesis of malignancies in humans (24). Thus, exploration of cancer-specific
miRNAs could help identify novel biomarkers and therapeutic targets
in cancer.
Human miR-212 (hsa-miR-212) shares close sequence
homology with hsa-miR-132; both are encoded from the same intron of
a small non-coding gene located on chromosome 17 in humans
(25). The miR-132/miR-212 cluster
was originally identified in neuronal cells and displayed important
functions in neuronal development (26,27).
Although the roles of miR-132/212 have been investigated mainly in
neurons, increasing evidence point to their involvement in
cancer.
Despite previous observation of significant
association between miR-212 and short-term survival and poor
outcome in patients with ESCC due to early metastasis (12), the function and underlying
mechanism of miR-212 remain unknown. In the present study, it was
demonstrated that miR-212 had no effect on ESCC cell proliferation,
but significantly promoted cell migration and invasion, possibly
through facilitating EMT and increasing the level of ECM-degrading
enzymes. Moreover, berberine may be used as an alternative therapy
to effectively inhibit invasion and metastasis in patients with
ESCC, who express high levels of miR-212.
Tumor cell metastasis is a complex multi-step
process. An important event of metastasis is EMT, by which
epithelial cells acquire mesenchymal phenotype, thereby obtaining
the ability to migrate and invade. EMT is characterized by loss of
epithelial markers and gain of mesenchymal markers. Several groups
of transcription factors, including Snail and Twist families,
repress the transcription of epithelial markers, such as
E-cadherin, and promote the transcription of mesenchymal markers,
such as β-catenin and vimentin. In the present study,
overexpression of miR-212 attenuated E-cadherin expression, but
augmented β-catenin, vimentin, and Twist1. This observation
indicates that miR-212 promotes ESCC migration by activation of the
EMT process.
Degradation of ECM and basement membrane is critical
for tumor invasion and metastasis. MMP2 and MMP9 are functionally
related gelatinase/collagenase IV proteins and their expression and
activity have been linked to metastasis or invasion in a majority
of cancer types (28,29). This led to the investigation of the
effect of miR-212 on the expression of MMP2, MMP9 and uPA in ESCC
cells. It was found that miR-212 significantly increased the
protein and mRNA levels of MMP9 and uPA in three ESCC cell lines,
with no alteration of MMP2 mRNA and protein levels. These findings
suggest that miR-212 increases the expression of ECM-degrading
enzymes to degrade the ECM, which allows ESCC cells to escape from
the primary tumor and invade adjacent tissues, and to migrate to
distant regions. It has been demonstrated that gene expression of
MMP2 and MMP9 is regulated mainly at the transcription level and
both are co-regulated to some extent in their expression due to
their promoters sharing common structural features (30). However, distinct compositions have
been identified between promoters of MMP2 and MMP9 (31). For example, a TATA box is present
in the cis element of the MMP9 promoter, but is not present in the
MMP2 promoter. The expression of MMP2 is mainly determined by the
ubiquitous Sp-1 family of transcription factors. Thus, this
diversity in the promoter compositions of MMP2 and MMP9 may be
associated with the mechanism by which miR-212 differently
regulated the expression of these proteins in ESCC cells.
Since increased miR-212 level was associated with
poor outcome in patients with ESCC (12), the potential of therapeutic drugs
to inhibit the role of miR-212 in promoting ESCC cell migration was
investigated. Cell migration is controlled by several signaling
cascades including PI3K/AKT, AMPK/ACC signaling, mTOR/p70S6K and
estrogen receptor stress. Of all the drugs tested in the present
study, berberine alone demonstrated an inhibitory effect on
miR-212-induced KYSE-450 migration and EMT. Berberine was reported
to attenuate cell migration and EMT in several types of cancer
cells by targeting a number of signaling molecules and processes,
including PI3K/AKT, p38, mTOR/p70S6K and AMPK-ERK (32–35).
As an oncogene, miR-212 targets multiple molecules in the
downstream signaling pathways to promote cell migration, invasion
and EMT. These target genes include PTEN, SOX4 and Patched-1
(36–38).
In conclusion, the results from the present study
demonstrate that miR-212 facilitates ESCC cell migration and
invasion rather than proliferation, and berberine is a potential
therapeutic drug against metastasis of ESCC in patients with high
miR-212 expression. Further investigation of the mechanisms and its
target gene by which miR-212 promotes ESCC cell migration and
invasion is required.
Acknowledgements
Not applicable.
Funding
The present study was supported by Key Technologies
R&D Program of Science and Technology Commission Foundation of
Henan Provence to BZ (grant no. 152102310110); Key Science and
Technique Fund of Xinxiang to BZ (grant no. ZG15018); Xinxiang
Medical University Student Innovation Fund to ZC (grant no.
YJSCX201709Y); The First Affiliated Hospital of Xinxiang Medical
University Foundation to YL (grant no. XYYFY2014BS-002); and The
First Affiliated Hospital of Xinxiang Medical University Youth
Foundation to BQ (grant no. QN-2017-A004).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZC and YL contributed equally to this work. ZC and
YL designed the methods and performed the majority of the
experiments; BQ, CG, XW, LG and WY performed the migration
experiments of the drug treatments and interpretation of the data;
ZC, YL and BZ contributed to the conception and design of the
study, and drafting the manuscript. All authors read and approved
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arnold M, Soerjomataram l, Ferlay J and
Forman D: Global incidence of oesophageal cancer by histological
subtype in 2012. Gut. 64:381–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Murphy G, McCormack V, Abedi-Ardekani B,
Arnold M, Camargo MC, Dar NA, Dawsey SM, Etemadi A, Fitzgerald RC,
Fleischer DE, et al: International cancer seminars: A focus on
esophageal squamous cell carcinoma. Ann Oncol. 28:2086–2093. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu
L and He J: Report of incidence and mortality in China cancer
registries, 2009. Chin J Cancer Res. 25:10–21. 2013.PubMed/NCBI
|
|
5
|
Cheng J, Jin H, Hou X, Lv J, Gao X and
Zheng G: Disturbed tryptophan metabolism correlating to progression
and metastasis of esophageal squamous cell carcinoma. Biochem
Biophys Res Commun. 486:781–787. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bracken CP, Scott HS and Goodall GJ: A
network-biology perspective of microRNA function and dysfunction in
cancer. Nat Rev Genet. 17:719–732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sakai NS, Samia-Aly E, Barbera M and
Fitzgerald RC: A review of the current understanding and clinical
utility of miRNAs in esophageal cancer. Semin Cancer Biol.
23:512–521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Harada K, Baba Y, Ishimoto T, Shigaki H,
Kosumi K, Yoshida N, Watanabe M and Baba H: The role of microRNA in
esophageal squamous cell carcinoma. J Gastroenterol. 51:520–530.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao BS, Liu SG, Wang TY, Ji YH, Qi B, Tao
YP, Li HC and Wu XN: Screening of microRNA in patients with
esophageal cancer at same tumor node metastasis stage with
different prognoses. Asian Pac J Cancer Prev. 14:139–143. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qi B, Liu SG, Qin XG, Yao WJ, Lu JG, Guo
L, Wang TY, Li HC and Zhao BS: Overregulation of microRNA-212 in
the poor prognosis of esophageal cancer patients. Genet Mol Res.
13:7800–7807. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li B, Li J, Xu WW, Guan XY, Qin XY, Zhang
LY, Law S, Tsao SW and Cheung AL: Suppression of esophageal tumor
growth and chemoresistance by directly targeting the PI3K/Akt
pathway. Oncotarget. 5:11576–11587. 2014.PubMed/NCBI
|
|
13
|
Lu Z, Peng K, Wang N, Liu HM and Hou G:
Downregulation of p70S6K enhances cell sensitivity to rapamycin in
esophageal squamous cell carcinoma. J Immunol Res.
2016:78289162016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Medina EA, Obertheu K, Polusani SR, Ortega
V, Velagaleti GV and Oyajobi BO: PKA/AMPK signaling in relation to
adiponectin's antiproliferative effect on multiple myeloma cells.
Leukemia. 28:2080–2089. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang F, Wang YG and Wang C: Metformin
inhibited growth, invasion and metastasis of esophageal squamous
cell carcinoma in vitro and in vivo. Cell Physiol Biochem.
51:1276–1286. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen YZ, Bai N, Bi JH, Liu XW, Xu Go,
Zhang LF, Li XQ and Huo R: Propranolol inhibits the proliferation,
migration and tube formation of hemangioma cells through HIF-1α
dependent mechanisms. Braz J Med Biol Res. 50:e61382017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang SX, Qi B, Yao WJ, Gu CW, Wei XF,
Zhao Y, Liu YZ and Zhao BS: Berberine displays antitumor activity
in esophageal cancer cells in vitro. World J Gastroenterol.
23:2511–2518. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Method. 5:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial- mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Baker EA, Stephenson TJ, Reed MW and Brown
NJ: Expression of proteinases and inhibitors in human breast cancer
progression and survival. Mol Pathol. 55:300–304. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rao JS, Gondi C, Chetty C, Chittivelu S,
Joseph PA and Lakka SS: Inhibition of invasion, angiogenesis, tumor
growth and metastasis by adenovirus-mediated transfer of antisense
uPAR and MMP-9 in non-small cell lung cancer cells. Mol Cancer
Ther. 4:1399–1408. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang YY, Tang LQ and Wei W: Berberine
attenuates podocytes injury caused by exosomes derived from high
glucose-induced mesangial cells through TGFβ1-PI3K/AKT pathway. Eur
J Pharmacol. 824:185–192. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mishan MA, Ahmadiankia N, Matin MM,
Heirani-Tabasi A, Shahriyari M, Bidkhori HR, Naderi-Meshkin H and
Bahrami AR: Role of berberine on molecular markers involved in
migration of esophageal cancer cells. Cell Mol Biol
(Noisy-le-grand). 61:37–43. 2015.PubMed/NCBI
|
|
24
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Remenyi J, vanden Bosch MW, Palygin O,
Mistry RB, McKenzie C, Macdonald A, Hutvagner G, Arthur JS,
Frenguelli BG and Pankratov Y: miR-132/212 Knockout mice reveal
roles for these miRNAs in regulating cortical synaptic transmission
and plasticity. PLoS One. 8:e625092013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Remenyi J, Hunter CJ, Cole C, Ando H,
Impey S, Monk CE, Martin KJ, Barton GJ, Hutvagner G and Arthur JS:
Regulation of the miR-212/132 locus by MSK1 and CREB in response to
neurotrophins. Biochem J. 428:281–291. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiping Z, Ming F, Lixiang W, Xiuming L,
Yuqun S, Han Y, Zhifang L, Yundong S, Shili L, Chunyan C and Jihui
J: MicroRNA-212 inhibits proliferation of gastric cancer by
directly repressing retinoblastoma binding protein 2. J Cell
Biochem. 114:2666–2272. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Roomi MW, Kalinovsky T, Niedzwiecki A and
Rath M: Modulation of uPA, MMPs and their inhibitors by a novel
nutrient mixture in human colorectal, pancreatic and hepatic
carcinoma cell lines. Int J Oncol. 47:370–376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yamashita K, Tanaka Y, Mimori K, Inoue H
and Mori M: Differential expression of MMP and uPA systems and
prognostic relevance of their expression in esophageal squamous
cell carcinoma. Int J Cancer. 110:201–207. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yan C and Boyd DD: Regulation of matrix
metalloproteinase gene expression. J Cell physiol. 211:19–26. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chakraborti S, Mandal M, Das S, Mandal A
and Chakraborti T: Regulation of matrix metalloproteinases: An
overview. Mol Cell Biochem. 253:269–285. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kou Y, Li L, Li H, Tan Y, Li B, Wang K and
Du B: Berberine suppressed epithelial mesenchymal transition
through cross-talk regulation of PI3K/AKT and RARα/RARβ in melanoma
cells. Biochem Biophys Res Commun. 479:290–296. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li L, Wang X, Sharvan R, Gao J and Qu S:
Berberine could inhibit thyroid carcinoma cells by inducing
mitochondrial apoptosis, G0/G1 cell cycle arrest and suppressing
migration via PI3K-AKT and MAPK signaling pathways. Biomed
Pharmacother. 95:1225–1231. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ai F, Chen M, Yu B, Yang Y, Xu G, Gui F,
Liu Z, Bai X and Chen Z: Berberine regulates proliferation,
collagen synthesis and cytokine secretion of cardiac fibroblasts
via AMPK-mTOR-p70S6K signaling pathway. Int J Clin Exp Pathol.
8:12509–12516. 2015.PubMed/NCBI
|
|
35
|
Kim HS, Kim MJ, Kim EJ, Yang Y, Lee MS and
Lim JS: Berberine-induced AMPK activation inhibits the metastatic
potential of melanoma cells via reduction of ERK activity and COX-2
protein expression. Biochem Pharmacol. 83:385–394. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xie M, Fu Z, Cao J, Liu Y, Wu J, Li Q and
Chen Y: MicroRNA-132 and microRNA-212 mediate doxorubicin
resistance by down-regulating the PTEN-AKT/NF-κB signaling pathway
in breast cancer. Biomed Pharmacother. 102:286–294. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fu W, Tao T, Qi M, Wang L, Hu J, Li X,
Xing N, Du R and Han B: MicroRNA-132/212 upregulation inhibits
TGF-β-mediated epithelial-mesenchymal transition of prostate cancer
cells by targeting SOX4. Prostate. 76:1560–1570. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ma C, Nong K, Wu B, Dong B, Bai Y, Zhu H,
Wang W, Huang X, Yuan Z and Ai K: miR-212 promotes pancreatic
cancer cell growth and invasion by targeting the hedgehog signaling
pathway receptor patched-1. J Exp Clin Cancer Res. 33:542014.
View Article : Google Scholar : PubMed/NCBI
|