Introduction
Semen abnormalities are a form of male infertility
which present in a variety of ways and may prevent the sperm from
achieving fertilization (1–4).
Previous studies have shown that there are several causes of
abnormal semen, including infection with sexually transmitted
diseases (STDs), retrograde ejaculation and an inability of the
ejaculate to clot properly, all of which can significantly affect
male fertility. In addition, sperm abnormalities may be inherited
or due to a hormone imbalance, medication or previous infection
(5). Narayana et al
indicated that O,O-dimethyl O-4-nitrophenyl phosphorothioate could
affect the sperm morphology and count in rats (6), and Padmalatha Rai et al
demonstrated that the anticancer drug tamoxifen citrate acts as a
germ cell mutagen by inducing sperm shape abnormalities in mice
in vivo (7). Additionally,
Calogero et al reported that a large proportion of patients
with oligoasthenoteratozoospermia and teratozoospermia have an
increased rate of sperm aneuploidy, and these patients also have
semen abnormalities (8). Studies
have also indicated that abnormal semen characteristics are induced
by testicular cancer (9,10). Although a number of the factors
which cause abnormal semen, including chemotherapeutic agents,
testicular tumors and microwave radiation (5,11,12)
have been identified, differences in epigenetic regulation between
normal and abnormal sperm have not been fully investigated.
microRNAs (miRNAs) are a class of naturally
occurring single-stranded short 21–23 nt non-coding RNAs (13,14)
which exist in a wide range of eukaryotic organisms (13–18).
Each mammalian miRNA can prevent the translation of a number of
downstream target mRNAs and ultimately lead to the inhibition of
target gene expression (19,20).
Therefore, a shift away from the targeting of crucial target genes
towards miRNA interference techniques may improve the effectiveness
of current gene-based diagnostic and therapeutic strategies
(15). However, most miRNA studies
have focused on the growth and development of stem cells,
differentiation, tumorigenesis and other pathological processes
(19,20) and have given little consideration
to the role of miRNAs in the development of abnormal semen and male
infertility.
Several methods, including northern blot analysis,
cloning and sequencing strategies, Invader assays, qRT-PCR and
sequencing-based assays have been used to determine the expression
of miRNAs in biological samples (21). However, miRNA microarrays have
become the method of choice for global miRNA profiling studies, as
large numbers of molecules can be screened simultaneously using a
flexible probe design strategy (21). Additionally, miRNA microarrays
provide a powerful tool for the analysis of miRNA expression
patterns and quantitative miRNA expression levels. Microarray
technology has become the most commonly utilized miRNA research
tool, as it is more efficient than time-consuming traditional
methods (22–27).
In this study, we used a miRNA microarray-based high
throughput approach to identify and quantify the miRNAs that were
differentially expressed between the total RNA isolated from the
normal semen from healthy males and the abnormal semen from
infertile males. The identification of differentially expressed
miRNAs in the abnormal semen of infertile males may support further
studies to elucidate the causes and characteristics of abnormal
semen.
Materials and methods
Patients
The present study involved 86 infertile males (B)
with abnormal semen and 86 normal healthy adult males (H) as the
control. The samples were collected from the inpatient clinic of
the International Peace Maternity and Child Health Hospital of the
China Welfare Institute (Shanghai, China) between February and
September 2010. All human materials were obtained according to
consent regulations and approved by the Ethical Review Committee of
the World Health Organization Collaborating Center for Research in
Human Reproduction in Shanghai, China as authorized by the Shanghai
Municipal Government. Due to material limitations, we could only
analyze a limited number of severely abnormal sperm samples.
Semen collection and assessment of semen
function
Semen samples were produced by masturbation,
collected in sterile containers and immediately transported to the
laboratory. A conventional semen profile was obtained for each
sample using the procedures described by the World Health
Organization (10).
Total RNA extraction
Total RNA was isolated from each semen sample using
the TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA,
USA) according to the manufacturer’s instructions (28,29).
The RNA samples were treated with DNase I (Sigma-Aldrich, St.
Louis, MO, USA) and then quantified.
miRNA microarray analysis
RNA labeling and hybridization were performed on
miRNA microarray chips as previously described (25,27,30).
Briefly, 50 μg of total RNA was purified using the mirVana miRNA
isolation kit (Ambion, Austin, TX, USA) to enrich the small RNA
fraction. The purified RNA was labeled with fluorescein and
hybridized using the CapitalBio mammalian miRNA array V3.0
(CapitalBio Corporation, Beijing, China) containing 2844 mature
miRNA gene oligonucleotide probes in triplicate, corresponding to
1823 human, 648 mouse and 373 rat miRNAs. Each individual’s semen
RNA was analyzed on a separate chip. Finally, scanned images of the
microarray were captured and the hybridization signals were
quantified. The signal intensity values were normalized to per-chip
mean values.
Total RNA extraction and reverse
transcription into cDNA
Following the detection of total RNA, we used a
Poly(A) Tailing kit (Ambion) to add a poly(A) tail to the RNA
products according to the kit’s instructions. The RNA samples were
treated with DNase I, quantified and reverse-transcribed into cDNA
using the ReverTra Ace-α First Strand cDNA Synthesis kit (Toyobo,
Osaka, Japan). Notably, this reverse transcription reaction uses
the oligo(dT) reverse transcription primer
5′-GCTGTCAACGATACGCTACCTAACGGCATGACAGTGTTTTTTTTTTTTTTT(C/G/A)-3′.
All reaction steps were carried out according to the manufacturer’s
instructions.
Quantitative real-time PCR validation
miRNA expression
In accordance with the manufacturer’s instructions
and as previously described (23),
qRT-PCR was conducted in the realplex4 real-time PCR
detection system from Eppendorf (Hamburg, Germany), using
SYBR® Green RealTime PCR Master mix (Toyobo) as the
detection dye. The qRT-PCR amplification process comprised 40
cycles of denaturation at 95°C for 10 sec and annealing at 57°C for
20 sec. The target cDNA was quantified using a relative
quantification method. A comparative threshold cycle (Ct) was used
to quantify the gene expression relative to the control
(calibrator). The steady-state mRNA levels were expressed as an
n-fold difference relative to the calibrator. For each sample, the
Ct values were normalized using the formula: ΔCt =
CtmiRNA − Ct18S rRNA. To detemine relative
expression levels, the following formula was used: ΔΔCt =
ΔCtB − ΔCtH. The values used to plot the
relative miRNA expression levels were calculated using the
expression 2−ΔΔCt. The miRNA levels were calibrated by
18S rRNA. The miRNA primers used in the cDNA amplification are
shown in Table I.
 | Table IThe miRNA qRT-PCR primers used in the
study. |
Table I
The miRNA qRT-PCR primers used in the
study.
| Accession no. | miRNA | qRT-PCR primers
(5′→3′) |
|---|
| MI0003581 | miR-574-5p |
5′-TGAGTGTGTGTGTGTGAGTGTGT-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0000063 | let-7b |
5′-CTATACAACCTACTGCCTTCCC-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0005775 | miR-297 |
5′-ATGTATGTGTGCATGTGCATG-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0000442 | miR-122 |
5′-AACGCCATTATCACACTAAATA-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0006415 | miR-1275 |
5′-GTGGGGGAGAGGCTGTC-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0006428 | miR-1281 |
5′-TCGCCTCCTCCTCTCCC-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0000781 | miR-373 |
5′-GAAGTGCTTCGATTTTGGGGTGT-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0000482 | miR-185 |
5′-AGGGGCTGGCTTTCCTCTGGTC-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0003137 | miR-193b |
5′-AACTGGCCCTCAAAGTCCCGCT-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0000461 | miR-145 |
5′-GGATTCCTGGAAATACTGTTCT-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0000290 | miR-214 |
5′-ACAGCAGGCACAGACAGGCAGT-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0000089 | miR-31 |
5′-TGCTATGCCAACATATTGCCAT-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0003513 | miR-455-3p |
5′-GCAGTCCATGGGCATATACAC-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0000102 | miR-100 |
5′-CAAGCTTGTATCTATAGGTATG-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0003161 | miR-517a |
5′-ATCGTGCATCCCTTTAGAGTGT-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0003140 | miR-512-3p |
5′-AAGTGCTGTCATAGCTGAGGTC-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0000070 | miR-16 |
5′-CCAGTATTAACTGTGCTGCTGA-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0003153 | miR-523 |
5′-GAACGCGCTTCCCTATAGAGGGT-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0000074 | miR-19b |
5′-TGTGCAAATCCATGCAAAACTGA-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0000439 | miR-23b |
5′-ATCACATTGCCAGGGATTACC-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
| MI0000083 | miR-26a |
5′-CCTATTCTTGGTTACTTGCACG-3′
(forward)
5′-GCTGTCAACGATACGCTACCTA-3′ (reverse) |
Northern blot analysis
All steps in the northern blotting process were
carried out as previously described (28,29).
For all samples, 20 μg good quality total RNA was analyzed on a 7.5
M urea 12% PAA denaturing gel and transferred to a Hybond-N+ nylon
membrane (Amersham, Freiburg, Germany). The membranes were
crosslinked using UV light for 30 sec at 1200 mJ/cm2.
Hybridization was performed using miRNA antisense StarFire probes
to detect the 22-nt miRNA fragments, according to the
manufacturer’s instructions. After washing, the membranes were
exposed for 20–40 h to Kodak XAR-5 films (Sigma-Aldrich). The
ethidium bromide-stained gels prior to the transfer of tRNA were
used as controls to ensure equal loading of the RNA samples.
Statistical analysis
Each experiment was performed at least three times
and data are the mean ± SE, where applicable. Differences were
evaluated using the Student’s t-test. p<0.05 was considered to
indicate a statistically significant result.
Results
Comparison of the characteristics and
semen parameters of the healthy males and infertile males
A total of 172 males were invited to participate in
this study: 86 healthy males with normal semen and 86 infertile
males with semen abnormalities. The only significant difference
between the populations was the percentage of progressive motile
(a+b) forms (p<0.001). The results of laboratory tests indicated
that asthenozoospermia was the most frequent finding in the 86
infertile males. The characteristics of the study participants are
presented in Table II.
 | Table IIComparison of the clinical
characteristics of the infertile males with semen abnormalities and
the healthy adult males. |
Table II
Comparison of the clinical
characteristics of the infertile males with semen abnormalities and
the healthy adult males.
| Parameter | Infertile males
(n=86) | Healthy males
(n=86) |
|---|
| Age (years) | 32±1 (27–41) | 32±2 (29–42) |
| Volume (ml) | 1.92±0.08
(1–2) | 2 |
| Concentration
(105/ml) | 97.59±18.05
(24.0–224.7) | 57.37±13.17
(39.0–117.4) |
| a+b (%) | 23.11±5.03
(0–46.2) | 45.17±6.34
(12.6–65.7) |
| a+b+c (%) | 32.85±5.52
(5.9–63.3) | 62.66±5.72
(49.3–82.6) |
Total RNA quality analysis
A 260 to 280 nm absorbance ratio (260/280)>1.8 is
usually considered to be an acceptable indicator of RNA purity for
miRNA microarrays and indicates an absence of detectable protein
contamination in the RNA sample (15). Following the extraction of total
RNA from the samples, the 260/280 ratio of each extract was
determined using a spectrophotometer (15,31).
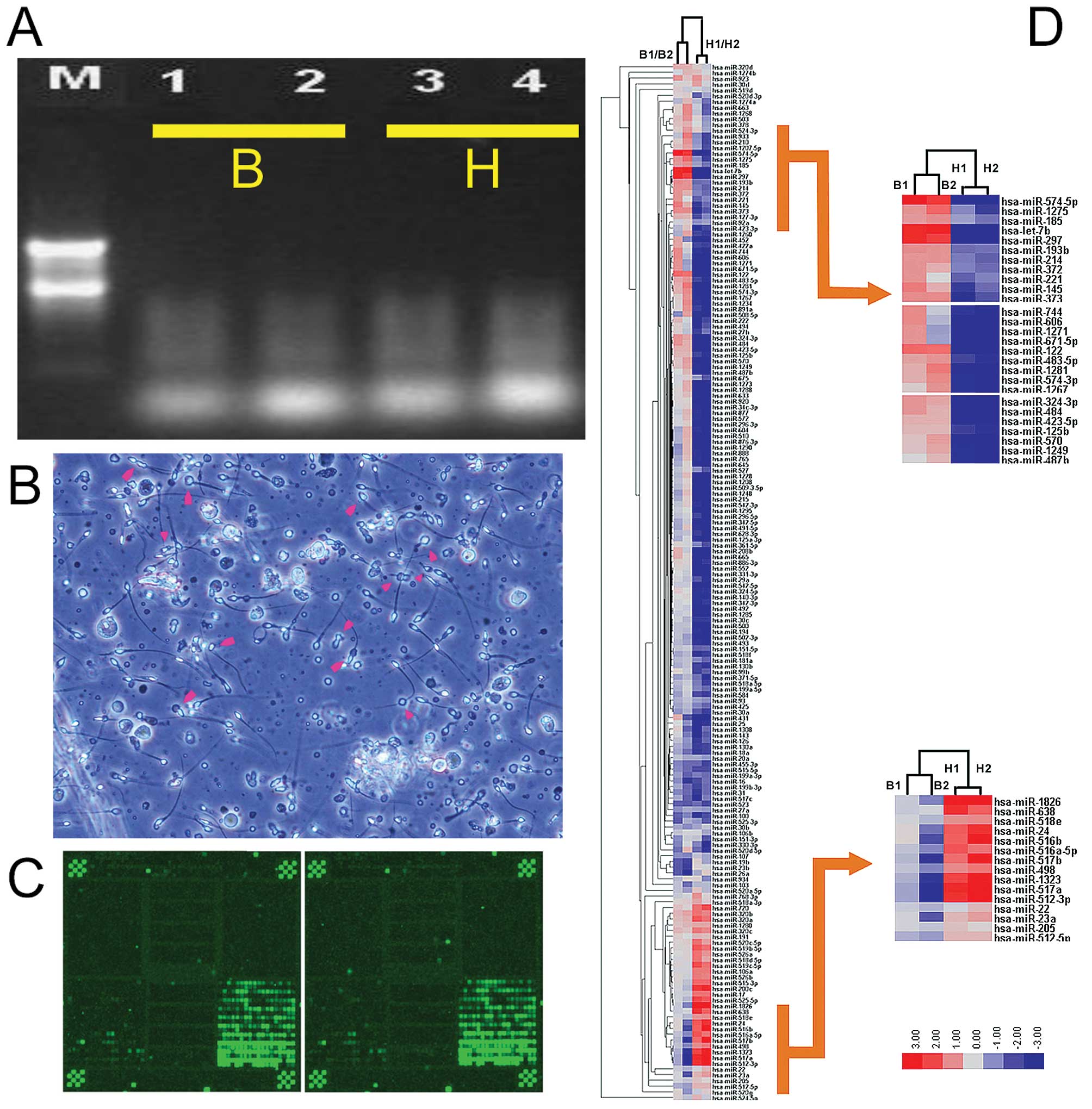
The 260/280 ratios ranged from 1.83 to 1.97. Formaldehyde
denaturing gel electrophoresis was used to confirm the presence of
clear 28S, 18S and 5S bands (Fig.
1) and the absence of marked RNA degradation. This analysis
indicated that the purity and integrity of each RNA sample met the
requirements of the miRNA microarray and qRT-PCR experiments
(15).
miRNA microarray quality control and
results analysis
In order to identify miRNAs which are differentially
expressed between the abnormal semen of infertile males and the
normal semen of healthy males, we prepared a miRNA microarray
containing 2844 oligonucleotide probes (1823 human, 648 mouse and
373 rat) complementary to known mammalian miRNAs (23,24,32).
All probes were repeated three times in each microarray and each
microarray contained 16 controls (Zip5, Zip13, Zip15, Zip21, Zip23,
Zip25, Y2, Y3, U6, New-U2-R, tRNA-R, has-let-7a, has-let-7b,
has-let-7c, 50% DMSO and Hex). In order to increase the reliability
of the results, each miRNA microarray assay was repeated twice
(24) and the scatter plots for
all spots indicated that a high reproducibility and reliability
were achieved (Fig. 2A).
The miRNA expression patterns for abnormal semen
from infertile males (B) and normal semen from healthy males (H)
were compared. Significance analysis of microarray (SAM) and a fold
change criterion (B/H ratio) >1.50 or <0.667 and p<0.001
were used to identify significant differences (32,33).
Using these criteria, we identified 52 miRNAs which were
differentially expressed between the semen of infertile males and
normal males. Analysis of the microarray expression levels
confirmed that 21 miRNAs (mi-574-5p, let-7b, miR-297, miR-122,
miR-1275, miR-1281, miR-373, miR-185, miR-193b, miR-145, miR-214,
miR-574-3p, miR-483-5p, miR-324-3p, miR-372, miR-484, miR-933,
miR-663, miR-1268, miR-923 and miR-1234) were significantly
overexpressed in the abnormal semen compared with the normal semen.
Conversely, 31 miRNAs (miR-1826, miR-493, miR-371-5p, miR-516a-5p,
miR-512-5p, miR-498, miR-30a, miR-23a, miR-130a, miR-103, miR-30b,
miR-27a, miR-18a, miR-525-3p, miR-517c, miR-199b-3p, miR-517b,
miR-107, miR-199a-3p, miR-1323, miR-515-5p, miR-31, miR-455-3p,
miR-100, miR-517a, miR-512-3p, miR-16, miR-523, miR-19b, miR-23b
and miR-26a) were significantly underexpressed in the abnormal
semen compared with the normal semen (Table III).
 | Table IIISummary of the SAM results for miRNA
expression in the abnormal semen of infertile males and the normal
semen of healthy adult males. |
Table III
Summary of the SAM results for miRNA
expression in the abnormal semen of infertile males and the normal
semen of healthy adult males.
| miRNA | Fold change
(B/H) | Mature miRNA
sequence | Chromosome
location | Sequence length
(nt) |
|---|
| miR-574-5p | 7.0715 |
UGAGUGUGUGUGUGUGAGUGUGU | 4 | 23 |
| let-7b | 5.7958 |
UGAGGUAGUAGGUUGUGUGGUU | 22 | 22 |
| miR-297 | 4.8753 |
AUGUAUGUGUGCAUGUGCAUG | 4 | 21 |
| miR-122 | 2.7916 |
UGGAGUGUGACAAUGGUGUUUG | 18 | 22 |
| miR-1275 | 2.3772 |
GUGGGGGAGAGGCUGUC | 6 | 17 |
| miR-1281 | 1.9876 |
UCGCCUCCUCCUCUCCC | 22 | 17 |
| miR-373 | 1.9799 |
GAAGUGCUUCGAUUUUGGGGUGU | 19 | 23 |
| miR-185 | 1.9584 |
UGGAGAGAAAGGCAGUUCCUGA | 22 | 22 |
| miR-193b | 1.9558 |
AACUGGCCCUCAAAGUCCCGCU | 16 | 22 |
| miR-145 | 1.9218 |
GUCCAGUUUUCCCAGGAAUCCCU | 5 | 23 |
| miR-214 | 1.9027 |
ACAGCAGGCACAGACAGGCAGU | 1 | 22 |
| miR-574-3p | 1.7689 |
CACGCUCAUGCACACACCCACA | 4 | 22 |
| miR-483-5p | 1.7640 |
AAGACGGGAGGAAAGAAGGGAG | 11 | 22 |
| miR-324-3p | 1.7295 |
ACUGCCCCAGGUGCUGCUGG | 17 | 20 |
| miR-372 | 1.7001 |
AAAGUGCUGCGACAUUUGAGCGU | 19 | 23 |
| miR-484 | 1.6988 |
UCAGGCUCAGUCCCCUCCCGAU | 16 | 22 |
| miR-933 | 1.6101 |
UGUGCGCAGGGAGACCUCUCCC | 2 | 22 |
| miR-663 | 1.6083 |
AGGCGGGGCGCCGCGGGACCGC | 20 | 22 |
| miR-1268 | 1.6016 |
CGGGCGUGGUGGUGGGGG | 15 | 18 |
| miR-923 | 1.5892 |
GUCAGCGGAGGAAAAGAAACU | 17 | 21 |
| miR-1234 | 1.5736 |
UCGGCCUGACCACCCACCCCAC | 8 | 22 |
| miR-1826 | 0.6548 |
AUUGAUCAUCGACACUUCGAACGCAAU | 16 | 27 |
| miR-493 | 0.6536 |
UGAAGGUCUACUGUGUGCCAGG | 14 | 22 |
| miR-371-5p | 0.6517 |
ACUCAAACUGUGGGGGCACU | 19 | 20 |
| miR-516a-5p | 0.6441 |
UUCUCGAGGAAAGAAGCACUUUC | 19 | 23 |
| miR-512-5p | 0.6322 |
CACUCAGCCUUGAGGGCACUUUC | 19 | 23 |
| miR-498 | 0.6191 |
UUUCAAGCCAGGGGGCGUUUUUC | 19 | 23 |
| miR-30a | 0.6070 |
UGUAAACAUCCUCGACUGGAAG | 6 | 22 |
| miR-23a | 0.6058 |
AUCACAUUGCCAGGGAUUUCC | 19 | 21 |
| miR-130a | 0.5913 |
CAGUGCAAUGUUAAAAGGGCAU | 11 | 22 |
| miR-103 | 0.5886 |
AGCAGCAUUGUACAGGGCUAUGA | 20 | 23 |
| miR-30b | 0.5771 |
UGUAAACAUCCUACACUCAGCU | 8 | 22 |
| miR-27a | 0.5577 |
UUCACAGUGGCUAAGUUCCGC | 19 | 21 |
| miR-18a | 0.4980 |
UAAGGUGCAUCUAGUGCAGAUAG | 13 | 23 |
| miR-525-3p | 0.4817 |
GAAGGCGCUUCCCUUUAGAGCG | 19 | 22 |
| miR-517c | 0.4783 |
AUCGUGCAUCCUUUUAGAGUGU | 19 | 22 |
| miR-199b-3p | 0.4700 |
ACAGUAGUCUGCACAUUGGUUA | 9 | 22 |
| miR-517b | 0.4672 |
UCGUGCAUCCCUUUAGAGUGUU | 19 | 22 |
| miR-107 | 0.4641 |
AGCAGCAUUGUACAGGGCUAUCA | 19 | 23 |
| miR-199a-3p | 0.4452 |
ACAGUAGUCUGCACAUUGGUUA | 19 | 22 |
| miR-1323 | 0.4352 |
UCAAAACUGAGGGGCAUUUUCU | 19 | 22 |
| miR-515-5p | 0.4279 |
UUCUCCAAAAGAAAGCACUUUCUG | 19 | 24 |
| miR-31 | 0.4137 |
AGGCAAGAUGCUGGCAUAGCU | 9 | 21 |
| miR-455-3p | 0.4117 |
GCAGUCCAUGGGCAUAUACAC | 9 | 21 |
| miR-100 | 0.3938 |
AACCCGUAGAUCCGAACUUGUG | 11 | 22 |
| miR-517a | 0.3889 |
AUCGUGCAUCCCUUUAGAGUGU | 19 | 22 |
| miR-512-3p | 0.3884 |
AAGUGCUGUCAUAGCUGAGGUC | 19 | 22 |
| miR-16 | 0.3455 |
UAGCAGCACGUAAAUAUUGGCG | 13 | 22 |
| miR-523 | 0.3075 |
GAACGCGCUUCCCUAUAGAGGGU | 19 | 23 |
| miR-19b | 0.2670 |
UGUGCAAAUCCAUGCAAAACUGA | 13 | 23 |
| miR-23b | 0.2616 |
AUCACAUUGCCAGGGAUUACC | 9 | 21 |
| miR-26a | 0.2221 |
UUCAAGUAAUCCAGGAUAGGCU | 12 | 22 |
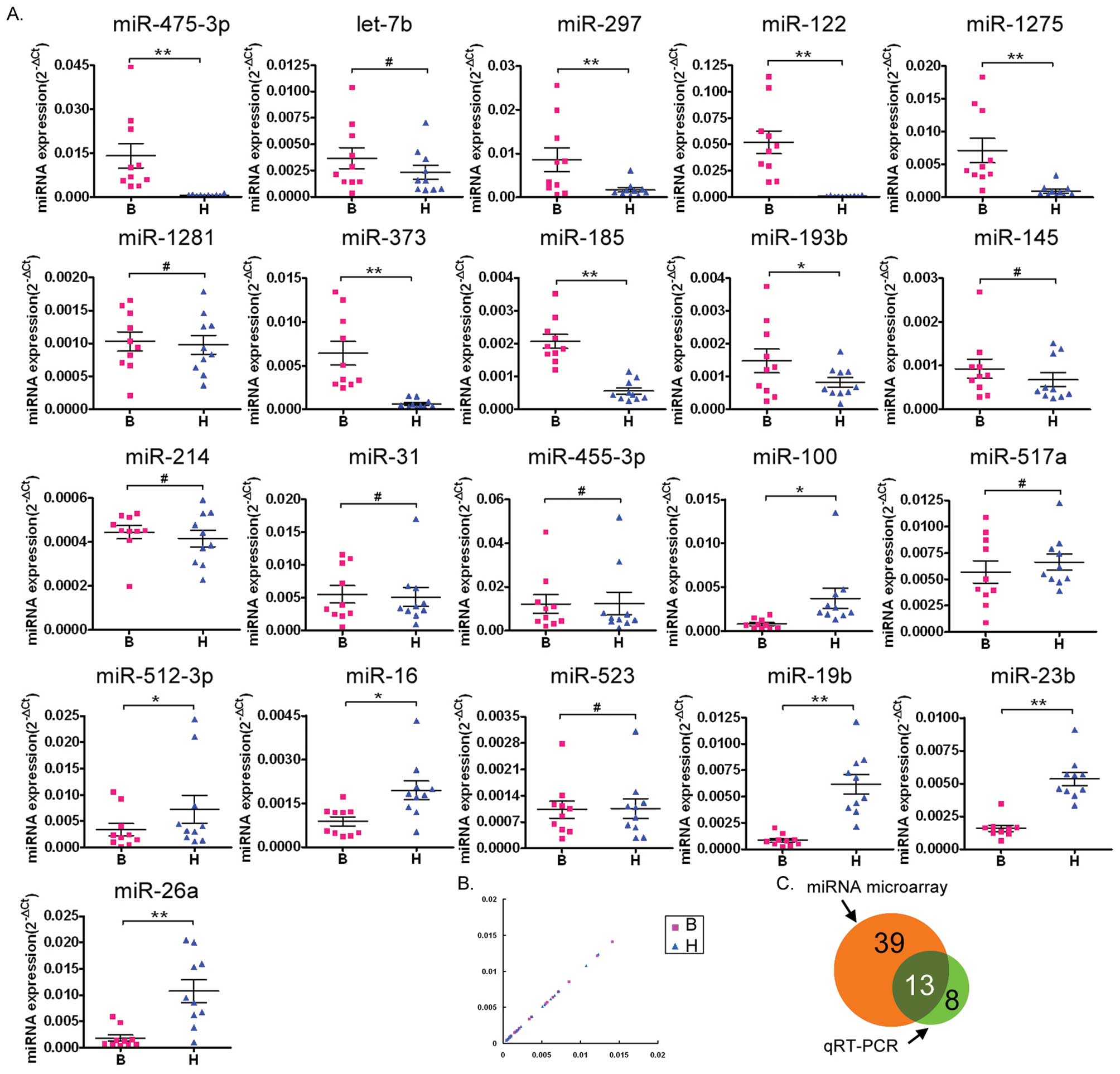
qRT-PCR confirmation of the miRNA
microarray results
Following common procedures for the confirmation of
microarray analysis (23,24,32–34),
qRT-PCR was used to confirm the results of the miRNA microarray
analysis. Of the 11 miRNAs identified by the microarray as being
the most overexpressed in the abnormal semen of infertile males
compared with normal semen (miR-574-5p, let-7b, miR-297, miR-122,
miR-1275, miR-1281, miR-373, miR-185, miR-193b, miR-145 and
miR-214), qRT-PCR confirmed that seven (miR-574-5p, miR-297,
miR-122, miR-1275, miR-373, miR-185 and miR-193b) were
overexpressed. Of the ten miRNAs identified as being underexpressed
in abnormal semen by the microarray (miR-31, miR-455-3p, miR-100,
miR-517a, miR-512-3p, miR-16, miR-523, miR-19b, miR-23b and
miR-26a), the qRT-PCR analysis confirmed that six of these
(miR-100, miR-512-3p, miR-16, miR-19b, miR-23b and miR-26a) were
underexpressed.
Scatter plot analysis of the qRT-PCR results
confirmed that seven miRNAs (miR-574-5p, miR-297, miR-122,
miR-1275, miR-373, miR-185 and miR-193b) were overexpressed and six
miRNAs (miR-100, miR-512-3p, miR-16, miR-19b, miR-23b and miR-26a)
were underexpressed in the semen of infertile males compared with
the normal semen (Fig. 2B).
A Venn diagram (Fig.
2C) was used to depict the correlation between the results of
the miRNA microarray and the 21 miRNAs tested by qRT-PCR. The
differential expression of 13 miRNAs (miR-574-5p, miR-297, miR-122,
miR-1275, miR-373, miR-185, miR-193b, miR-100, miR-512-3p, miR-16,
miR-19b, miR-23b and miR-26a) in the abnormal semen of the
infertile males was confirmed by qRT-PCR (indicated by the overlap
in the diagram). The expression levels of the other miRNAs
correlated in some or other methods. Overall, the qRT-PCR analysis
indicated that the miRNA microarray results had some small errors,
however, it confirmed that a significant number of miRNAs are
differentially regulated in the abnormal semen of infertile
males.
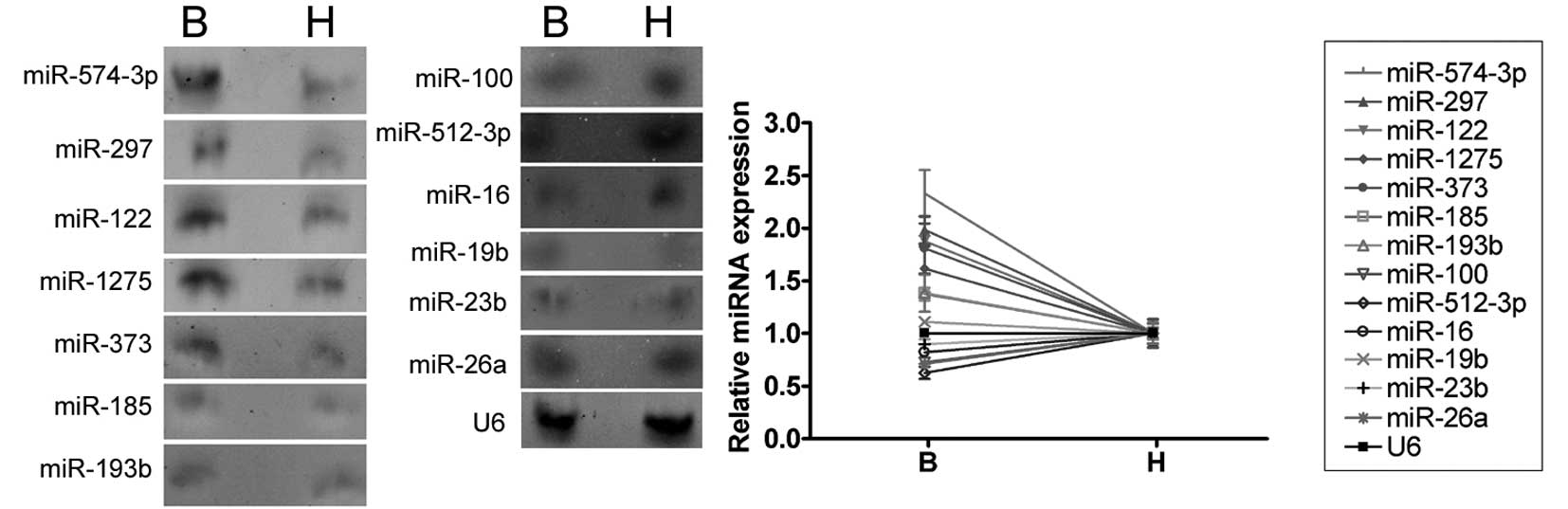
Northern blot validation of miRNA
expression
The expression levels of the 13 miRNAs which were
confirmed to be differently expressed by qRT-PCR were further
investigated by northern blotting of the RNA isolated from the
abnormal semen of three infertile males and the normal semen of
three healthy adult males. Anti-sense miRNA locked nucleic acid
probes were used for each miRNA (Fig.
3). The northern blotting hybridization signals for miR-574-5p,
miR-297, miR-122, miR-1275, miR-373, miR-185 and miR-193b were
weaker in the semen of healthy adult controls than that of the
infertile males, confirming that these miRNAs are upregulated in
the abnormal semen. The miR-100, miR-512-3p, miR-16, miR-19b,
miR-23b and miR-26a hybridization bands were barely detectable and,
therefore, we could not confirm the differential regulation of
these miRNAs using northern blotting.
Discussion
Mature miRNAs are an abundant class of 21–23 nt
non-coding RNAs which regulate the expression of their target genes
and are involved in many biological processes (15,21,23,24,32–34).
To date, more than 1600 miRNAs have been identified in plants,
animals and viruses (16,21,35,36).
It is currently estimated that miRNAs account for approximately 1%
of all predicted genes and that up to 30% of the genes in higher
eukaryotic genomes may be regulated by miRNAs (21); therefore, many miRNAs remain to be
identified in mammalian genomes. Little is known concerning the
patterns or levels of miRNA expression in the abnormal semen of
infertile males (24,33).
The aim of the study was to identify which miRNAs
are differentially expressed between abnormal and normal sperm, in
order to provide a foundation for future studies on the function
and role of miRNAs in semen abnormalities. We profiled the
expression of a number of miRNAs using a miRNA microarray and
demonstrated that the expression of 52 miRNAs was significantly
different in the abnormal semen of infertile males compared with
the semen of healthy males. These results suggest that miRNAs are
involved in the development of male infertility associated with
semen abnormalities.
We used qRT-PCR to confirm the expression levels of
21 of the 52 miRNAs which were differentially expressed in the
microarray. In total, 13 of the 21 miRNAs tested were identified as
being differentially expressed in abnormal semen by the microarray
and qRT-PCR. Although, there were some discrepancies in the results
of the microarray and the qRT-PCR analysis, the miRNA microarray
provided a rapid method for identifying a large number of
differentially expressed miRNAs in abnormal semen which could then
be confirmed by qRT-PCR.
This study describes the global expression patterns
of miRNAs in the abnormal semen from infertile males and
contributes to the growing understanding of the role of miRNAs in
the development of semen abnormalities. Moreover, the differential
expression patterns of miRNAs between normal and abnormal semen may
enable the direct diagnosis of semen abnormalities or provide novel
therapeutic targets for infertile males.
Acknowledgements
This study was supported by a grant from the
Shanghai Committee Medical Science Foundation of China (No.
10411967100) to Te Liu.
References
|
1
|
Zenzmaier C, Gerth R, Gruschwitz M,
Lindner H, Plas E and Berger P: Decreased levels of genuine large
free hCG alpha in men presenting with abnormal semen analysis.
Reprod Biol Endocrinol. 9:1142011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hu W, Yang H, Sun J, et al: Polymorphisms
in CYP1B1 modify the risk of idiopathic male infertility with
abnormal semen quality. Clin Chim Acta. 412:1778–1782. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chatzimeletiou K, Sioga A, Oikonomou L, et
al: Semen analysis by electron and fluorescence microscopy in a
case of partial hydatidiform mole reveals a high incidence of
abnormal morphology, diploidy, and tetraploidy. Fertil Steril.
95:e2431–e2435. 2011. View Article : Google Scholar
|
|
4
|
Moretti E, Castellini C, Mourvaki E, et
al: Distribution of α- and δ-tocopherols in seminal plasma and
sperm fractions of men with normal and abnormal semen parameters. J
Androl. 32:232–239. 2011.
|
|
5
|
Kowalczuk CI, Saunders RD and Stapleton
HR: Sperm count and sperm abnormality in male mice after exposure
to 2.45 GHz microwave radiation. Mutat Res. 122:155–161. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Narayana K, Prashanthi N, Nayanatara A,
Kumar HH, Abhilash K and Bairy KL: Effects of methyl parathion
(O,O-dimethyl O-4-nitrophenyl phosphorothioate) on rat sperm
morphology and sperm count, but not fertility, are associated with
decreased ascorbic acid level in the testis. Mutat Res. 588:28–34.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Padmalatha Rai S and Vijayalaxmi KK:
Tamoxifen citrate induced sperm shape abnormalities in the in vivo
mouse. Mutat Res. 492:1–6. 2001.PubMed/NCBI
|
|
8
|
Calogero AE, De Palma A, Grazioso C, et
al: Aneuploidy rate in spermatozoa of selected men with abnormal
semen parameters. Hum Reprod. 16:1172–1179. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jacobsen R, Bostofte E, Engholm G, et al:
Risk of testicular cancer in men with abnormal semen
characteristics: cohort study. BMJ. 321:789–792. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Torra R, Sarquella J, Calabia J, et al:
Prevalence of cysts in seminal tract and abnormal semen parameters
in patients with autosomal dominant polycystic kidney disease. Clin
J Am Soc Nephrol. 3:790–793. 2008. View Article : Google Scholar
|
|
11
|
Ravnborg TL, Jensen TK, Andersson AM,
Toppari J, Skakkebaek NE and Jørgensen N: Prenatal and adult
exposures to smoking are associated with adverse effects on
reproductive hormones, semen quality, final height and body mass
index. Hum Reprod. 26:1000–1011. 2011. View Article : Google Scholar
|
|
12
|
Barratt CL, Björndahl L, Menkveld R and
Mortimer D: ESHRE special interest group for andrology basic semen
analysis course: a continued focus on accuracy, quality, efficiency
and clinical relevance. Hum Reprod. 26:3207–3212. 2011. View Article : Google Scholar
|
|
13
|
Sumazin P, Yang X, Chiu HS, et al: An
extensive microRNA-mediated network of RNA-RNA interactions
regulates established oncogenic pathways in glioblastoma. Cell.
147:370–381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Poulton JS, Huang YC, Smith L, et al: The
microRNA pathway regulates the temporal pattern of Notch signaling
in Drosophila follicle cells. Development. 138:1737–1745. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lei P, Li Y, Chen X, Yang S and Zhang J:
Microarray based analysis of microRNA expression in rat cerebral
cortex after traumatic brain injury. Brain Res. 1284:191–201. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function (Review). Cell. 116:281–297.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yoo AS, Sun AX, Li L, et al:
MicroRNA-mediated conversion of human fibroblasts to neurons.
Nature. 476:228–231. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dai Y, Diao Z, Sun H, Li R, Qiu Z and Hu
Y: MicroRNA-155 is involved in the remodelling of
human-trophoblast-derived HTR-8/SVneo cells induced by
lipopolysaccharides. Hum Reprod. 26:1882–1891. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He L and Hannon GJ: MicroRNAs: small RNAs
with a big role in gene regulation (Review). Nat Rev Genet.
5:522–531. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
El Ouaamari A, Baroukh N, Martens GA,
Lebrun P, Pipeleers D and van Obberghen E: miR-375 targets
3′-phosphoinositide-dependent protein kinase-1 and regulates
glucose-induced biological responses in pancreatic beta-cells.
Diabetes. 57:2708–2717. 2008.
|
|
21
|
Yang Y, Bai W, Zhang L, et al:
Determination of microRNAs in mouse preimplantation embryos by
microarray. Dev Dyn. 237:2315–2327. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Calin GA, Liu CG, Sevignani C, et al:
MicroRNA profiling reveals distinct signatures in B cell chronic
lymphocytic leukemias. Proc Natl Acad Sci USA. 101:11755–11760.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bloomston M, Frankel WL, Petrocca F, et
al: MicroRNA expression patterns to differentiate pancreatic
adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA.
297:1901–1908. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yan N, Lu Y, Sun H, et al: A microarray
for microRNA profiling in mouse testis tissues. Reproduction.
134:73–79. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang LL, Zhang Z, Li Q, et al: Ethanol
exposure induces differential microRNA and target gene expression
and teratogenic effects which can be suppressed by folic acid
supplementation. Hum Reprod. 24:562–579. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barshack I, Meiri E, Rosenwald S, et al:
Differential diagnosis of hepatocellular carcinoma from metastatic
tumors in the liver using microRNA expression. Int J Biochem Cell
Biol. 42:1355–1362. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li W, Xie L, He X, et al: Diagnostic and
prognostic implications of microRNAs in human hepatocellular
carcinoma. Int J Cancer. 123:1616–1622. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheng W, Liu T, Jiang F, et al:
microRNA-155 regulates angiotensin II type 1 receptor expression in
umbilical vein endothelial cells from severely pre-eclamptic
pregnant women. Int J Mol Med. 27:393–399. 2011.PubMed/NCBI
|
|
29
|
Zhang L, Liu T, Huang Y and Liu J:
microRNA-182 inhibits the proliferation and invasion of human lung
adenocarcinoma cells through its effect on human cortical
actin-associated protein. Int J Mol Med. 28:381–388.
2011.PubMed/NCBI
|
|
30
|
Li S, Chen X, Zhang H, et al: Differential
expression of microRNAs in mouse liver under aberrant energy
metabolic status. J Lipid Res. 50:1756–1765. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang WX, Wilfred BR, Baldwin DA, et al:
Focus on RNA isolation: obtaining RNA for microRNA (miRNA)
expression profiling analyses of neural tissue. Biochim Biophys
Acta. 1779:749–757. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu HH, Tian X, Li YJ, Wu CA and Zheng CC:
Microarray-based analysis of stress-regulated microRNAs in
Arabidopsis thaliana. RNA. 14:836–843. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu H, Neilson JR, Kumar P, et al: miRNA
profiling of naive, effector and memory CD8 T cells. PLoS One.
2:e10202007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kuhn DE, Nuovo GJ, Martin MM, et al: Human
chromosome 21-derived miRNAs are overexpressed in down syndrome
brains and hearts. Biochem Biophys Res Commun. 370:473–477. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ambros V: The functions of animal
microRNAs (Review). Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Du T and Zamore PD: microPrimer: the
biogenesis and function of microRNA (Review). Development.
132:4645–4652. 2005. View Article : Google Scholar : PubMed/NCBI
|