Introduction
Rheumatoid arthritis (RA) is a systemic autoimmune
disease which affects 0.5–1% of the general population worldwide
(1). A chronic and deforming
arthritis, RA is characterized by accelerated inflammatory joint
destruction of articular cartilage and bone and synovial
hyperplasia, which ultimately leads to severe disabilities and a
poor quality of life (2,3). The etiology of RA is unknown, but
genetic factors are thought to be important in the pathogenesis and
progress of RA (1,4). One class of genetic variants that
have recently been the center of attention are DNA polymorphisms
that affect microRNA (miRNA) binding (5). miRNAs are approximately 22-nucleotide
(nt) non-coding RNAs that are involved in the post-transcriptional
regulation of gene expression by affecting the stability and
translation of mRNAs (6).
Compelling evidence indicates that miRNAs act as key regulators of
various processes, including early development, cell proliferation,
differentiation, stress resistance, cell fate determination,
apoptosis, signal transduction and organ development (7–10).
miRNAs are present in dried biological fluids, including semen,
saliva, vaginal secretions and menstrual blood and are expected to
be diagnostic and prognostic biomarkers of various diseases,
including cancer and autoimmune diseases such as RA (11,12).
Abnormal expression of several miRNAs has been
detected in RA in various cell types and these miRNAs regulate
specific pathways, thus leading to the inflammatory milieu
occurring in RA (2). Hsa-mir-499
is involved in autoimmune and inflammatory disease. The targets of
hsa-mir-499 include IL-17Rβ, IL-23α, IL-2R, IL-6, IL-2 and IL-18R.
IL-6 activates the production of CRP (C-reactive protein) and
fibrinogen through the liver and IL-17Rβ, IL-23α, IL-2R, IL-6, IL-2
and IL-18R contribute to the progress and pathogenesis of RA
(8). Findings of recent studies
have shown that the rs3746444 polymorphism in the pre-miR-499 is
correlated with several diseases, including breast cancer (13), cervical squamous cell carcinoma
(CSCC) (14), hepatocellular
carcinoma (15), RA (8), coronary artery disease (CAD)
(16), chronic obstructive
pulmonary disease (COPD) (17) and
tuberculosis (17).
More recently the miRNA hsa-mir-146a has also
received considerable attention, as it was reported to be
overexpressed in synovial fibroblasts, synovial tissue, synovial
fluid monocytes, peripheral blood mononuclear cells (PBMCs) and
serum plasma of RA patients (18).
This polymorphism was principally studied for its association with
several diseases, including psoriatic arthritis (19), multiple sclerosis (MS) (20), tuberculosis (17), ulcerative colitis, RA (8,21),
SLE (16) and various types of
cancer (22–29).
Overall, these two single nucleotide polymorphisms
(SNPs; rs3746444 and rs2910164) are located at the pre-miRNA
regions of hsa-mir-499 and hsa-mir-146a (21), respectively. Given the evidence
that SNPs located in the mature miRNA regions affect binding to
target mRNAs and the pre-miRNA maturation process (30), the aim of the present study was to
evaluate the impact of the rs3746444 and rs2910164 polymorphisms on
risk of RA in a sample of the Iranian population.
Materials and methods
Patients
We evaluated the possible association between
polymorphisms of hsa-mir-146a and hsa-mir-499 genes (rs2910164 G/C
and rs3746444 T/C, respectively) and RA susceptibility in 104
patients meeting the American College of Rheumatology criteria for
RA (31). The patients were
selected from the Rheumatology Clinic at Zahedan University of
Medical Sciences (4,32,33).
The control group involved 110 healthy individuals with no
relationship to RA patients. The Ethics Committee of Zahedan
University of Medical Sciences approved the project and informed
consent was obtained from all patients and healthy individuals.
Genomic DNA was extracted from peripheral blood samples as
previously described (33).
Tetra amplification refractory mutation
system-polymerase chain reaction (T-ARMS-PCR)
T-ARMS-PCR is a simple and rapid method with a high
level of accuracy for the detection of SNPs (34–36).
This system was used for genotyping of the rs2910164 G/C and
rs3746444 T/C polymorphisms. Genotyping of rs2910164 and rs3746444
was performed using two outer primers (FO and RO) and two inner
allele-specific primers (FI and RI) for each SNP, as listed in
Table I. PCR was performed using
commercially available PCR premix (AccuPower PCR PreMix; Bioneer,
Daejeon, South Korea) according to the manufacturer’s instructions.
The PCR cycling conditions were 95°C for 5 min followed by 30
cycles of 30 sec at 95°C, 25 sec at 61°C for rs2910164, 27 sec at
60°C for rs3746444 and 25 sec at 72°C, with a final extension of
72°C for 10 min. The PCR products were verified on 2% agarose gels
containing 0.5 μg/ml ethidium bromide and observed under UV light.
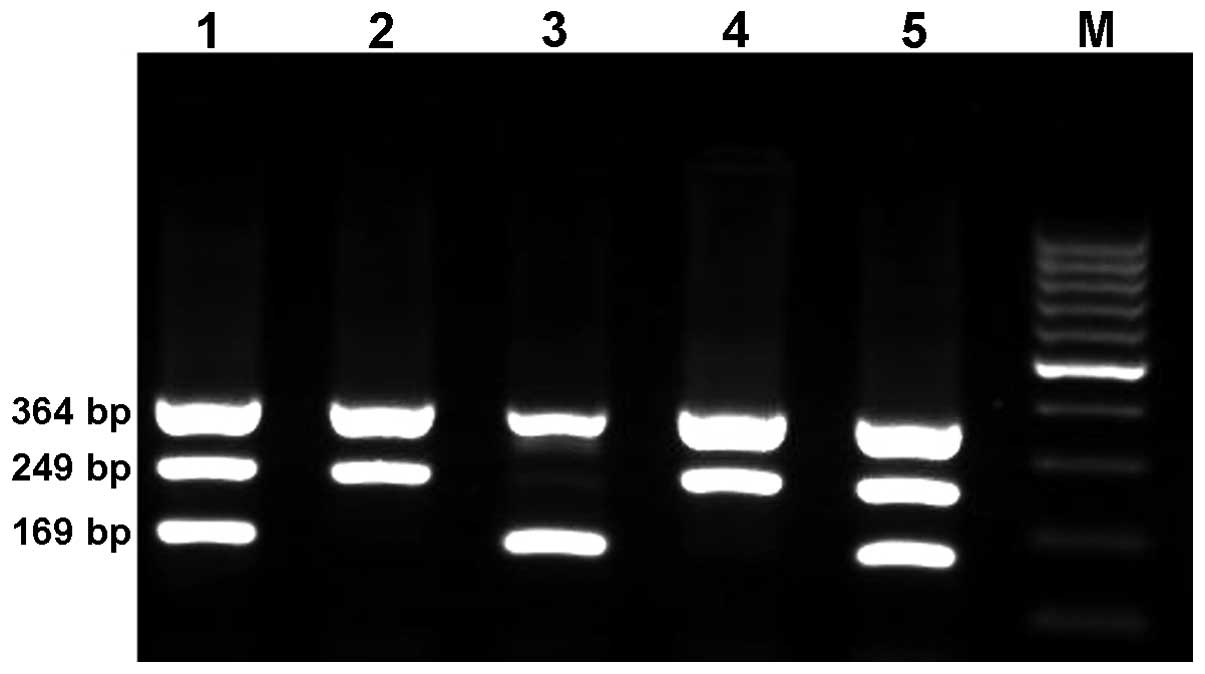
The product sizes for rs2910164 were 169 bp for the C allele and
249 bp for the G allele, while the product size for the two outer
primers (control band) was 364 bp (Fig. 1). The amplicon sizes for the
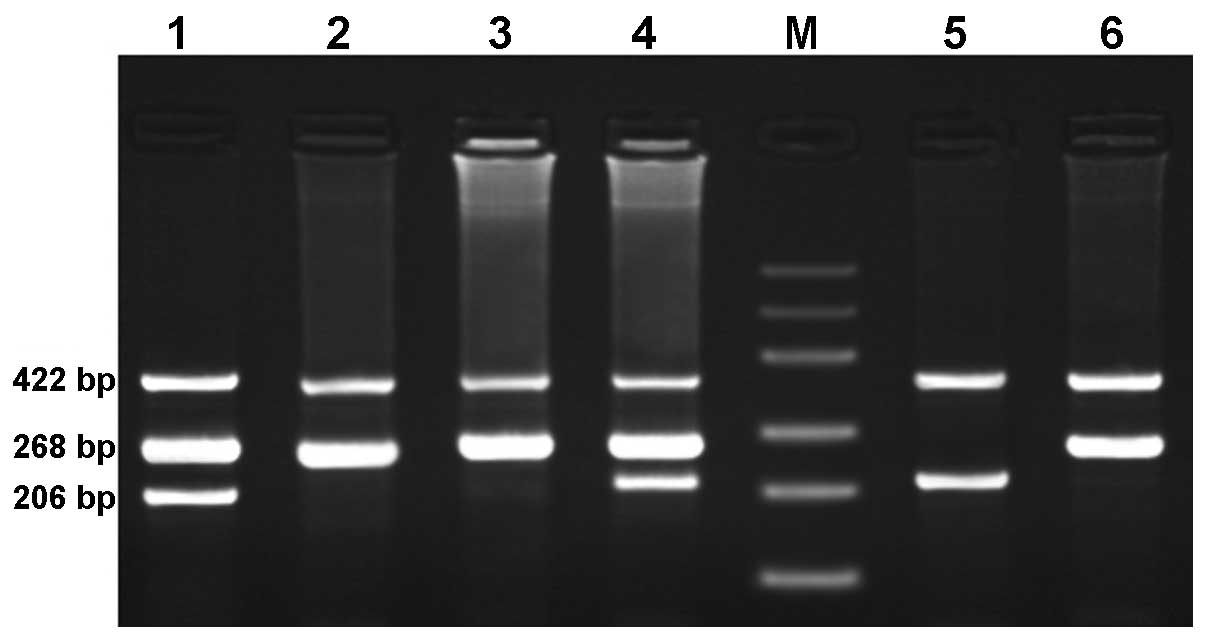
rs3746444 were 422 bp for the control band, 206 bp for the C allele
and 268 bp for the T allele (Fig
2). To ensure genotyping quality, we regenotyped 20% of the
random samples and found no genotyping errors.
 | Table IPrimer sequences for the detection of
hsa-mir-146a and hsa-miR-499 gene polymorphisms. |
Table I
Primer sequences for the detection of
hsa-mir-146a and hsa-miR-499 gene polymorphisms.
| Gene | Primer | Sequence
(5′-3′) | Amplicon size
(bp) |
|---|
| hsa-mir-146a;
rs2910164 G>C | FO |
GGCCTGGTCTCCTCCAGATGTTTAT | 364 |
| RO |
ATACCTTCAGAGCCTGAGACTCTGCC | 364 |
| FI (C allele) |
ATGGGTTGTGTCAGTGTCAGACGTC | 169 |
| RI (G allele) |
GATATCCCAGCTGAAGAACTGAATTTGAC | 249 |
| hsa-miR-499;
rs3746444 T>C | FO |
GAGTGACCAGGCCCCTTGTCTCTATTAG | 422 |
| RO |
TTGCTCTTTCACTCTCATTCTGGTGATG | 422 |
| FI (C allele) |
ATGTTTAACTCCTCTCCACGTGACCG | 206 |
| RI (T allele) |
GGGAAGCAGCACAGACTTGCTGTTAT | 268 |
Statistical analysis
Statistical analysis was calculated using SPSS 18
software. Data were analyzed by independent sample t-test and
χ2 test. The association between genotypes of
hsa-mir-146a and hsa-mir-499 genes and RA were assessed by
computing the odds ratio (OR) and 95% confidence intervals (95%
CIs) from logistic regression analyses. P<0.05 was considered to
indicate a statistically significant difference.
Results
The study group consisted of 104 RA patients (91
females and 13 males) with an average age of 44.7±13.3 years, and
110 healthy subjects (70 females and 40 males) with a mean age of
43.5±10.2 years. No significant difference was found between the
groups with respect to age (P=0.458), but a significant difference
was observed between the groups with respect to gender
(P=0.0001).
The frequency distribution of hsa-mir-499 rs3746444
T/C genotypes in RA patients and normal subjects are demonstrated
in Table II. A significant
difference was found between case and control groups with regard to
rs3746444 T/C polymorphism (χ2=13.32, P=0.001). The
hsa-mir-499 rs3746444 T/C polymorphism was a risk factor for
predisposition to RA in codominant (TT vs. TC: OR, 2.11; 95% CI,
1.08–4.11; P=0.029; TT vs. CC: OR, 3.88; 95% CI, 1.68–8.98;
P=0.002), dominant (TT vs. TC-CC: OR, 2.64; 95% CI, 1.48–4.72;
P=0.001) and recessive (TC-CC vs. CC: OR, 3.05; 95% CI, 1.36–6.33;
P=0.007) tested inheritance models (Table II). Furthermore, the rs3746444 C
allele was identified as a risk factor for susceptibility to RA
(OR, 2.49; 95% CI, 1.63–3.81; P<0.0001).
 | Table IIFrequency distribution of hsa-mir-499
genotypes in RA patients and normal subjects. |
Table II
Frequency distribution of hsa-mir-499
genotypes in RA patients and normal subjects.
| Model | rs3746444
T>C | RA n (%) | Control n (%) | aOR (95% CI) | P-value |
|---|
| Codominant | TT | 46 (44.2) | 74 (67.3) | Ref. | - |
| TC | 32 (30.8) | 25 (22.7) | 2.11
(1.08–4.11) | 0.029 |
| CC | 26 (25.0) | 11 (10.0) | 3.88
(1.68–8.98) | 0.002 |
| Dominant | TT | 46 (44.2) | 74 (67.3) | Ref. | - |
| TC+CC | 58 (58.8) | 36 (32.7) | 2.64
(1.48–4.72) | 0.001 |
| Recessive | TT+TC | 78 (75.0) | 99 (90.0) | Ref. | - |
| CC | 26 (25.0) | 11 (10.0) | 3.05
(1.36–6.83) | 0.007 |
| Alleles | T | 124 (59.6) | 173 (78.6) | Ref. | - |
| C | 84 (40.4) | 47 (21.4) | 2.49
(1.63–3.81) | <0.0001 |
As demonstrated in Table III, the genotypes and allele
frequencies of mir-146a rs2910164 were not found to be as
significantly different between RA patients and control subjects
(χ2=0.348, P=0.841).
 | Table IIIFrequency distribution of hsa-mir-499
genotypes in RA patients and normal subjects. |
Table III
Frequency distribution of hsa-mir-499
genotypes in RA patients and normal subjects.
| Model | rs3746444
T>C | RA n (%) | Control n (%) | aOR (95%CI) | P-value |
|---|
| Codominant | GG | 57 (54.8) | 64 (59.2) | Ref. | - |
| GC | 39 (37.5) | 37 (33.6) | 1.06
(0.58–1.94) | 0.840 |
| CC | 8 (7.7) | 9 (8.2) | 0.89
(0.31–2.58) | 0.835 |
| Dominant | GG | 57 (54.8) | 64 (58.2) | Ref. | - |
| GC+CC | 47 (45.2) | 46 (41.8) | 1.03
(0.58–1.81) | 0.918 |
| Recessive | GG+GC | 96 (92.3) | 101 (91.8) | Ref. | - |
| CC | 8 (7.7) | 9 (8.2) | 0.87
(0.31–2.46) | 0.796 |
| Alleles | G | 139 (71.6) | 165 (75.0) | Ref. | - |
| C | 55 (28.4) | 55 (28.4) | 55 (28.4) | 0.504 |
Discussion
In the present study, we analyzed the correlation
between genetic polymorphisms in hsa-mir-146a rs2910164 and
hsa-mir-499 rs3746444 and susceptibility to RA in a sample of the
Iranian population. The hsa-mir-499 rs3746444 polymorphism was
revealed to be associated with an overall increased risk of RA in
codominant, dominant and recessive tested inheritance models. The
prevalence of rs3746444 TC (30.8%) and CC (25%) variants in RA
patients were identified as significantly higher than that in the
healthy individuals (22.7 and 10%, respectively) and the C allele
(minor allele) of rs3746444 was found to be as more frequent in
patients with RA than that of controls (40.4 vs. 21.4%,
respectively). However, no association was detected between the
hsa-mir-146a rs2910164 polymorphism and the risk of RA in our
population. In contrast to our findings, Yang et al(21) did not detect a significant
association between hsa-mir-499 rs3746444 polymorphisms and RA.
However, the authors observed that carriers of the CT genotype in
rs3746444 had a higher level of anti-cyclic citrullinated protein
(CCP) antibody. In agreement with results of the present study, no
significant difference was detected between the groups with respect
to the hsa-mir-146a rs2910164 polymorphism.
Polymorphisms in miRNA genes may alter a wide
spectrum of biological processes by affecting the processing and/or
target selection of miRNAs (30).
A growing number of studies have revealed that polymorphisms in
miRNA target sites affect the pathogenesis of several human
diseases, including inflammatory bowel (11) and Crohn’s disease (37), ulcerative colitis (11), psoriasis (38), COPD (39), SLE (40), MS (20) and RA (41–44).
The most severe consequence of RA is joint damage, which leads to
disability, deformity, morbidity and mortality (1,45).
The principal benefit of the detection of hsa-mir-146a and
hsa-mir-499 in circulating PBMCs is for utilization as biomarkers
to monitor the course of the disease, without the difficulty of
invasive surgical procedures to obtain joint tissues and cells for
miRNA analysis (2).
The polymorphism in the hsa-mir-499 rs3746444 has
been reported to have a marked correlation with a variety of
diseases including breast cancer (13), CSCC (14), hepatocellular carcinoma (15), RA (8), CAD (16), COPD (17) and tuberculosis (17). In their study, Yang et al
have demonstrated that carriers of the heterozygote genotype (CT)
of rs3746444 in RA exhibit higher levels of CRP and erythrocyte
sedimentation rate compared to the homozygote carriers (CC and TT),
indicative of an important role for rs3746444 in the progress and
inflammatory reaction of RA (8).
Additional investigation by the same group detected no positive
correlation between the SNPs (rs3746444 and rs2910164) and RA.
Those authors found that carriers of the genotype CT in rs3746444
had a higher level of anti-CCP antibody in RA and that the SNP
rs3746444 may be a candidate biomarker for predicting joint damage
in RA patients (21). However, no
significant association was observed between rs3746444 and risk of
several diseases, including SLE (16), schizophrenia (46), asthma (47) and colorectal (32), gallbladder (48), breast (23) and lung cancer (49).
No association was found between rs2910164 and
predisposition to RA in our population. This common hsa-mir-146a
polymorphism, rs2910164, involves a G>C nucleotide replacement
that causes change from a G:U pair to a C:U mismatch in the stem
structure of the hsa-mir-146a precursor, which affects the
specificity of mature hsa-mir-146a in binding to its targets,
resulting in an elevated expression of hsa-mir-146a (5). The hsa-mir-146a was found to be
upregulated in psoriasis (50) and
circulating PBMCs of RA patients (18), but is downregulated in SLE
(50). Hsa-mir-146a binds several
targets, including IRAK2, FADD, IRF-5, Stat-1, PTC1 and FAF1,
highlighting the important role of this miRNA in inflammation and
apoptosis processes (5).
In conclusion, the present study has shown a marked
correlation between the hsa-mir-499 rs3746444 polymorphism and
susceptibility to RA in a sample of the Iranian population.
However, no association was revealed between the hsa-mir-146a
rs2910164 variant and RA susceptibility. Consistent with growing
evidence, the present study has demonstrated that miRNA
polymorphisms may be suitable for use as diagnostic biomarkers for
RA in future.
Acknowledgements
This study was partly supported by a research grant
from the Zahedan University of Medical Sciences. The authors would
like to thank all subjects who willingly participated in the
study.
References
|
1
|
Perricone C, Ceccarelli F and Valesini G:
An overview on the genetic of rheumatoid arthritis: a never-ending
story. Autoimmun Rev. 10:599–608. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ceribelli A, Nahid MA, Satoh M and Chan
EK: MicroRNAs in rheumatoid arthritis. FEBS Lett. 585:3667–3674.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tobon GJ, Youinou P and Saraux A: The
environment, geo-epidemiology, and autoimmune disease: rheumatoid
arthritis. J Autoimmun. 35:10–14. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hashemi M, Moazeni-Roodi AK, Fazaeli A, et
al: The L55M polymorphism of paraoxonase-1 is a risk factor for
rheumatoid arthritis. Genet Mol Res. 9:1735–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chatzikyriakidou A, Voulgari PV, Georgiou
I and Drosos AA: miRNAs and related polymorphisms in rheumatoid
arthritis susceptibility. Autoimmun Rev. 11:639–641. 2012.
|
|
6
|
Hua Z, Chun W and Fang-Yuan C:
MicroRNA-146a and hemopoietic disorders. Int J Hematol. 94:224–229.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Roy S and Sen CK: MiRNA in innate immune
responses: novel players in wound inflammation. Physiol Genomics.
43:557–565. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang B, Chen J, Li Y, et al: Association
of polymorphisms in pre-miRNA with inflammatory biomarkers in
rheumatoid arthritis in the Chinese Han population. Hum Immunol.
73:101–106. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stein MT, Elias ER, Saenz M, Pickler L and
Reynolds A: Autistic spectrum disorder in a 9-year-old girl with
macrocephaly. J Dev Behav Pediatr. 31:632–634. 2010.PubMed/NCBI
|
|
10
|
Sonkoly E, Ståhle M and Pivarcsi A:
MicroRNAs: novel regulators in skin inflammation. Clin Exp
Dermatol. 33:312–315. 2008. View Article : Google Scholar
|
|
11
|
Zwiers A, Kraal L, van de Pouw Kraan TC,
Wurdinger T, Bouma G and Kraal G: Cutting edge: a variant of the
IL-23R gene associated with inflammatory bowel disease induces loss
of microRNA regulation and enhanced protein production. J Immunol.
188:1573–1577. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Murata K, Yoshitomi H, Tanida S, et al:
Plasma and synovial fluid microRNAs as potential biomarkers of
rheumatoid arthritis and osteoarthritis. Arthritis Res Ther.
12:R862010. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu Z, Liang J, Wang Z, et al: Common
genetic variants in pre-microRNAs were associated with increased
risk of breast cancer in Chinese women. Hum Mutat. 30:79–84. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhi H, Wang L, Ma G, et al: Polymorphisms
of miRNAs genes are associated with the risk and prognosis of
coronary artery disease. Clin Res Cardiol. 101:289–296. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xiang Y, Fan S, Cao J, Huang S and Zhang
LP: Association of the microRNA-499 variants with susceptibility to
hepatocellular carcinoma in a Chinese population. Mol Biol Rep.
39:7019–7023. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang J, Yang B, Ying B, et al:
Association of pre-microRNAs genetic variants with susceptibility
in systemic lupus erythematosus. Mol Biol Rep. 38:1463–1468. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li D, Wang T, Song X, et al: Genetic study
of two single nucleotide polymorphisms within corresponding
microRNAs and susceptibility to tuberculosis in a Chinese Tibetan
and Han population. Hum Immunol. 72:598–602. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pauley KM, Satoh M, Chan AL, Bubb MR,
Reeves WH and Chan EK: Upregulated miR-146a expression in
peripheral blood mononuclear cells from rheumatoid arthritis
patients. Arthritis Res Ther. 10:R1012008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chatzikyriakidou A, Voulgari PV, Georgiou
I and Drosos AA: The role of microRNA-146a (miR-146a) and its
target IL-1R-associated kinase (IRAK1) in psoriatic arthritis
susceptibility. Scand J Immunol. 71:382–385. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fenoglio C, Cantoni C, De Riz M, et al:
Expression and genetic analysis of miRNAs involved in
CD4+ cell activation in patients with multiple
sclerosis. Neurosci Lett. 504:9–12. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang B, Zhang JL, Shi YY, et al:
Association study of single nucleotide polymorphisms in pre-miRNA
and rheumatoid arthritis in a Han Chinese population. Mol Biol Rep.
38:4913–4919. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Akkiz H, Bayram S, Bekar A, Akgollu E,
Uskudar O and Sandikci M: No association of pre-microRNA-146a
rs2910164 polymorphism and risk of hepatocellular carcinoma
development in Turkish population: a case-control study. Gene.
486:104–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Catucci I, Yang R, Verderio P, et al:
Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as
low-penetrance alleles in German and Italian familial breast cancer
cases. Hum Mutat. 31:E1052–1057. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
George GP, Gangwar R, Mandal RK, Sankhwar
SN and Mittal RD: Genetic variation in microRNA genes and prostate
cancer risk in North Indian population. Mol Biol Rep. 38:1609–1615.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo H, Wang K, Xiong G, et al: A
functional variant in microRNA-146a is associated with risk of
esophageal squamous cell carcinoma in Chinese Han. Fam Cancer.
9:599–603. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jazdzewski K, Liyanarachchi S, Swierniak
M, et al: Polymorphic mature microRNAs from passenger strand of
pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci USA.
106:1502–1505. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Z, Li G, Wei S, et al: Genetic
variants in selected pre-microRNA genes and the risk of squamous
cell carcinoma of the head and neck. Cancer. 116:4753–4760. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Min KT, Kim JW, Jeon YJ, et al:
Association of the miR-146aC>G, 149C>T, 196a2C>T, and
499A>G polymorphisms with colorectal cancer in the Korean
population. Mol Carcinog. Dec 7–2011.(Epub ahead of print).
|
|
29
|
Mittal RD, Gangwar R, George GP, Mittal T
and Kapoor R: Investigative role of pre-microRNAs in bladder cancer
patients: a case-control study in North India. DNA Cell Biol.
30:401–406. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Duan R, Pak C and Jin P: Single nucleotide
polymorphism associated with mature miR-125a alters the processing
of pri-miRNA. Hum Mol Genet. 16:1124–1131. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Arnett FC, Edworthy SM, Bloch DA, et al:
The American Rheumatism Association 1987 revised criteria for the
classification of rheumatoid arthritis. Arthritis Rheum.
31:315–324. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sandoughi M, Fazaeli A, Bardestani G and
Hashemi M: Frequency of HLA-DRB1 alleles in rheumatoid arthritis
patients in Zahedan, southeast Iran. Ann Saudi Med. 31:171–173.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hashemi M, Moazeni-Roodi AK, Fazaeli A, et
al: Lack of association between paraoxonase-1 Q192R polymorphism
and rheumatoid arthritis in southeast Iran. Genet Mol Res.
9:333–339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hashemi M, Moazeni-Roodi A, Bahari A and
Taheri M: A tetra-primer amplification refractory mutation
system-polymerase chain reaction for the detection of rs8099917
IL28B genotype. Nucleosides Nucleotides Nucleic Acids. 31:55–60.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hashemi M, Eskandari-Nasab E, Fazaeli A,
et al: Association of genetic polymorphisms of
glutathione-S-transferase genes (GSTT1, GSTM1, and GSTP1) and
susceptibility to nonalcoholic fatty liver disease in Zahedan,
southeast Iran. DNA Cell Biol. 31:672–677. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hashemi M, Hoseini H, Yaghmaei P, et al:
Association of polymorphisms in glutamate-cysteine ligase catalytic
subunit and microsomal triglyceride transfer protein genes with
nonalcoholic fatty liver disease. DNA Cell Biol. 30:569–575. 2011.
View Article : Google Scholar
|
|
37
|
Chatzikyriakidou A, Voulgari PV, Georgiou
I and Drosos AA: A polymorphism in the 3′-UTR of interleukin-1
receptor-associated kinase (IRAK1), a target gene of miR-146a, is
associated with rheumatoid arthritis susceptibility. Joint Bone
Spine. 77:411–413. 2010.
|
|
38
|
Wu LS, Li FF, Sun LD, et al: A miRNA-492
binding-site polymorphism in BSG (basigin) confers risk to
psoriasis in central south Chinese population. Hum Genet.
130:749–757. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Adcock IM, Caramori G and Barnes PJ:
Chronic obstructive pulmonary disease and lung cancer: new
molecular insights. Respiration. 81:265–284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Brest P, Lapaquette P, Souidi M, et al: A
synonymous variant in IRGM alters a binding site for miR-196 and
causes deregulation of IRGM-dependent xenophagy in Crohn’s disease.
Nat Genet. 43:242–245. 2011.PubMed/NCBI
|
|
41
|
Apparailly F: Looking for microRNA
polymorphisms as new rheumatoid arthritis risk loci? Joint Bone
Spine. 77:377–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Glinsky GV: Disease phenocode analysis
identifies SNP-guided microRNA maps (MirMaps) associated with human
‘master’ disease genes. Cell Cycle. 7:3680–3694. 2008.PubMed/NCBI
|
|
43
|
Glinsky GV: SNP-guided microRNA maps
(MirMaps) of 16 common human disorders identify a clinically
accessible therapy reversing transcriptional aberrations of nuclear
import and inflammasome pathways. Cell Cycle. 7:3564–3576. 2008.
View Article : Google Scholar
|
|
44
|
Zhao ZZ, Croft L, Nyholt DR, et al:
Evaluation of polymorphisms in predicted target sites for micro
RNAs differentially expressed in endometriosis. Mol Hum Reprod.
17:92–103. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Egerer K, Feist E and Burmester GR: The
serological diagnosis of rheumatoid arthritis: antibodies to
citrullinated antigens. Dtsch Arztebl Int. 106:159–163.
2009.PubMed/NCBI
|
|
46
|
Zou M, Li D, Lv R, et al: Association
between two single nucleotide polymorphisms at corresponding
microRNA and schizophrenia in a Chinese population. Mol Biol Rep.
39:3385–3391. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Okubo M, Tahara T, Shibata T, et al:
Association study of common genetic variants in pre-microRNAs in
patients with ulcerative colitis. J Clin Immunol. 31:69–73. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Okubo M, Tahara T, Shibata T, et al:
Association between common genetic variants in pre-microRNAs and
gastric cancer risk in Japanese population. Helicobacter.
15:524–531. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tian T, Shu Y, Chen J, et al: A functional
genetic variant in microRNA-196a2 is associated with increased
susceptibility of lung cancer in Chinese. Cancer Epidemiol
Biomarkers Prev. 18:1183–1187. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li L, Chen XP and Li YJ: MicroRNA-146a and
human disease. Scand J Immunol. 71:227–231. 2010. View Article : Google Scholar : PubMed/NCBI
|