Introduction
Esophageal cancer, a type of gastrointestinal
malignancy, is common in China. Its morbidity and mortality rank
first in the world, particularly esophageal squamous cell carcinoma
(1). In esophageal cancer there is
usually high expression of cyclin E, indicating that high
expression of cyclin E is associated with esophageal cancer
(2). We speculate that cyclin
E gene silencing by small interfering RNA (siRNA) is likely to
inhibit esophageal cancer cell proliferation, which may provide a
new method for inhibiting the growth and metastasis of esophageal
cancer cells. In this study, siRNA vectors targeting the cyclin
E gene were transfected into EC9706, Eca109 and KYSE30 human
esophageal cancer cell lines, and the effects of cyclin E
gene silencing on growth, proliferation and invasion of esophageal
cancer cells were observed.
Materials and methods
Construction of siRNA vectors targeting
cyclin E
The double-stranded oligonucleotide siCE951 encoding
the corresponding shRNA was synthesized by Shanghai Sangon
Biological Engineering Co., Ltd. (China) according to the
nucleotide fragment of the cyclin E mRNA sequence (NM
057182.1) 951–969 with BamHI and HindIII incision
enzyme residues on both ends, respectively. The synthetic
oligonucleotide chain was inserted into the pRNA-U6.1/Neo vector
(GenScript, Piscataway, NJ, USA) between the BamHI and
HindIII sites, and then transformed into E. coli DH5α
for incubation. The recombinant plasmid was extracted followed by
sequencing to confirm the insertion sequence. The recombinant
vector pRNA-U6.1/Neo-siCE951 was obtained.
Cell culture and transfection
EC9706, Eca109 and KYSE30 esophageal cancer cell
lines were placed in a 24-well plate at 2×105 cells per
well in order to be incubated in RPMI-1640 medium (Gibco, Carlsbad,
CA, USA) containing 10% fetal bovine serum. When cell adhesion
reached 90–95%, pRNA-U6.1/Neo-siCE951 and pRNA-U6.1/Neo-Con
plasmids were respectively transfected into EC9706, Eca109 and
KYSE30 cells using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA,
USA). The study included experimental groups transfected with
pRNA-U6.1/Neo-siCE951 plasmid, negative control groups transfected
with pRNA-U6.1/Neo-Con plasmid and blank control groups transfected
only with liposomes. Twelve hours after transfection, the medium
was replaced by complete medium containing 600 μg/ml G418 for
4-week culture at 37°C in an atmosphere of 5% CO2 in
order to obtain stably transfected cells for future use.
Cyclin E mRNA expression detected by
RT-PCR
Total RNA from each group was extracted using Qiagen
RNeasy Mini kit (Qiagen, Hilden, Germany). The cyclin E mRNA
in each group was amplified with a One Step SYBP
PrimeScript® RT-PCR kit (Takara Biotechnology Co., Ltd.,
Dalian, China). The amplification primers of cyclin E were
5′-CGGGTCCACAGGGATGCGAAGGA-3′ and 5′-CAG GTGTGGGGATCAGGGAGCA-3′.
The amplification primers of internal control GAPDH were
5′-GCCTTCCGTGTC CCCACTGC-3′ and 5′-CAATGCCAGCCCCAGCGTCA-3′.
Amplification reaction conditions were: pre-denaturing at 94°C for
20 sec and 60°C for 60 sec, for 40 cycles. Amplification was
performed 5 times for each sample. The relative expression level of
cyclin E mRNA was indicated with the ratio of cyclin
E to internal control GAPDH.
Cyclin E protein expression detected by
western blotting
Cell disruption was performed with RIPA for protein
extraction. The protein concentration in the supernatant was
determined using the Bradford method to adjust the protein load.
The protein sample underwent SDS-PAGE, and was then transferred to
a membrane. The membrane was sealed in 25 ml of fresh nonfat dry
milk for 1 h followed by the addition of 1:800 mouse anti-cyclin
E monoclonal antibody and mouse anti-GAPDH (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA), for a 1- or 2-h
incubation at room temperature. Horseradish peroxidase-conjugated
goat anti-mouse IgG antibody (1:2000, Santa Cruz Biotechnology,
Inc.) was added for 1-h incubation followed by coloration. The gray
values of western blot bands were determined with Kodak Digital ID
Image analysis software. The relative expression level of cyclin
E protein was indicated with the gray value ratio of cyclin
E to internal control GAPDH.
Cell proliferation
EC9706, Eca109 and KYSE30 esophageal cancer cell
lines were placed in a 96-well plate at 1×104 cells per
well with 5 wells for each group. The absorbance value of each well
was determined using a cell counting kit-8 (CCK-8, Dojindo,
Shanghai, China), and then the average absorbance value was
calculated. The cell growth curve in each group was drawn with
incubation time as the horizontal axis and with OD value as the
vertical axis.
Analysis of the cell cycle using flow
cytometry
Cells in the logarithmic growth phase were placed in
a 6-well plate at a density of 1×106 cells per well.
After trypsinization, the cells were centrifuged at 800 rpm for 15
min for cell collection. The cells were resuspended in 0.4 ml PBS
followed by addition of 0.7 ml absolute alcohol containing 3% serum
for fixation at 4°C for 24 h. RNase-A was added to a final
concentration of 50 μg/ml for digestion in a water bath for 30 min
at 37°C. Propidium iodide (PI, Sigma, St. Louis, MO, USA) was added
to a final concentration of 65 μg/ml for staining in an ice bath
for 30 min away from light. After filtration with 300-screen nylon
mesh, the cells were observed with a flow cytometer. The amount of
cells in the G0/G1, S and G2/M phases were calculated. Testing was
performed in triplicate in each group.
Apoptosis detected with Annexin V/PI
double-staining flow cytometry
The cells were collected by centrifugation, and then
washed three times with 1 ml cold PBS. After centrifugation, the
cells were stained with Annexin V-FITC at room temperature in the
dark for 20 min. After cell collection, the cells were resuspended.
PI was added for staining in an ice bath for 5 min away from light.
The cells were washed with PBS and then observed with a flow
cytometer.
Soft agar colony formation assay
Agarose (1.2%) and 2X RPMI-1640 medium were mixed
(at a ratio of 1:1) in a petri dish, and then 2X antibiotics and
20% of fetal calf serum were added. Agarose (0.7%) and 2X DMEM
medium were mixed (1:1), followed by addition of 0.2 ml cells
(5×103/ml), which were added into the petri dish
mentioned above for a 10- to 14-day incubation. Ten fields were
selected to count the colony-forming units (number of cells >50
was counted as a colony-forming unit) under an inverted microscope
in each group.
Transwell culture system
The cells were adjusted at 2×105/ml in
each group 48 h following transfection. The upper chamber of
24-well Transwell Permeable Supports with 8-μm pores (Corning Cat.
no. 3422) was loaded with 200 μl cell suspension, and the lower
chamber was loaded with 500 μl medium containing 10% serum for
incubation in an atmosphere of 5% CO2 at 37°C for 48 h.
Five wells were set up for each group. The cells on the Matrigel
and in the upper chamber were collected using cotton swabs. Ten
fields were selected to be observed under a microscope and the mean
was calculated.
Statistical analysis
Statistical treatment was performed with SPSS 16.0
software. All data are expressed as the means ± SD. One-way ANOVA
was used for data analysis. Statistical significance was
established at P<0.05.
Results
Construction of siRNA vectors targeting
cyclin E
Recombinant vectors were transformed and transfected
bacteria were obtained. Sequencing for the target gene in the
recombinant plasmid was performed. Results of sequencing were
consistent with the hairpin single-strand DNA that had been
designed, demonstrating that recombinant vectors were successfully
constructed.
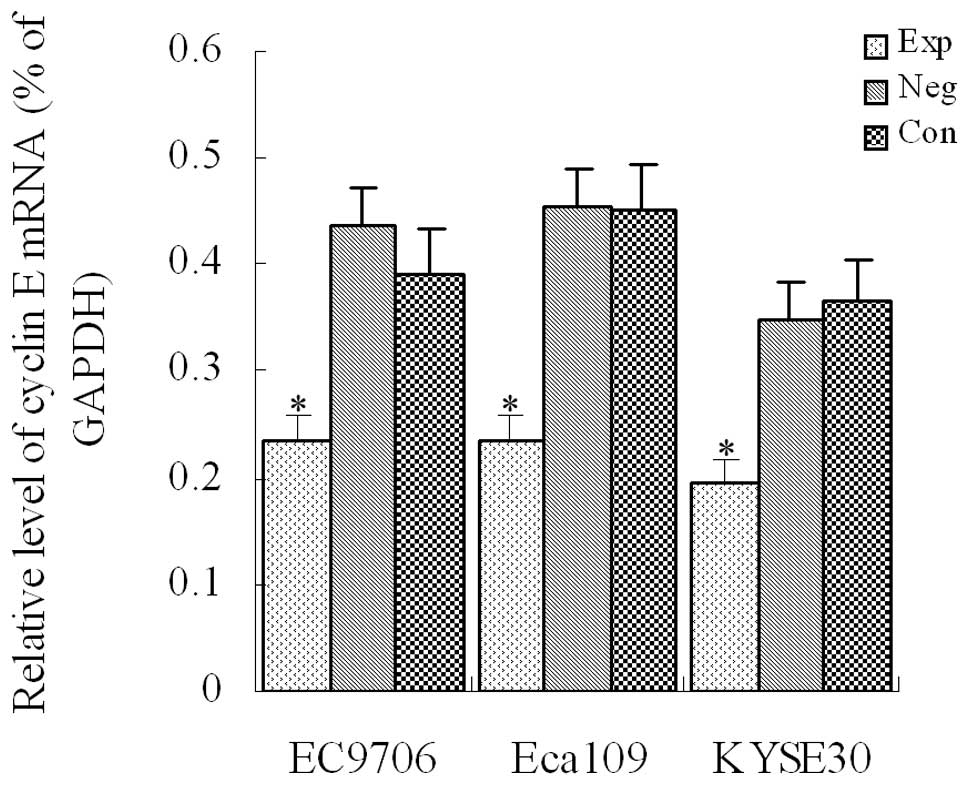
Cyclin E mRNA expression detected using
RT-PCR in transfected cells of each group
After siRNA vectors were transfected into EC9706,
Eca109 and KYSE30 cells, the level of cyclin E mRNA
expression was detected with fluorescence-quantitative PCR in each
group. Results indicated that compared with the control groups, the
level of cyclin E mRNA expression was significantly
decreased in each experimental group (P<0.01, Fig. 1), demonstrating that cyclin
E mRNA expression was significantly inhibited in EC9706, Eca109
and KYSE30 esophageal cancer cells.
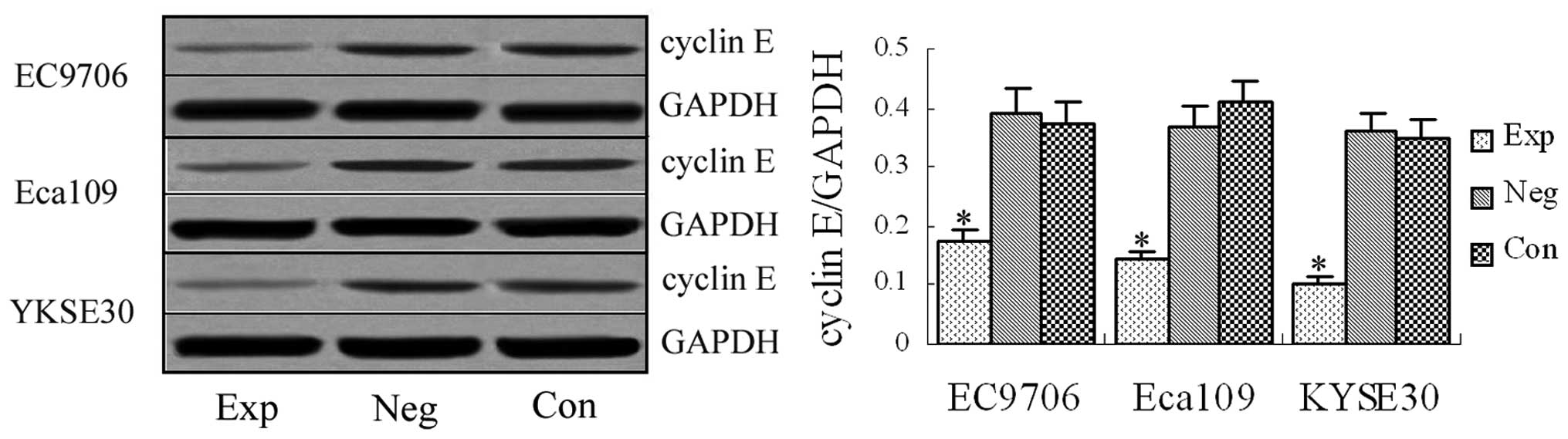
Cyclin E protein expression detected by
western blotting
Western blotting revealed the specific band at 54
kDa in the experimental groups and blank and negative control
groups (Fig. 2). The western blot
was significantly stronger in the blank and negative control groups
than in the experimental groups, demonstrating that the designed
siRNA vectors targeting cyclin E gene effectively interfered
with cyclin E protein expression in EC9706, Eca109 and
KYSE30 esophageal cancer cells.
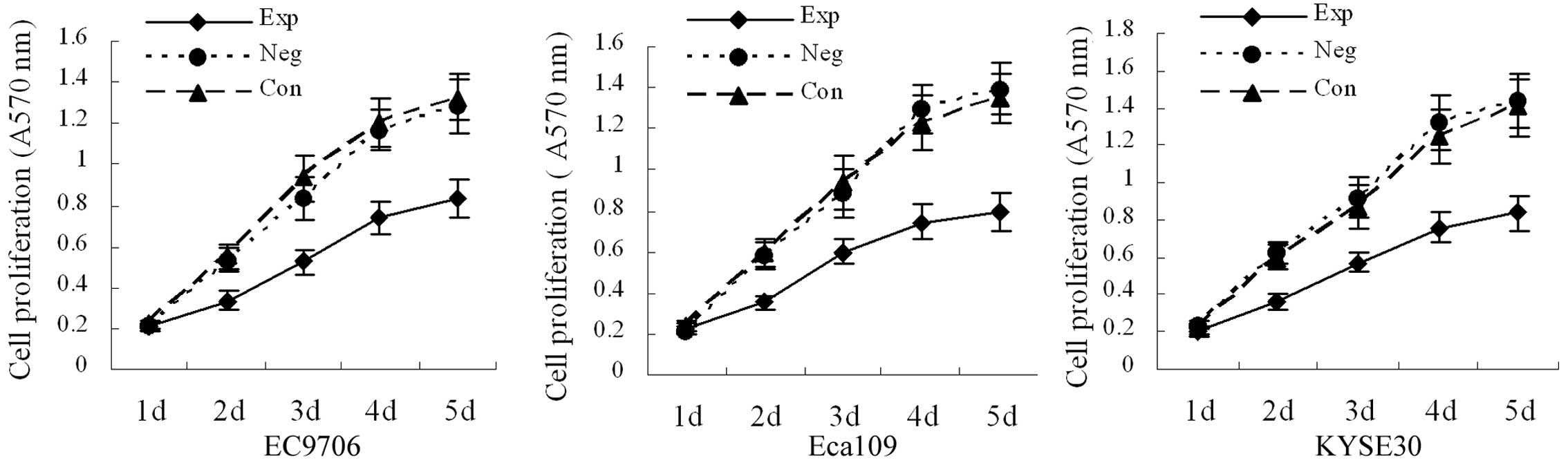
Cell proliferation
Cell growth curves are shown in Fig. 3. There was no significant
difference in absorbance values between the blank and negative
control groups (P>0.05). Compared with the blank and negative
control groups, the absorbance values on the third, fourth and
fifth day were significantly decreased in the experimental groups
(P<0.05), demonstrating that inhibition of cyclin E
expression can decrease the growth velocity of EC9706, Eca109 and
KYSE30 esophageal cancer cells.
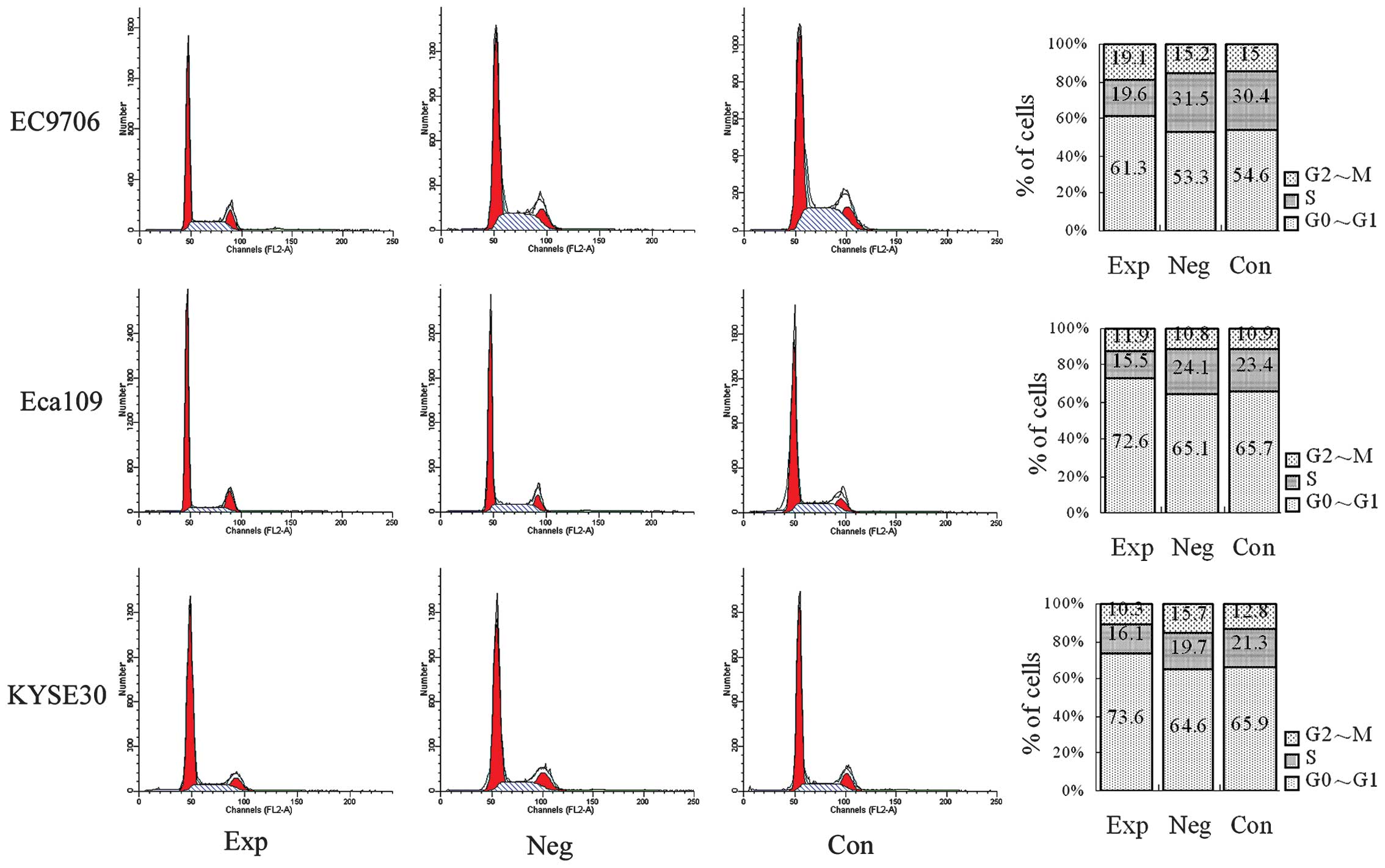
Cell cycle detected with flow
cytometry
The status of the cell cycle is shown in Fig. 4. Compared with the blank and
negative control groups, the number of cells in the S and G2/M
phases were reduced and the number of cells in the G0/G1 phase were
increased in experimental groups (P<0.05), demonstrating that
inhibiting cyclin E expression inhibits the cell cycle
process, namely that the number of cells in the S and G2/M phases
are reduced and the number of cells in the G0/G1 phase are
increased in EC9706, Eca109 and KYSE30 esophageal cancer cells.
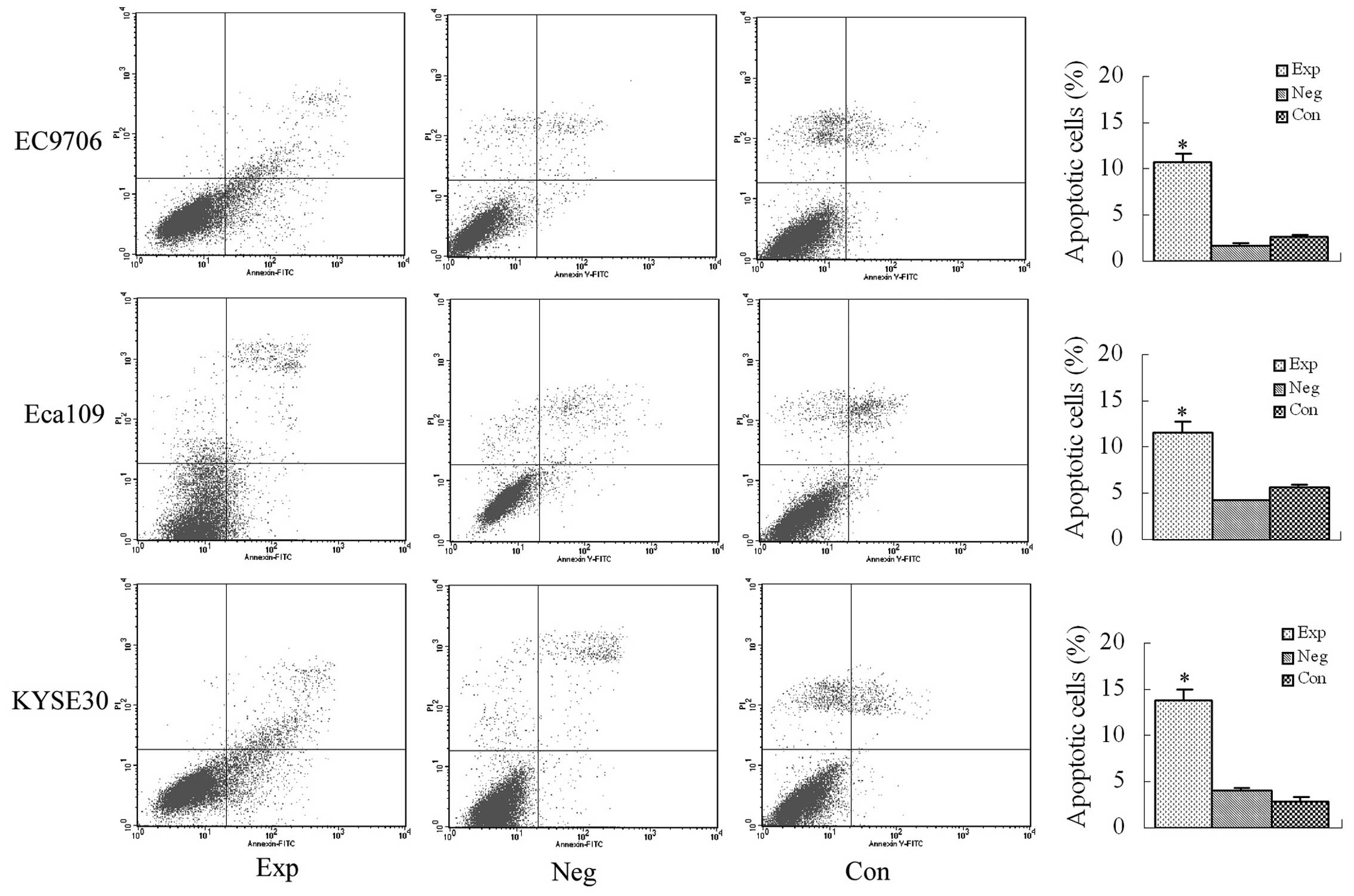
Apoptosis detected using Annexin V/PI
double-staining flow cytometry
The early apoptosis rates were 10.67% in EC9706
cells, 11.7% in Eca109 cells and 13.83% in KYSE30 cells, and they
were all significantly higher than that in the blank and negative
control groups (P<0.01, Fig.
5).
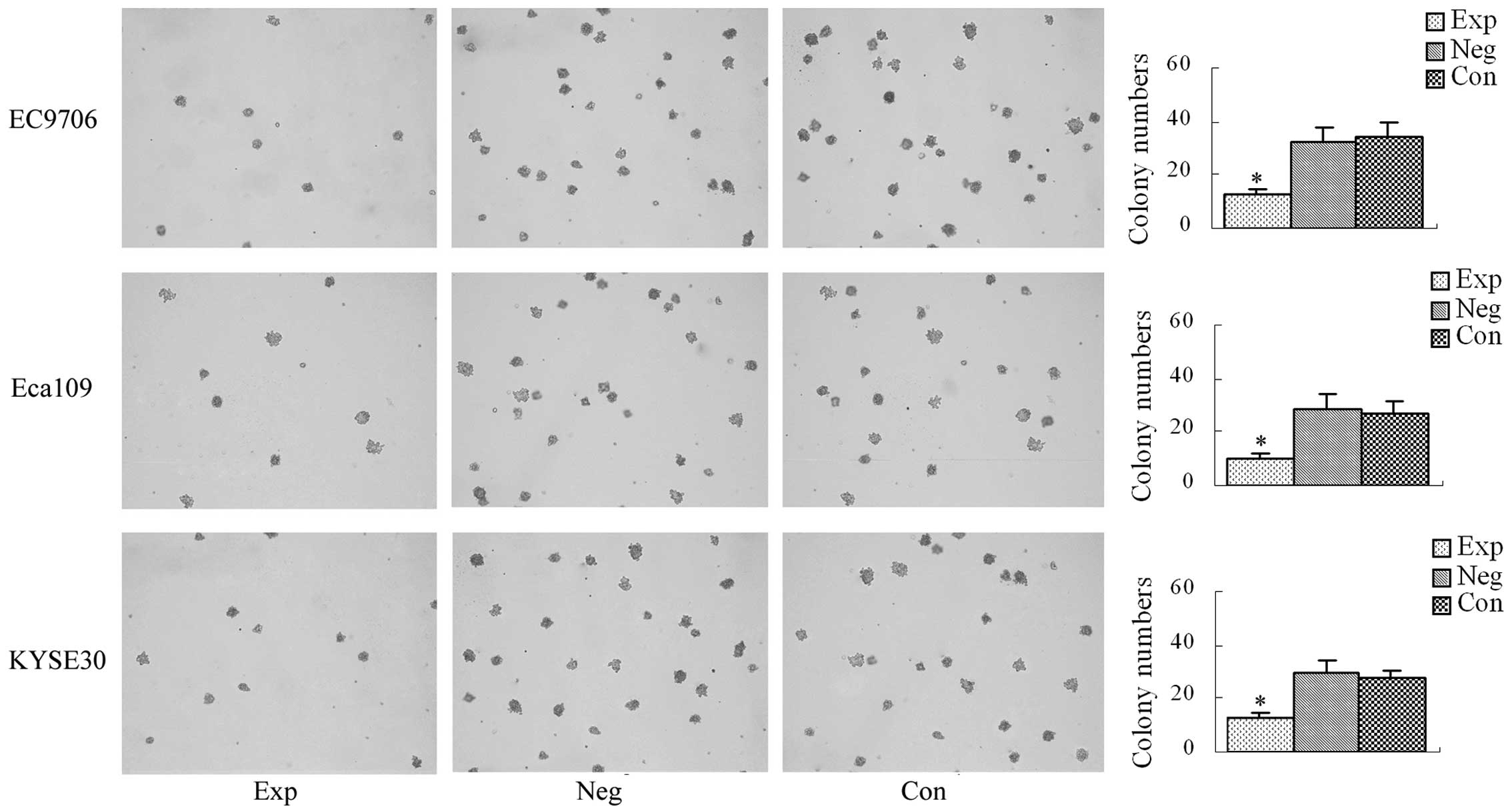
Soft agar colony formation assay
The average number of colony-forming units in the
experimental groups and the blank and negative control groups are
shown in Fig. 6. There was no
significant difference in the average number of colony-forming
units between the blank and negative control groups (P>0.05).
Compared with the blank and negative control groups, the average
number of colony-forming units was significantly decreased in the
experimental groups (P<0.05), demonstrating that inhibition of
cyclin E expression decreases the number of colony-forming
units in EC9706, Eca109 and KYSE30 esophageal cancer cells.
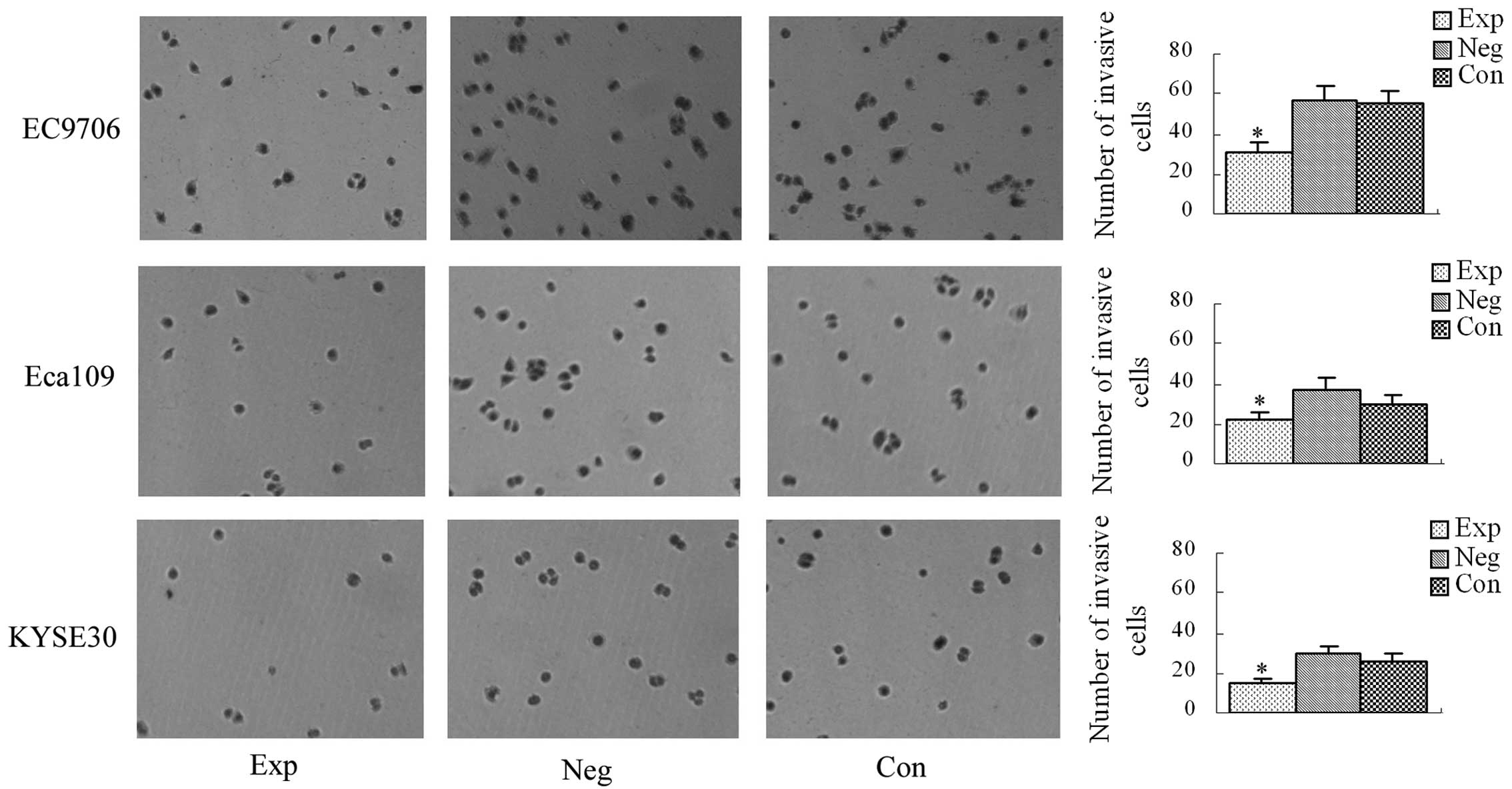
Results of the transwell culture
system
The average numbers of cells penetrating the
transwell membrane in the experimental groups and the blank and
negative control groups are shown in Fig. 7. There was no significant
difference in the average number of cells penetrating the transwell
membrane between the blank and negative control groups (P>0.05).
Compared with the blank and negative control groups, the average
number of cells penetrating the transwell membrane was
significantly decreased in the experimental groups (P<0.05),
demonstrating that inhibition of cyclin E expression
decreases the invasive ability of EC9706, Eca109 and KYSE30
esophageal cancer cells.
Discussion
Carcinogenesis is a complex, multi-factor,
multi-stage and multi-step process (3–5). One
of the important mechanisms of carcinogenesis involves disorder of
the cell cycle leading to uncontrolled cell proliferation (6,7). The
cell cycle, a highly ordered process, is regulated by a variety of
proteins, and cyclins are important in ensuring a normal cell cycle
occurs (8,9). Previously, studies on the
pathogenesis of esophageal cancer have mainly focused on
environmental, nutritional and genetic factors (10–12).
In recent years, great progress has been made in studies on the
pathogenesis of esophageal cancer at the molecular level. It has
been confirmed that the genes related to esophageal cancer include
P53, cyclin D1, VEGF, GPR39,
Wnt-1 and cyclin E(13–18).
Cyclin E, a type of G1 cyclin, was first discovered by Koff
et al in 1991 (19).
Cyclin E promotes cell cycle G1/S transition and cell
division along with CDK2 (20).
Under normal conditions, cyclin E begins to be synthesized
in G1 metaphase, reaches a peak in the G1/S phase, and rapidly
descends in the S phase; and cyclin E expression is strictly
regulated by cells. In abnormal conditions, a variety of factors
lead to cyclin E overexpression, which activates its
downstream protein to allow cell over-proliferation, leading to
tumor formation (21–23).
RNA interference technology can specifically block
or reduce the expression of the target gene and plays a role in
inhibiting tumor growth (24–27).
It has been reported that siRNA targeting cyclin E gene
silencing effectively inhibits cancer cell growth (28–31).
Based on our results, we believe that cyclin
E silencing significantly decreases the expression of cyclin
E mRNA and protein in EC9706, Eca109 and KYSE30 esophageal
cancer cells, which further inhibits overactivity of its downstream
proteins and induces cell cycle arrest at the G0/G1 phase,
inhibiting cancer cell division and retarding cancer cell growth.
The transwell assay indicated that cyclin E silencing
decreased the invasive ability of esophageal cancer cells, which
may be associated with the fact that cyclin E silencing
decreases the expression of cancer cell metastasis-related
molecules. The correlation between cyclin E and cancer cell
invasion remains to be further studied. This study provides an
experimental basis for exploring the pathogenesis and targeted
therapy of esophageal cancer.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
2
|
Fujii S, Tominaga O, Nagawa H, Tsuno N,
Nita ME, Tsuruo T and Muto T: Quantitative analysis of the cyclin
expression in human esophageal cancer cell lines. J Exp Clin Cancer
Res. 7:491–496. 1998.
|
|
3
|
Meng X, Zhong J, Liu S, Murray M and
Gonzalez-Angulo AM: A new hypothesis for the cancer mechanism.
Cancer Metastasis Rev. 31:247–268. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meng X and Riordan NH: Cancer is a
functional repair tissue. Med Hypotheses. 66:486–490. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Erdman SE and Poutahidis T: Cancer
inflammation and regulatory T cells. Int J Cancer. 127:768–779.
2010.PubMed/NCBI
|
|
6
|
Satyanarayana A and Kaldis P: Mammalian
cell-cycle regulation: several Cdks, numerous cyclins and diverse
compensatory mechanisms. Oncogene. 28:2925–2939. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sánchez I and Dynlacht BD: New insights
into cyclins, CDKs, and cell cycle control. Semin Cell Dev Biol.
16:311–321. 2005.PubMed/NCBI
|
|
8
|
Stamatakos M, Palla V, Karaiskos I,
Xiromeritis K, Alexiou I, Pateras I and Kontzoglou K: Cell cyclins:
triggering elements of cancer or not? World J Surg Oncol.
8:1112010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Malumbres M and Barbacid M: To cycle or
not cycle: a critical decision in cancer. Nat Rev Cancer.
1:222–231. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Berretta M, Lleshi A, Fisichella R,
Berretta S, Basile F, Li Volti G, Bolognese A, Biondi A, De Paoli
P, Tirelli U and Cappellani A: The role of nutrition in the
development of esophageal cancer: what do we know? Front Biosci.
4:351–357. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen YK, Lee CH, Wu IC, Liu JS, Wu DC, Lee
JM, Goan YG, Chou SH, Huang CT, Lee CY, Hung HC, Yang JF and Wu MT:
Food intake and the occurrence of squamous cell carcinoma in
different sections of the esophagus in Taiwanese men. Nutrition.
25:753–761. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Islami F, Boffetta P, Ren JS, Pedoeim L,
Khatib D and Kamangar F: High-temperature beverages and foods and
esophageal cancer risk - a systematic review. Int J Cancer.
125:491–524. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Taghavi N, Biramijamal F, Sotoudeh M,
Khademi H, Malekzadeh R, Moaven O, Memar B, A’rabi A and
Abbaszadegan MR: p16INK4a hypermethylation and p53, p16 and MDM2
protein expression in esophageal squamous cell carcinoma. BMC
Cancer. 10:1382010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shimada H, Matsushita K and Tagawa M:
Recent advances in esophageal cancer gene therapy. Ann Thorac
Cardiovasc Surg. 14:3–8. 2008.
|
|
15
|
Shimada H, Matsubara H and Ochiai T: p53
gene therapy for esophageal cancer. J Gastroenterol. 37(Suppl 14):
87–91. 2002. View Article : Google Scholar
|
|
16
|
Xie F, Liu H, Zhu YH, Qin YR, Dai Y, Zeng
T, Chen L, Nie C, Tang H, Li Y, Fu L and Guan XY: Overexpression of
GPR39 contributes to malignant development of human esophageal
squamous cell carcinoma. BMC Cancer. 11:862011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tanaka T, Ishiguro H, Kuwabara Y, Kimura
M, Mitsui A, Katada T, Shiozaki M, Naganawa Y, Fujii Y and Takeyama
H: Vascular endothelial growth factor C (VEGF-C) in esophageal
cancer correlates with lymph node metastasis and poor patient
prognosis. J Exp Clin Cancer Res. 29:832010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He J, Sheng T, Stelter AA, Li C, Zhang X,
Sinha M, Luxon BA and Xie J: Suppressing Wnt signaling by the
hedgehog pathway through sFRP-1. J Biol Chem. 281:35598–35602.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Koff A, Cross F, Fisher A, Schumacher J,
Leguellec K, Philippe M and Roberts JM: Human cyclin E, a new
cyclin that interacts with two members of the CDC2 gene family.
Cell. 66:1217–1228. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sauer K and Lehner CF: The role of cyclin
E in the regulation of entry into S phase. Prog Cell Cycle Res.
1:125–139. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mazumder S, Plesca D and Almasan A: A
jekyll and hyde role of cyclin E in the genotoxic stress response:
switching from cell cycle control to apoptosis regulation. Cell
Cycle. 6:1437–1442. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schildkraut JM, Moorman PG, Bland AE,
Halabi S, Calingaert B, Whitaker R, Lee PS, Elkins-Williams T,
Bentley RC, Marks JR and Berchuck A: Cyclin E overexpression in
epithelial ovarian cancer characterizes an etiologic subgroup.
Cancer Epidemiol Biomarkers Prev. 17:585–593. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou YJ, Xie YT, Gu J, Yan L, Guan GX and
Liu X: Overexpression of cyclin E isoforms correlates with poor
prognosis in rectal cancer. Eur J Surg Oncol. 37:1078–1084. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sharp PA: RNA interference: 2001. Genes
Dev. 15:485–490. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hannon GJ: RNA interference. Nature.
418:244–251. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Elbashir SM, Harborth J, Lendeckel W,
Yalcin A, Weber K and Tuschl T: Duplexes of 21-nucleotide RNAs
mediate RNA interference in cultured mammalian cells. Nature.
411:494–498. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
McCaffrey AP, Meuse L, Pham TT, Conklin
DS, Hannon GJ and Kay MA: RNA interference in adult mice. Nature.
418:38–39. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim DH and Rossi JJ: Strategies for
silencing human disease using RNA interference. Nat Rev Genet.
8:173–184. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jankovic R, Radulovic S and
Brankovic-Magic M: siRNA and miRNA for the treatment of cancer. J
BUON. 14(Suppl 1): S43–S49. 2009.
|
|
30
|
Gondi CS and Rao JS: Concepts in in vivo
siRNA delivery for cancer therapy. J Cell Physiol. 220:285–291.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang Y, Gao H, Lin SY, Goss JA,
Brunicardi FC and Li K: siRNA-based targeting of cyclin E
overexpression inhibits breast cancer cell growth and suppresses
tumor development in breast cancer mouse model. PLoS One.
5:e128602010. View Article : Google Scholar : PubMed/NCBI
|