Introduction
Defensins are cationic host defense polypeptides
observed in numerous organisms, including plants (1,2),
invertebrates (3), vertebrates
(4) and insects (5). The predominant functions of defensins
serve to protect the host from the invasion of various pathogens.
According to the spatial pattern of six conserved cysteine residues
they are divided into α-, β- and θ-defensin (6). β-defensins are widely distributed in
a number of organs, particularly on the epithelial surfaces of male
reproductive organs, suggesting they are involved in the resistance
to microbial colonization and possibly in male fertility. Thus far,
>40 putative β-defensins have been predicted by comprehensive
searches of the human genome. However, due to their antibacterial
activity, low molecular weight and the complicated disulfide bonds,
few β-defensins have been expressed and purified in
vitro.
Human β-defensin 6, also termed DEFB106, was
initially identified and cloned by Yamaguchi et al in 2002
(7). Two copies of DEFB106 are
located in 8p23-p22 with head-to-head orientation. The
transcription of the DEFB106 gene was highly detected in the
epididymis, testis and lung (7–9), and
was downregulated in the epididymides of non-obstructive
azoospermic men (10). Obtaining
active DEFB106 peptide was an essential step in determining the
involvement of DEFB106 in immunity and fertility. Although DEFB106
fused with thioredoxin A was expressed and purified in 2008
(11), the molecular weight,
structure, and homogeneity of DEFB106 remained poorly
characterized. In our previous study, several soluble β-defensins
(e.g., HBD1-3 and rBin1b) were successfully expressed by a
promising intein-mediated auto-cleavage expression system and the
purified recombinant mature protein products were all bioactive
with good homogeneity and structure (12,13).
This approach, not only improved the solubility and host
cytotoxicity of defensins, but also omitted exogenous proteases
required to remove the fusion tag. In the present study, the
recombinant DEFB106 protein was expressed by the intein-mediated
expression system and was well characterized. Purified DEFB106
peptides showed wide antimicrobial spectra against not only
Escherichia coli (E. coli) and Candida
albicans SC5314 (C. albicans), but also
Staphylococcus aureus CMCC26003 (S. aureus) and
demonstrated a high affinity for heparin and lipopolysaccharide
(LPS). In addition, the tissue distribution of DEFB106 was
determined by immunohistochemical staining (IHC).
Materials and methods
Plasmids and strains
pGEM-T Easy Vector systems (Promega Corporation,
Madison, WI, USA) were utilized to clone DEFB106. pTWIN1 (New
England Biolabs, Inc., Beijing, China) was used as the expression
vector. E. coli BL21 (DE3) [F-ompT hsdSB
(rB-mB)-gal dcm (DE3); Stratagene, La Jolla,
CA, USA] was used as a host for heterologous protein expression.
E. coli K12D31, S. aureus CMCC26003 and C.
albicans SC5314 were used for antimicrobial assays.
Human tissue array and epididymis
slice
Human tissue array was purchased from Shanxi
Chaoying Biotechnology Co., Ltd., (Xi’an, China). The epididymis
slice was preserved by our laboratory, and was obtained from a
healthy donor (age, 25 years) who died in a car accident. All the
procedures were approved by the Ethics Committee of Shanghai
Institutes for Biological Sciences Chinese Academy of Sciences
(Shanghai, China) and informed written consent was obtained from
the donor’s family.
Construction of expression vectors
The nucleotide sequence encoding the putative mature
DEFB106 peptide containing 45 residues (FFDEKCNKLKGTCKNNCGKNE
ELIALCQKSLKCCRTIQPCGSIID) was sub-cloned by PCR from the
pGEM-DEFB106 plasmid and inserted into the pTWIN1 vector and
digested with NcoI and XhoI (Fig. 1). The forward primer was
5′-GCGCCATGGACTTTTTTGATGAGAA-3′,
(NcoI restriction site underlined) and the reverse primer
was 5′-GGCCTCGAGTTAATCTATAATGCTC-3′
(XhoI restriction site underlined). Enzymes used in this
study were purchased from Takara Co., Ltd. (Shiga, Japan).
Protein expression, purification and
optimal pH for cleavage
All the experiments were conducted as in our
previous study, but with minor modifications. (12). The vectors were confirmed with DNA
sequencing and transformed into competent E. coli BL21 (DE3)
cells. A single colony was incubated in 3 ml LB medium
(Luria-Bertani broth) with 100 μg/ml ampicillin and cultured at
37°C and 83 × g in a shaking incubator overnight. The culture was
transferred into 30 ml LB medium at a ratio of 2% (v/v). When the
cell cultures (37°C, 83 × g) reached OD600 nm = 0.8, different
concentrations of IPTG (0.3, 0.5 and 1.0 mM) were added to induce
protein expression at different temperatures (37, 22 and 16°C). For
a large scale culture, 250 ml LB medium was induced with 0.3 mM
IPTG at OD600 = 0.8, 37°C. The cells were harvested by
centrifugation (5,000 × g for 10 min at 4°C) and were re-suspended
in 5 ml lysis buffer [20 mM sodium phosphate buffer, pH 8.5, 0.5 M
NaCl, 0.1 mM EDTA and 0.1% Triton X-100 (v/v)] and lysed by
sonication on ice. Following centrifugation at 13,000 × g for 30
min the supernatant was isolated and incubated with 1 ml chitin
beads at 4°C for 30 min with gentle shaking. Subsequent to washes
with 20 volumes of washing buffer (20 mM sodium phosphate buffer pH
8.0, 0.5 M NaCl, 0.1 mM EDTA and 0.1% Triton X-100), beads
affiliated with protein samples were equally divided into 1.5 ml
Eppendorf tubes, each containing 1.0 ml cleavage buffer (20 mM
sodium phosphate buffer, 0.5 M NaCl and 0.1 mM EDTA) with varying
pHs of 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0 and 7.5. Following
incubation overnight (>18 h) at room temperature, beads were
washed twice with cleavage buffer at the corresponding pHs, and the
supernatant was collected after centrifugation at 500 × g for 5
min. A total quantity of 10 μl chitin beads were mixed with 10 μl
2X sodium dodecyl sulphate-polyacrylamide gel electrophoresis
(SDS-PAGE) loading buffer and bathed in boiling water for 10 min.
Cleavage efficiency was estimated by SDS-PAGE and the Gel Image
system version 4.00 (Tanon Science and Technology Co., Shanghai,
China). For large-scale preparation of DEFB106, the culture volume
was expanded to 1 liter and 5 ml beads were used. Following
cleavage and elution, protein was concentrated by ultrafiltration
with an Amicon Ultra 3K device (Millipore, Billerica, MA, USA) and
applied to a Superdex-75 column by the Fast Protein Liquid
Chromatography (FPLC) system (GE Healthcare, Wasukesha, WI, USA).
Protein was eluted with 20 mM phosphate buffer, pH 7.2.
Mass spectrometry and circular dichroism
spectroscopy analysis
The molecular weight of the purified protein was
determined by matrix-assisted laser desorption ionization
time-of-flight (MALDI-TOF) mass spectra recorded on a Bruker
microFlex MALDI-TOF-MS spectrometer (ABI, Foster City, CA, USA).
The peptide sample was mixed with an equal volume of matrix 2,
5-dihydroxybenzoic acid. The instrument was operated in the
positive ion/linear mode with an accelerating voltage of 20 kV and
scanning m/z range of 3,000–12,000.
Circular dichroism spectroscopy was performed on a
Jasco J-810 spectropolarimeter (Tokyo, Japan) using a quartz cell
of 1 mm path length at 25°C with a scanning range of 200–250 nm and
scanning speed of 100 nm/min.
Antimicrobial activity assays
The antimicrobial activity of DEFB106 was determined
using the colony forming unit (CFU) assay, as described previously
(12,14). E. coli K12D31 grown in LB
medium with 50 μg/ml streptomycin at 37°C, S. aureus
CMCC26003 grown in Mueller-Hinton (MH) broth at 37°C and C.
albicans SC5314 grown in YPD (2% tryptone, 1% yeast extract and
2% glucose, at 30°C) were used as the sensitive strains. When grown
to the mid-phase, the strains were diluted to a concentration of
106 CFU/ml in the cold sterile 10 mM sodium phosphate
buffer (E. coli/S. aureus, pH 7.4; C. albican
pH 6.8). When added to recombinant DEFB106 (10 mM PBS served as the
negative control) of different concentrations, the strains were
incubated for 3 h at 37°C (E. coli and S. aureus) and
30°C (C. albicans). The cultures were then serially diluted
and cultured on LB (E. coli), MH (S. aureus) or YPD
(C. albicans) agar plates in triplicate, respectively.
Following incubation (E. coli and S. aureus at 37°C
overnight, and C. albicans at 30°C for 24 h), the surviving
colonies were manually counted, and the antimicrobial activity was
calculated using the formula: % survival = (number of surviving
colonies treatment with β-defensins/surviving colonies of control)
× 100 (13).
Heparin and LPS-binding assays
Binding assays were performed in 96-well microplates
at room temperature using the Octet Red 96 system (Fortebio, Menlo
Park, CA, USA) (15,16). Prior to executing the binding
assay, the recombinant protein was labeled with biotin at a molar
ratio of 1:1 (biotin:protein) in 20 mM PBS at pH 7.2, and the
unbound biotin was removed by Zeba™ spin desalting columns (Thermo
Fisher Scientific, Waltham, MA, USA) according to the
manufacturer’s instructions. In addition, Biosensor tips (Fortebio)
were pre-wetted with PBS for 10 min. Subsequent to this, 50 μg/ml
DEFB106-biotin, PBS and heparin (GE Healthcare Bio-Sciences AB,
Uppsala, Sweden) or lipopolysaccharide [LPS, from E. coli
O111:B4 (Sigma-Aldrich, St. Louis, MO, USA)] with serial dilutions
were added to the 96-well microtiter plates at a volume of 200
μl/well. The subsequent measurement processes were all under
computer control. Program procedures were established as follows:
For the initial step, biosensors were washed in PBS buffer for 60
sec to form a baseline; DEFB106 labeled with biotin was loaded into
biosensors for 450 sec; biosensors were moved into PBS buffer in
another line of wells for 420 sec to remove any unbound
DEFB106-biotin; the biosensors labeled with DEFB106-biotin were
exposed to heparin or LPS of different concentrations for
association, and were monitored for 300 sec; and then, the
biosensors were moved back into PBS buffer to disassociate for
another 300 sec. Data were fit globally and generated automatically
by Octet User software (version 7.0; Fortebio).
To investigate whether the affinity of DEFB106 was
based on its charges, 700 μg purified protein was incubated with
700 μl Heparin Sepharose CL-6B (GE Healthcare, Wasukesha, WI, USA)
in 20 mM Tris-HCl, pH 7.4 at 4°C for 1 h. After washing three times
with 0.5 ml buffer, heparin beads were divided into seven equal
parts, and eluted by NaCl solution of different concentration
ranging from 0 to 3.0 M. Then the beads were added to 100 μl
SDS-PAGE loading buffer and analysed on 15% SDS-PAGE gels.
IHC of DEFB106
IHC was performed with human tissue arrays and human
epididymis tissue sections to identify the distribution of DEFB106.
Primary goat antibody against the C-terminus of DEFB106 was
purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA)
and diluted with PBS containing 10% normal rabbit serum at a ratio
of 1:50 and incubated with slices overnight at 4°C. The slices were
washed five times every 5 min. The secondary antibody horseradish
peroxidase-conjugated rabbit anti-goat (dilution 1:100; Boster
Biological Technology, Ltd., Wuhan, China) was added and incubated
with slices for 1 h at room temperature. The normal goat IgG served
as a negative control. When stained with DAB (brown), the sections
were counterstained with hematoxylin (blue).
Results
Expression and purification of
recombinant DEFB106
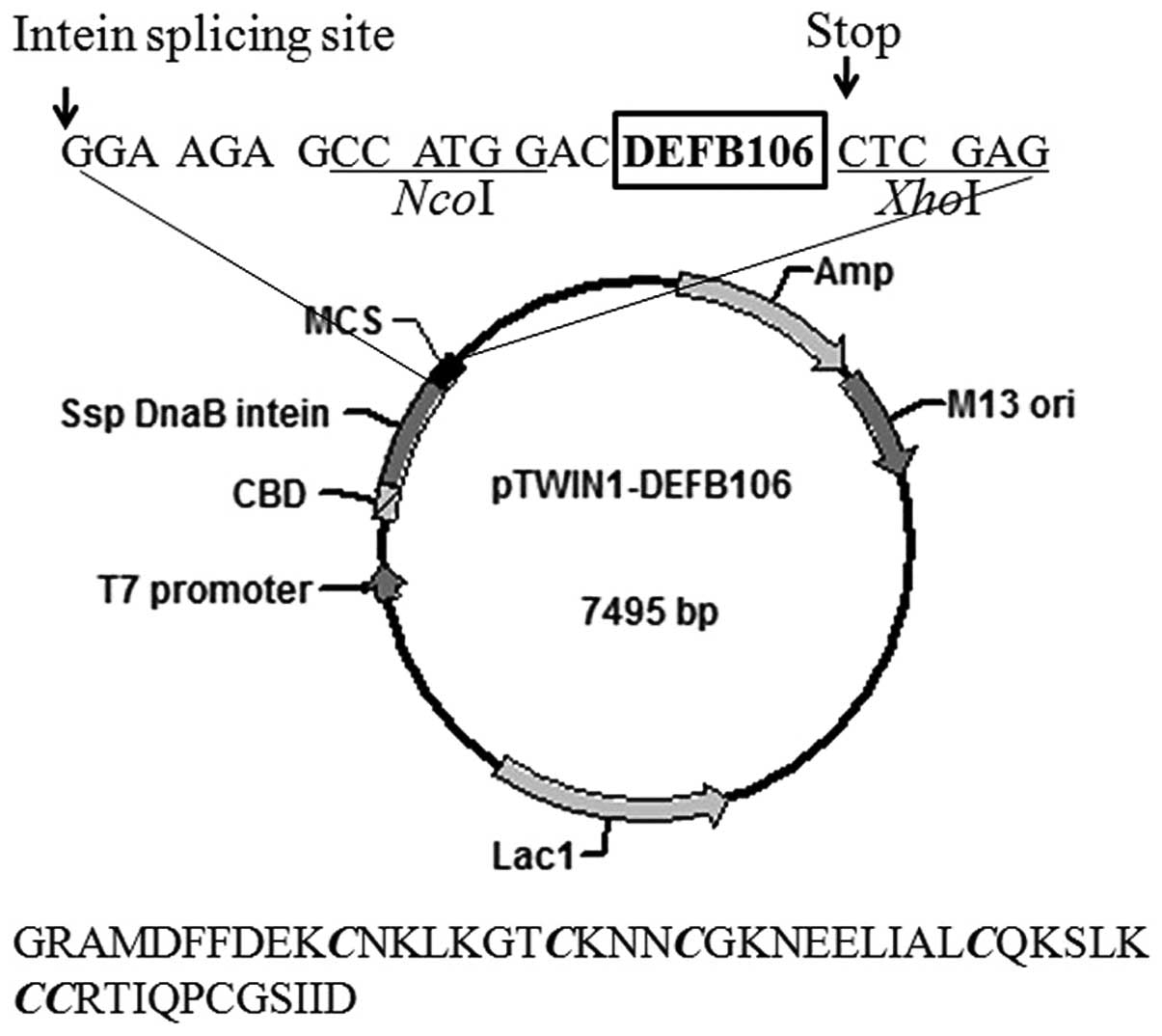
The structure of the expression plasmid, pTWIN1 is
shown in Fig. 1. The nucleotide
sequence encoding mature DEFB106 was sub-cloned and inserted into
the C-terminus of Ssp DnaB intein. The amino acid sequence of the
final eluted polypeptide is shown beneath the plasmid map in
Fig. 1. The first four amino
acids, GRAM (glycine, arginine, alanine, methionine), were derived
from pTWIN1 and the first glycine among them, adjacent to the
intein, increased the cleavage efficiency of the fusion protein. In
addition, one aspartate was added at the N-terminum of the mature
DEFB106 to avoid frame shift.
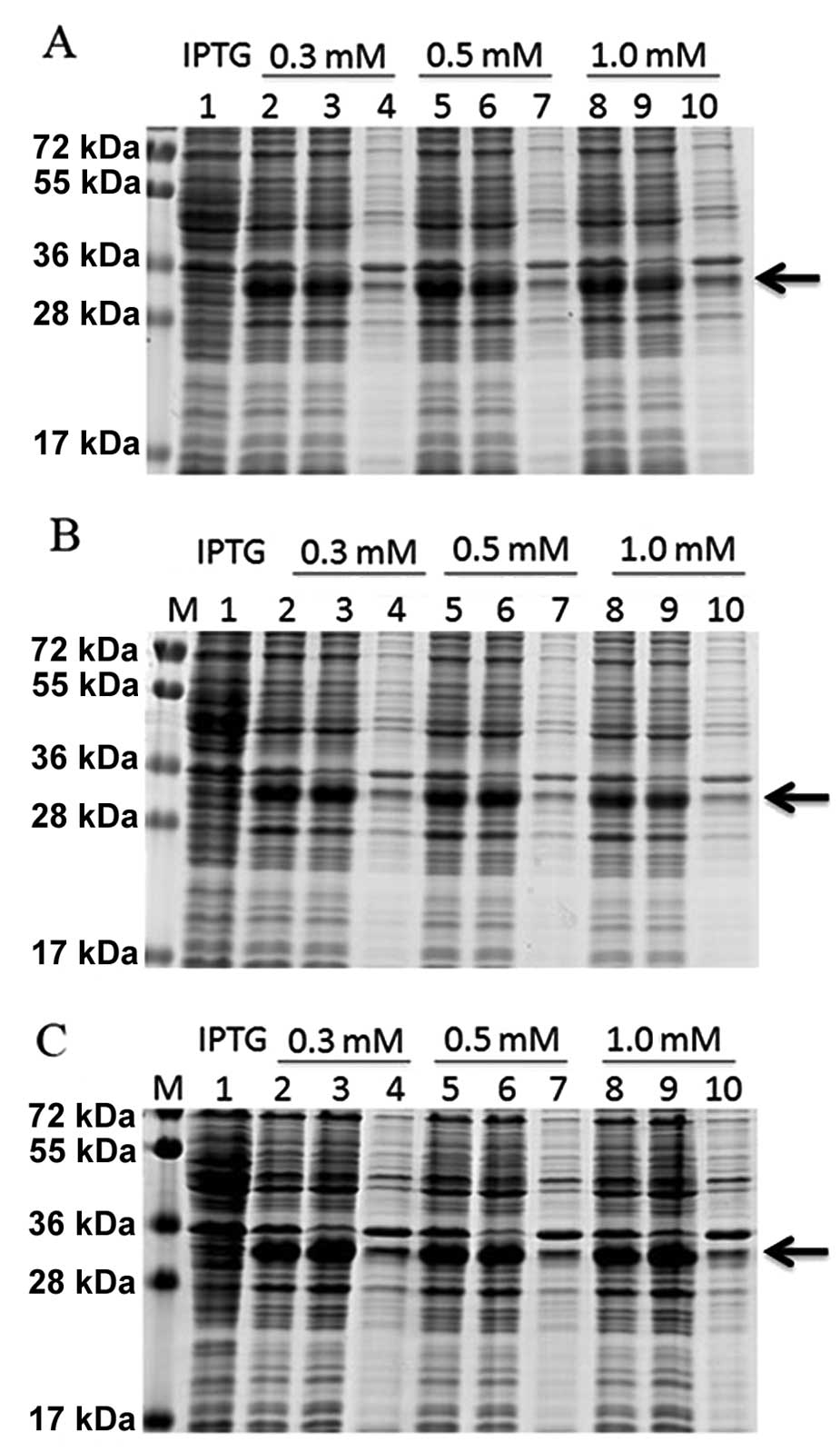
The concentration of IPTG and the temperature were
optimized to maximize the expression of the DEFB106 fusion protein.
SDS-PAGE analysis showed that the DEFB106 fusion protein was highly
soluble independent of the IPTG concentration and culture
temperature (16, 22 and 37°C). The maximum expression yield was
induced with 0.3 mM IPTG at 37°C (Fig.
2).
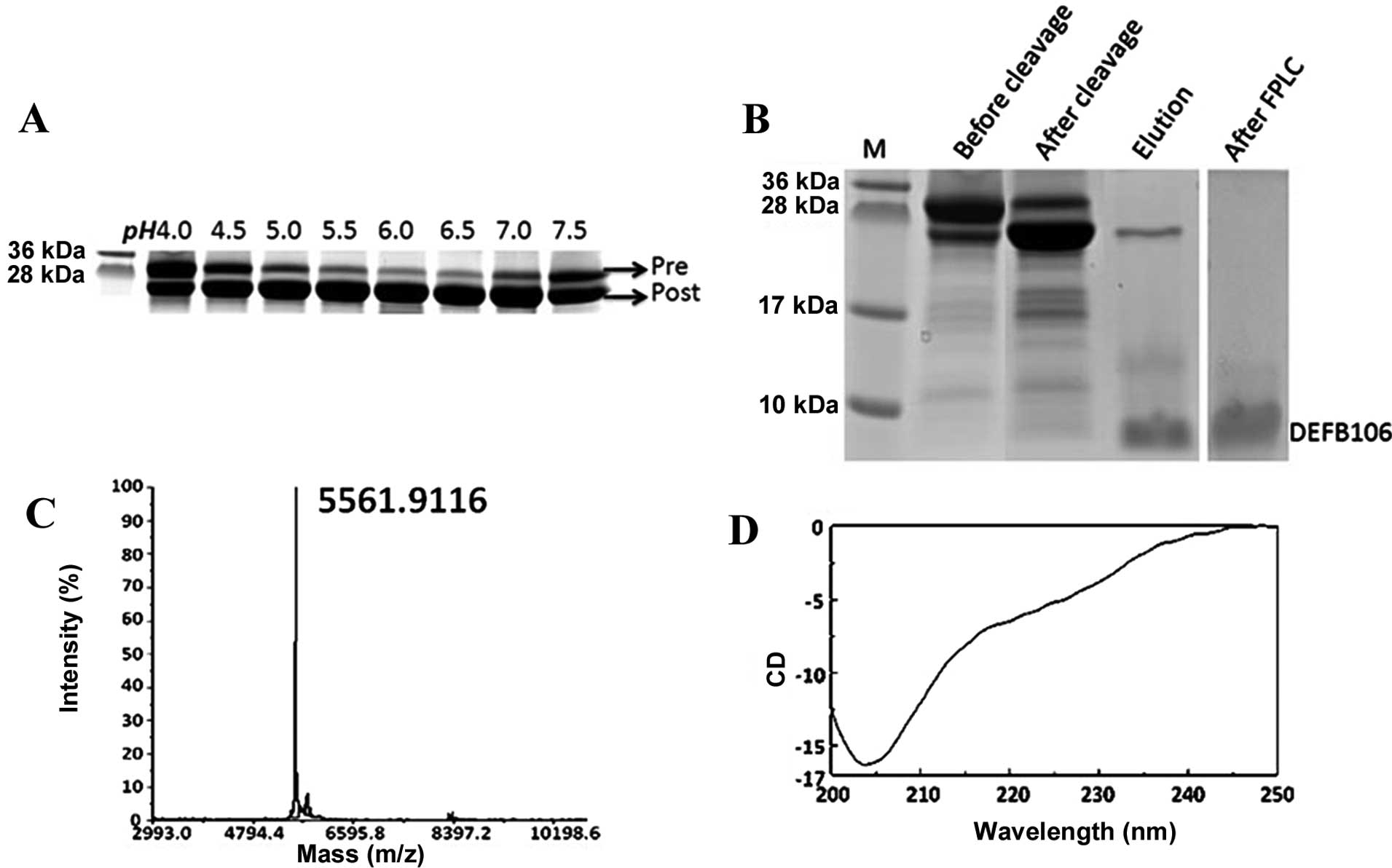
As the auto-cleavage mediated by intein is pH
sensitive (17), it was determined
that the optimal pH for auto-cleavage of intein-DEFB106 ranged from
6.0 to 6.5 (Fig. 3A). This finding
was consistent with findings of previous studies, but different
from that of other β-defensins, which range from pH 5.0 to 6.0
(12,13). The PAGE gel image analysis showed
that ~80% of DEFB106 fusion protein was cleaved (Fig. 3B). The purity of the released
DEFB106 protein was determined by Tricine-SDS-PAGE (Fig. 3B), a useful tool for the analysis
of small molecular proteins with different charges (18). Following one step affinity
purification, the purity of DEFB106 was up to 80%, and reached 95%
with a yield of 3–5 mg/l after FPLC, analyzed by computer gray scan
(Fig. 3B).
Identification of recombinant
DEFB106
To analyze the quality of the recombinant protein,
the molecular mass of the purified DEFB106 was identified by the
MALDI-TOF mass spectrometry. As shown in Fig. 3C, the measured molecular mass was
5561.9, which was in accordance with the predicted theoretical
molecular mass of 5562.6 Da. The predominant structure of DEFB106
in 20 mM phosphate buffer at pH 6.5 was estimated by Circular
dichroism spectra (Fig. 3D) to be
β-sheet (approximately accounted for 66.5%), which conformed to the
typical β-sheet structure of β-defensins.
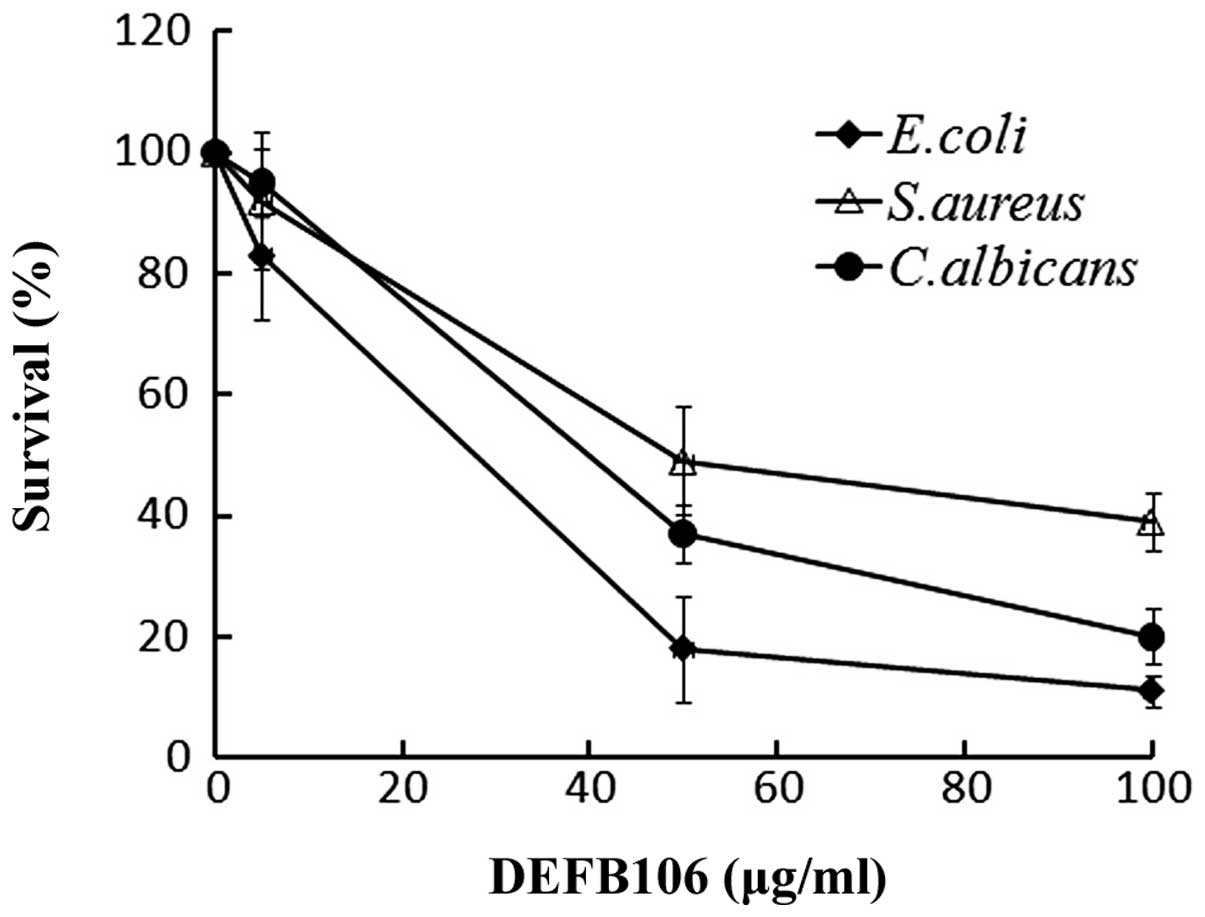
Antimicrobial activity
A broad spectrum of antimicrobial activity is a
common feature of active β-defensins. The antimicrobial activity of
the purified DEFB106 against E. coli K12D31, S.
aureus and C. albicans was determined. The growth of all
three bacterial strains was suppressed with increasing
concentrations of DEFB106 protein (Fig. 4). E. coli K12D31
(LD50 = 30 μg/ml) exhibited increased sensitivity to
DEFB106 compared with C. albicans (LD50 = 50
μg/ml) and S. aureus (LD50 = 60 μg/ml).
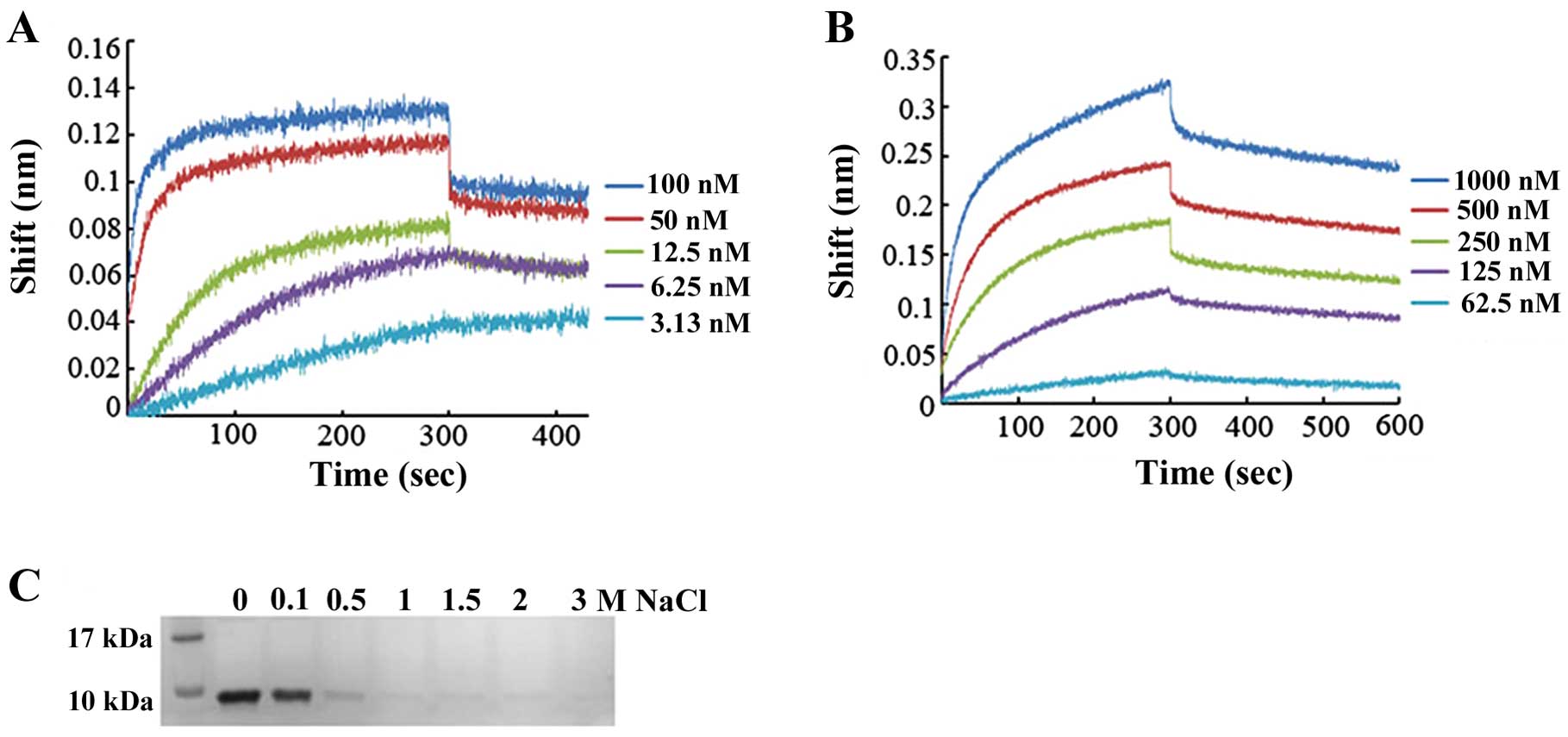
Affinity of DEFB106 for heparin and
LPS
Certain antimicrobial peptides have been shown to
bind heparin and LPS (19).
Moreover, it has been hypothesized that the affinity of several
β-defensins and related peptides for heparin-derived disaccharide
may be correlated with their antimicrobial activity (20). As defensins are cationic peptides,
whereas heparin and LPS have negative charges, there may be high
affinity between peptides and polysaccharides. The affinity of
DEFB106 for heparin and LPS was measured based on the Bio-layer
interferomertry (BLI) instrument (Octet; ForteBio). To deduce the
binding affinity via kinetic rate constants (KD =
koff/kon, where KD is the
equilibrium dissociation constant, kon the association
rate constant, and koff the dissociation rate constant),
biosensor arrays were loaded with 50 μg/ml DEFB106-biotin and moved
to heparin (concentrations ranged from 100 to 3.125 nM) or LPS
(concentrations ranged from 1,000 to 62.5 nM), respectively. As
shown in Fig. 5A and B, the
affinity of DEFB106 binding with heparin and LPS was determined to
be KD = 5.08E-11 M and KD = 1.90E-08 M. The
affinity of DEFB106 for heparin was higher than that for LPS. The
affinity of DEFB106 protein for heparin beads was also approved by
its binding and elution from heparin beads with NaCl solution as
shown in Fig. 5C, which
demonstrated that the negative charges may be predominantly
responsible for the affinity.
Localization of DEFB106 by IHC
The localization of native DEFB106 protein was
important in the investigation of the physiological and
pathological functions. By using an antibody that recognized the
C-terminus of DEFB106, IHC was used to identify the distribution of
DEFB106 protein on human tissue arrays. Notably, native DEFB106
signals were observed in the bone marrow and skin, and particularly
in the nuclei of epididymal cells (Fig. 6). In addition, DEFB106 signals in
the skin appeared to be in the nuclei of hair follicle peripheral
cells. As the high intensity of DEFB106 signals in the epididymis
were consistent with the high mRNA level, IHC of human epididymis
tissue slides were further stained to analyze the distribution of
DEFB106. The positive signals were detected through the whole
tissue from caput to cauda regions. The signals in the cauda region
were more intensive than in the caput and corpus.
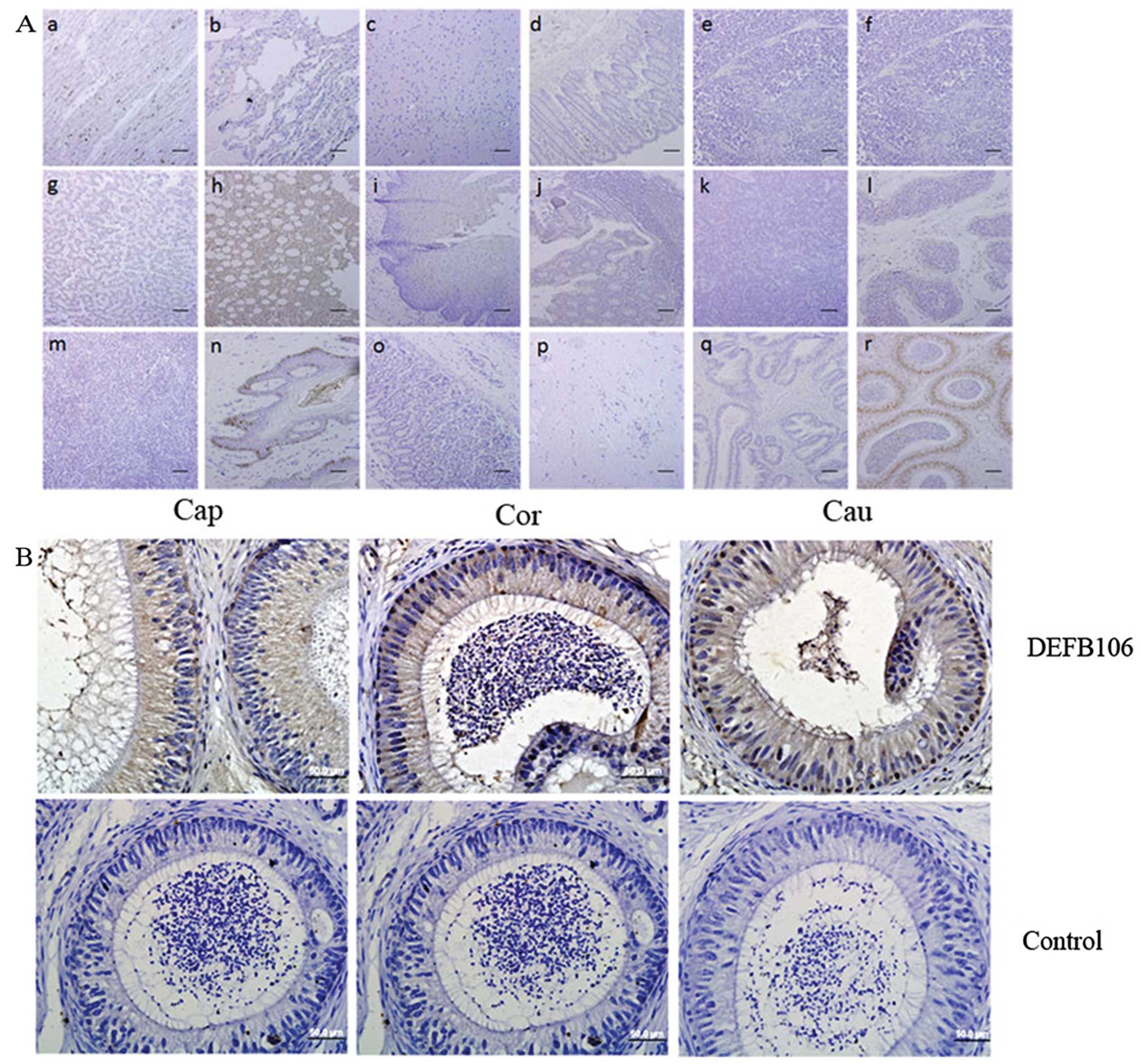 | Figure 6Distribution of native DEFB106
determined by immunohistochemistry. (A) Human tissue arrays (scale
bar, 200 μm). (Aa) Heart, (Ab) lung, (Ac) brain, (Ad) rectum, (Ae)
thymus, (Af) ovary, (Ag) liver, (Ah) bone marrow, (Ai) esophagus,
(Aj) colon, (Ak) pancreas, (Al) testis, (Am) spleen, (An) skin,
(Ao) stomach, (Ap) fibrous connective tissue, (Aq) prostate and
(Ar) epididymis. (B) Human epididymis paraffin sections (bars, 50
μm). Brown indicates DEFB106 signal and blue counterstained with
hematoxylin shows the nuclei. Cap, caput; Cor, corpus; Cau,
cauda. |
Discussion
Currently, only 13 protein products have been
confirmed among >40 open reading frames (ORFs) of putative human
β-defensins. Using the intein-mediated auto-cleavage expression
system, we not only obtained active DEFB106 protein with high
yield, but also successfully expressed and purified numerous
β-defensins, including Bin1b, HBD1, HBD2 and HBD3 (12,13).
This system allowed easy purification under mild conditions
(21) with high yield and low
cost, which renders it an option for the investigation of other
human defensins.
Although the recombinant expression of DEFB106
protein has been suggested (11),
the protein has not been well characterized. In the present study,
the molecular weight of the predominant peak of recombinant DEFB106
protein recorded by mass spectrometry, was consistent with its
calculated value. In addition, the major structure of the
recombinant DEFB106 protein in solution was the β-sheet, which was
in accordance with the common features of other β-defensins. The
DEFB106 protein produced by the intein-mediated auto-cleavage
system showed a wide antimicrobial spectrum (including E.
coli K12D31, S. aureus and C. albicans), similar
with that of other β-defensins including Bin1b (22), Defb15 (23), HBD1, HBD2 and HBD3 (24). However, Huang et al(11) demonstrated that purified DEFB106
showed antimicrobial activity against E. coli but not S.
aureus. The difference may derive from the different quality of
proteins and secondary structures in solution.
DEFB106 showed novel interaction with heparin with
high affinity, suggesting its possible involvement in innate and
adaptive immunity. Heparin is a highly sulfated glycosaminoglycan
(GAG), produced in high abundance by mammalian cells and has the
highest negative charge density. By interacting with heparin,
defensin appears to be involved in neutralizing the heparin
anticoagulation activity and the inhibition activity of
non-enzymatic fibrinolysis (25).
Additionally, defensins are involved in the release of GAGs close
to the sites of bacterial pathogen infection, suggesting that they
are involved in innate immunity (20). As with chemokine/glycosaminoglycan
(GAG) interactions, defensin-GAG complexes are likely to be
important in chemotaxis (26). In
addition, β-defensins have been shown to link innate and adaptive
immune systems (27). For example,
HBD1-4 possesses chemotactic properties by recruiting immature
dendritic cells, memory T cells and/or mast cells (28–30).
As a novel human β-defensin, DEFB106 may serve as an important
component in local tissues in combating infectious pathogens and
inhibiting non-enzymatic fibrinolysis.
DEFB106 interacted with LPS, a predominant component
of the surface of Gram-negative bacteria and led to bacterial
death, possibly by disrupting the lipid bilayers as demonstrated in
previous studies (31–33). As antimicrobial peptides are known
to bind with the negatively charged LPS-leaflet of Gram-negative
bacteria and studies have shown that β-defensin has the potential
to neutralize LPS-induced immune responses (34–36),
DEFB106 may also participate in regulating the inflammatory
response.
Mammalian β-defensins undergo rapid evolution and in
certain cases orthologous genes evolve to adapt to species-specific
functions. Native DEFB106 was localized in the nuclei of
epididymis, bone marrow and skin, which was inconsistent with its
orthologous gene Defb15 (7,23).
This finding suggests that DEFB106 may have other unknown functions
in addition to antimicrobial activity.
In conclusion, recombinant DEFB106 was prepared and
characterized with broad antimicrobial spectrum and high affinity
for heparin and LPS. The distribution of native DEFB106 was
determined predominantly in the nuclei of epididymal cells, bone
marrow and skin. The results of the present study may therefore aid
in determining the physiological and pathological functions of
DEFB106.
Acknowledgements
The authors greatly acknowledge Professor Xiangfu Wu
(Chinese Academy of Sciences, China) for the provision of E.
coli K12D31, professor Guanghua Huang (Chinese Academy of
Sciences, China) for C. albicans and professor YuanKang Ye
(Tongji Hospital, China) for S. aureus, and also thank Aihua
Liu for technical assistance in tissue section preparation. This
study was supported by grants from the Natural Science Foundation
of China (project nos. 31101030, 81270744 and 30930053) and the
Chinese Academy of Sciences [Knowledge Innovation Program Grant
(grant no. KSCX2-EW-R-07)].
References
|
1
|
Thomma BP, Cammue BP and Thevissen K:
Plant defensins. Planta. 216:193–202. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lay FT and Anderson MA: Defensins -
components of the innate immune system in plants. Curr Protein Pept
Sci. 6:85–101. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rodríguez de la Vega RC and Possani LD: On
the evolution of invertebrate defensins. Trends Genet. 21:330–332.
2005.
|
|
4
|
Lehrer RI and Ganz T: Defensins of
vertebrate animals. Curr Opin Immunol. 14:96–102. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hoffmann JA and Hetru C: Insect defensins:
inducible antibacterial peptides. Immunol Today. 13:411–415. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tang YQ, Yuan J, Osapay G, et al: A cyclic
antimicrobial peptide produced in primate leukocytes by the
ligation of two truncated alpha-defensins. Science. 286:498–502.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamaguchi Y, Nagase T, Makita R, et al:
Identification of multiple novel epididymis-specific beta-defensin
isoforms in humans and mice. J Immunol. 169:2516–2523. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kao CY, Chen Y, Zhao YH and Wu R:
ORFeome-based search of airway epithelial cell-specific novel human
β-defensin genes. Am J Respir Cell Mol Biol. 29:71–80.
2003.PubMed/NCBI
|
|
9
|
Semple CA, Rolfe M and Dorin JR:
Duplication and selection in the evolution of primate beta-defensin
genes. Genome Biol. 4:R312003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dubé E, Hermo L, Chan PT and Cyr DG:
Alterations in gene expression in the caput epididymides of
nonobstructive azoospermic men. Biol Reprod. 78:342–351.
2008.PubMed/NCBI
|
|
11
|
Huang L, Ching CB, Jiang R and Leong SS:
Production of bioactive human beta-defensin 5 and 6 in
Escherichia coli by soluble fusion expression. Protein Expr
Purif. 61:168–174. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Diao H, Guo C, Lin D and Zhang Y:
Intein-mediated expression is an effective approach in the study of
beta-defensins. Biochem Biophys Res Commun. 357:840–846. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dong J, Yu H, Zhang Y, Diao H and Lin D:
Soluble fusion expression and characterization of human
beta-defensin 3 using a novel approach. Protein Pept Lett.
18:1126–1132. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yenugu S, Chintalgattu V, Wingard CJ,
Radhakrishnan Y, French FS and Hall SH: Identification, cloning and
functional characterization of novel beta-defensins in the rat
(Rattus norvegicus). Reprod Biol Endocrinol. 4:72006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Abdiche Y, Malashock D, Pinkerton A and
Pons J: Determining kinetics and affinities of protein interactions
using a parallel real-time label-free biosensor, the Octet. Anal
Biochem. 377:209–217. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Do T, Ho F, Heidecker B, Witte K, Chang L
and Lerner L: A rapid method for determining dynamic binding
capacity of resins for the purification of proteins. Protein Expr
Purif. 60:147–150. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mathys S, Evans TC, Chute IC, et al:
Characterization of a self-splicing mini-intein and its conversion
into autocatalytic N- and C-terminal cleavage elements: facile
production of protein building blocks for protein ligation. Gene.
231:1–13. 1999. View Article : Google Scholar
|
|
18
|
Schägger H and von Jagow G: Tricine-sodium
dodecyl sulfate-polyacrylamide gel electrophoresis for the
separation of proteins in the range from 1 to 100 kDa. Anal
Biochem. 166:368–379. 1987.PubMed/NCBI
|
|
19
|
Andersson E, Rydengård V, Sonesson A,
Mörgelin M, Björck L and Schmidtchen A: Antimicrobial activities of
heparin-binding peptides. Eur J Biochem. 271:1219–1226. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
McCullough BJ, Kalapothakis JM, Chin W, et
al: Binding a heparin derived disaccharide to defensin inspired
peptides: insights to antimicrobial inhibition from gas-phase
measurements. Phys Chem Chem Phys. 12:3589–3596. 2010. View Article : Google Scholar
|
|
21
|
Banki MR, Feng L and Wood DW: Simple
bioseparations using self-cleaving elastin-like polypeptide tags.
Nat Methods. 2:659–661. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo C, Diao H, Lian Y, et al: Recombinant
expression and characterization of an epididymis-specific
antimicrobial peptide BIN1b/SPAG11E. J Biotechnol. 139:33–37. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao Y, Diao H, Ni Z, Hu S, Yu H and Zhang
Y: The epididymis-specific antimicrobial peptide β-defensin 15 is
required for sperm motility and male fertility in the rat
(Rattus norvegicus). Cell Mol Life Sci. 68:697–708.
2011.
|
|
24
|
Pazgier M, Hoover DM, Yang D, Lu W and
Lubkowski J: Human beta-defensins. Cell Mol Life Sci. 63:1294–1313.
2006. View Article : Google Scholar
|
|
25
|
Liapina LA, Kondashevskaia MV, Kokriakov
VN and Shamova OV: Interaction of heparin with defensin, a
nonenzymatic cationic protein from neutrophils. Vopr Med Khim.
38:39–42. 1992.(In Russian).
|
|
26
|
Seo ES, Blaum BS, Vargues T, et al:
Interaction of human β-defensin 2 (HBD2) with glycosaminoglycans.
Biochemistry. 49:10486–10495. 2010.
|
|
27
|
Yang D, Chertov O, Bykovskaia SN, et al:
Beta-defensins: linking innate and adaptive immunity through
dendritic and T cell CCR6. Science. 286:525–528. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen X, Niyonsaba F, Ushio H, et al:
Antimicrobial peptides human beta-defensin (hBD)-3 and hBD-4
activate mast cells and increase skin vascular permeability. Eur J
Immunol. 37:434–444. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hoover DM, Boulegue C, Yang D, et al: The
structure of human macrophage inflammatory protein-3alpha/CCL20.
Linking antimicrobial and CC chemokine receptor-6-binding
activities with human beta-defensins. J Biol Chem. 277:37647–37654.
2002. View Article : Google Scholar
|
|
30
|
Vargues T, Morrison GJ, Seo ES, et al:
Efficient production of human beta-defensin 2 (HBD2) in
Escherichia coli. Protein Pept Lett. 16:668–676. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ouellet M, Otis F, Voyer N and Auger M:
Biophysical studies of the interactions between 14-mer and 21-mer
model amphipathic peptides and membranes: insights on their modes
of action. Biochim Biophys Acta. 1758:1235–1244. 2006. View Article : Google Scholar
|
|
32
|
Shai Y: Mechanism of the binding,
insertion and destabilization of phospholipid bilayer membranes by
alpha-helical antimicrobial and cell non-selective membrane-lytic
peptides. Biochim Biophys Acta. 1462:55–70. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Buffy JJ, Hong T, Yamaguchi S, Waring AJ,
Lehrer RI and Hong M: Solid-state NMR investigation of the depth of
insertion of protegrin-1 in lipid bilayers using paramagnetic
Mn2+. Biophys J. 85:2363–2373. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Motzkus D, Schulz-Maronde S, Heitland A,
et al: The novel beta-defensin DEFB123 prevents
lipopolysaccharide-mediated effects in vitro and in vivo. FASEB J.
20:1701–1702. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu H, Yu H, Gu Y, et al: Human
beta-defensin DEFB126 is capable of inhibiting LPS-mediated
inflammation. Appl Microbiol Biotechnol. 97:3395–3408. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu H, Dong J, Gu Y, Liu H, Xin A, et al:
The novel human β-defensin 114 regulates lipopolysaccharide
(LPS)-mediated inflammation and protects sperm from motility loss.
J Biol Chem. 288:12270–12282. 2013.
|