Introduction
Breast cancer is the most common malignancy
affecting females worldwide (1),
accounting for 25% of all new cases of cancer. One in eight to one
in 12 females are likely to suffer from breast cancer during their
lifetime in developed countries and one in 22 is likely to develop
the disease in developing countries. Breast cancer incidence rate
varies at least ten-fold worldwide (2), largely due to a range of
socio-economic, reproductive, hormonal, nutritional and genetic
factors (3). Breast cancer is the
second most common cancer in females following cervical cancer in
India and has replaced cervical cancer among females in Indian
cities (4,5,6). It
is estimated that there are ~85,000 new cases of breast cancer in
India annually constituting ~25% of total cancer cases (4). Although epidemiologic investigations
have identified numerous risk factors for breast cancer, including
early menopause (7), alcohol and
tobacco consumption (8), radiation
exposure (9), obesity (10), physical activity (11,12),
urbanization (13), sedentary
lifestyle (12), dietary fat
(14), changes in reproductive
patterns (15,16) including delayed childbearing and
having fewer children, nulliparity (17,18),
lack of breast-feeding (16,18,19),
multiple abortions (19) and
post-menopausal hormone replacement therapy (20), genetic factors have also been
identified as a major contributor to the risk of developing this
disease.
Breast cancer 2, early onset (BRCA2) was the second
breast cancer susceptibility gene to be discovered and is important
in the error-free repair of DNA double strand breaks as well as
transcriptional regulation (21–23).
In normal cells, BRCA2 ensures the stability of the cell’s genetic
material (DNA) and prevents uncontrolled cell growth. Germline
mutations in BRCA2 are predicted to account for ~35% of families
with multiple cases of early onset female breast cancer and are
also associated with an increased risk of male breast cancer and
ovarian cancer (24,25). The spectrum of BRCA2 mutations has
been characterized in different populations worldwide, with
significant variation of the relative contribution of this gene to
breast cancer between populations (26,27).
A database of the mutation spectra of the BRCA2 gene has been
established (Breast Cancer Information Core, BIC; database;
http://research.nhgri.nih.gov/projects/bic/) and to
date a large number of distinct variations in this gene have been
registered in the BIC database. However, studies on BRCA2 mutations
have mainly involved hereditary breast and ovarian cancer families
which represent only a small proportion of total cases, whereas
studies focusing on sporadic breast cancer representing the
majority of total breast cancer cases remain relatively sparse.
Kashmir constitutes an ethnically pure population as
it is distinct from other regions due to intra-community marriages,
food habits, location, local environment and culture, thus
providing a genetically pure group of people, which reduces the
risk of multiple alleles that mixed populations tend to harbor. The
present study has therefore been set out to examine the frequency
of mutations in BRCA2 in a panel of 100 breast cancer patients from
the geographically and ethnically distinct population of Kashmir.
Studies that are population based may give a more representative
estimate, however only a few relatively small studies of this type
have been published from the Kashmir population.
Materials and methods
Patients and samples
After obtaining approval from the Ethics Committee
of Sher-I-Kashmir Institute of Medical Sciences (Srinagar, Jammu
and Kashmir, India), patients presenting for treatment of breast
cancer for the first time at the Sher-I-Kashmir Institute of
Medical Sciences were recruited for the study with prior informed
consent. Patients underwent fine needle aspiration cytology and
histopathological examination to establish the clinical profile.
Blood samples and surgically resected breast tissue (which included
tumor tissue, normal tissue and lymph node/s wherever involved)
were collected from 245 sporadic breast cancer cases. All samples
were snap-frozen at −70°C until analysis. A questionnaire was used
to collect the information on clinico-epidemiological
characteristics, including age, family history of disease, body
mass, menopause status, site of tumor, marital status, provisional
diagnosis, lymph node/s involved and the clinical tumor stage of
patients.
DNA extraction and PCR amplification
High molecular weight genomic DNA from tissue
samples of breast cancer patients and blood samples were isolated
by proteinase K digestion and phenol-chloroform extraction.
Air-dried DNA pellet was resuspended in Tris-EDTA and the DNA
concentration was measured by a spectrophotometric method.
PCR amplification using 11 sets of primer pairs was
used to amplify exons 2, 9, 11.1, 11.2, 11.3, 11.4, 11.5, 11.6 (for
6174delT), 18, 20 and 25 of BRCA2 as follows. Exon 2: F, CTC AGT
CAC ATA ATA AGG AAT; R,ACA CTG TGA CGT ACT GGG TTT T. Exon 9: F,
CTA GTG ATT TTA AAC TAT AAT TTT G, R, GTT CAA CTA AAC AGA GGA CT.
Exon 11.1: F, ATT TAG TGA ATG TGA TTG ATG G; R, TGA TTC TTT GCC TCT
AGA AA. Exon 11.2: F, CAA AAG TGG AAT ACA GTG ATA C; R, TCT GTT TCA
TGA AGT TCC TT. Exon 11.3: F, TTC AAA AAT AAC TGT CAA TCC; R, TCT
TTG AAG AAC ATT TTG CT. Exon 11.4: F, ACA AAT GGG CAG GAC TCT TAG
G; R, TCT GCA TTC CTC AGA AGT GG; Exon 11.5: F, GAA TCA GGA AGT CAG
TTT GA; R, TAT CAG TTG GCA TTT ATT ATT TTT. Exon 11.6: F, GGG AAG
CTT CAT AAG TCA GTC; R, TTT GTA ATG AAG CAT CTG ATA CC; Exon 18: F,
GTG ACT TGT TTA AAC AGT GGA A; R, ATT GAG CAT CCT TAG TAA GCA; Exon
20: F, AAG TGA ATA TTT TTA AGG CAG TT; R, TAT ATG GTA AGT TTC AAG
AAT; Exon 25: F, TTA GAG TTT CCT TTC TTG CAT C; R, AAG CTA TTT CCT
TGA TAC TGG A. PCR was performed in an Applied Biosysytem
Minicycler at a respective annealing temperature. PCR assays were
conducted in a reaction volume of 50 μl containing 100–120 ng of
genomic DNA, 0.2 mM of dNTPs (Sigma, St. Louis, MO, USA), 1X PCR
buffer (Sigma), 0.5 U of Taq DNA Polymerase (Sigma) and 10 pmol of
each primer (Sigma). Amplification was performed for 30–35 cycles
in a thermal cycler (Applied Biosystems Inc., Foster City, CA, USA)
with each cycle consisting of 30 sec at 95°C, 45 sec at the optimal
annealing temperature and 45 sec at 72°C, with a 10 min extension
at 72°C following the last cycle.
Statistical analysis
All statistical analyses were performed using the
SPSS program. P≤0.05 was considered to indicate a statistically
significant difference. The prevalence of BRCA2 mutations obtained
in Kashmiri patients was compared with compiled data for breast
cancer in the BIC database (www.ngri.nih.gov/intramuraresearch/BIC) and other
areas, including India.
Results
Mutational screening of commonly mutated exons
(2,9,11,18,20,25)
of BRCA2 in 242 tumor and associated normal samples of Kashmiri
breast cancer patients revealed a total of five variations, out of
which four were somatic and one was germline in nature. All somatic
variations were located in exon 11 and the germline variation was
observed in the UTR region of exon 2.
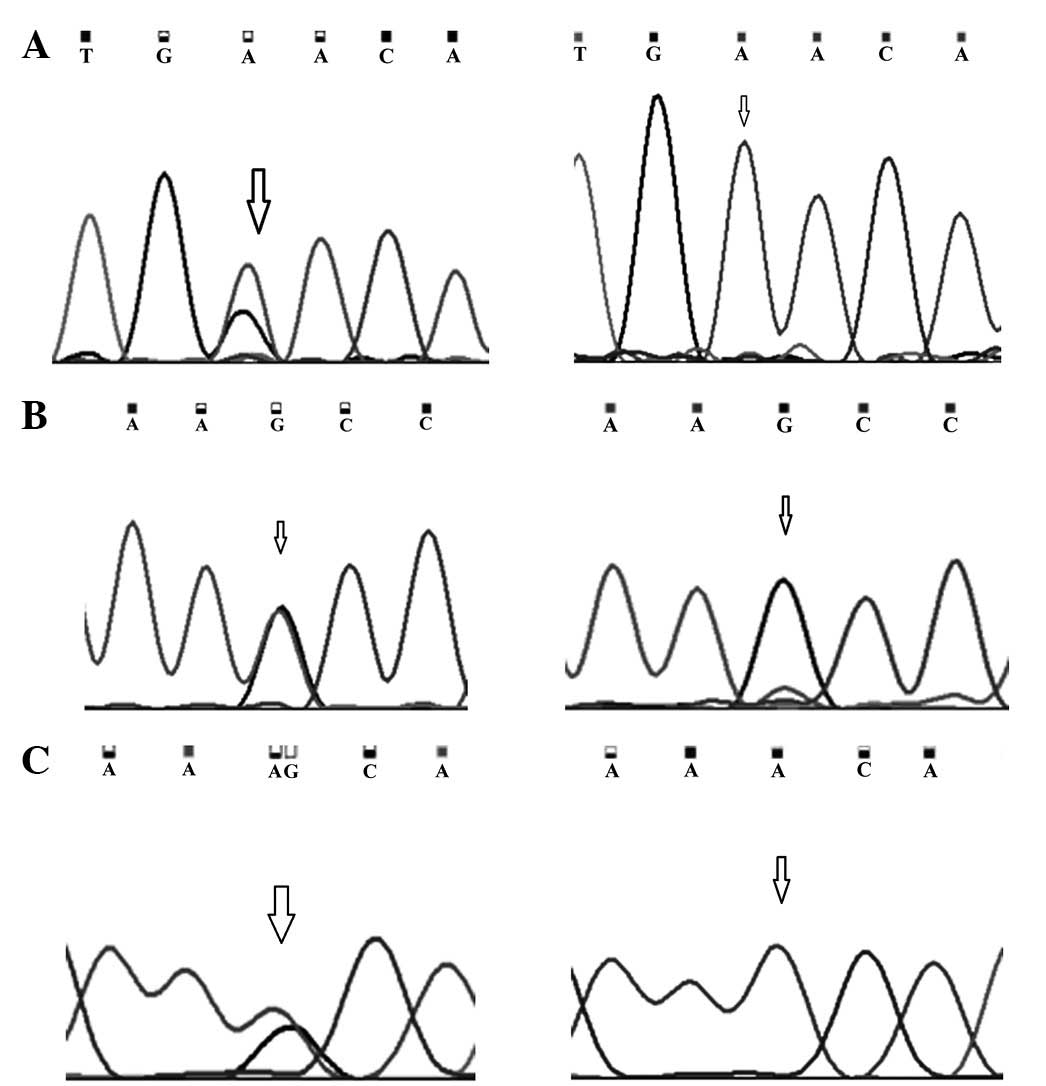
Genotype analysis revealed the presence of the
heterozygous genotype at codon positions 846 (TCC/TCA) and 868
(CCT/ACT) in 62% (150/242) of the normal breast tissue samples,
whereas the remaining 38% (92/242) had homozygous genotype (TCC/TCC
at codon 846 and CCT/CCT at codon 868). It was observed that the
heterozygous variants were replaced by homozygous genotype TCA/TCA
at codon position 846 and ACT/ACT at codon position 868, dissimilar
from the one present in normal samples in 88% of the associated
tumour samples. It was also observed that the duo change was linked
and none of the two were individually replaced in any of the
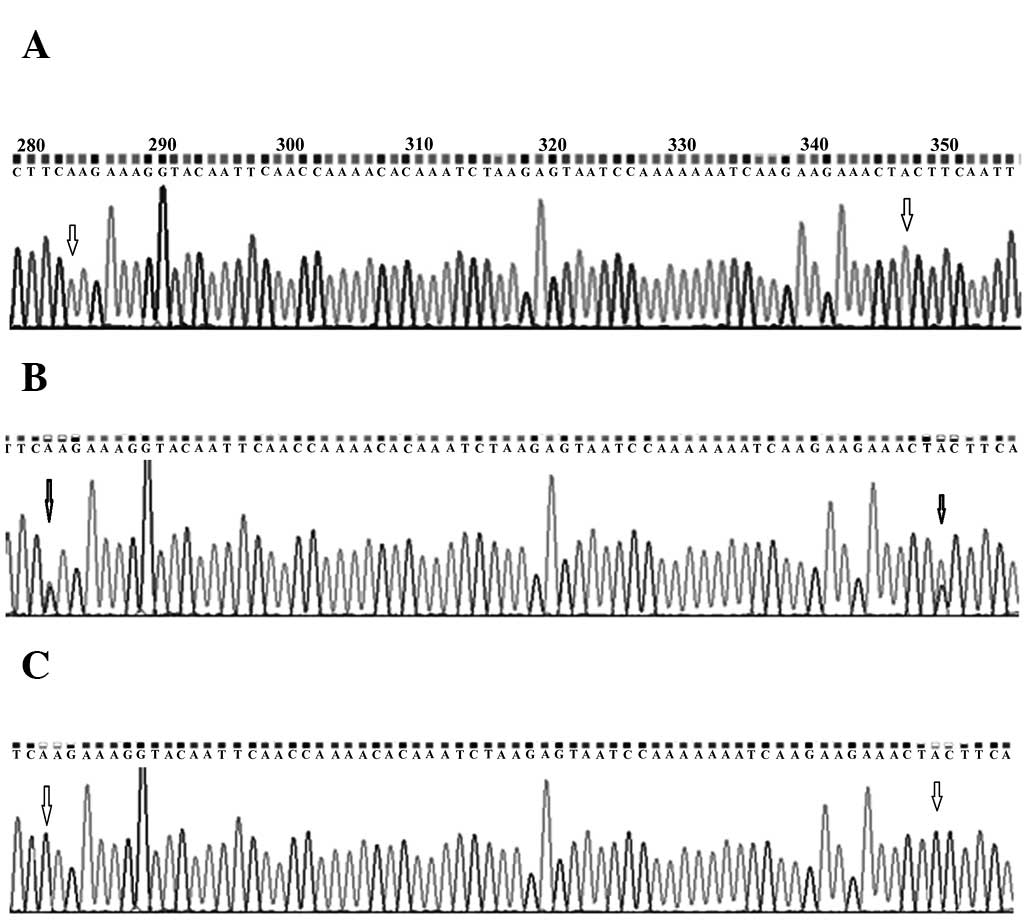
samples (Fig. 1). C>A variation
at amino acid position 846 is a silent variation and has already
been reported (rs11571654), whereas the missense variation at amino
acid position 868 leading to the replacement of proline by
threonine appeared to be novel.
Mutations observed in exon 11 at codon position 991
and 1131 are A:T>G:C transitions (Fig. 2). A somatic mutation at amino acid
position 991 (AAC/GAC) leading to the replacement of asparagine
with aspartic acid was present in 24% (58/242) of all cancer cases,
whereas the A to G (AAA/AAG) transition at amino acid position 1131
was a silent mutation and was demonstrated in 20% (48/242) of all
cancer patients. The germline variation G:C>A:T transition
observed in the UTR region of exon 2 was observed in 32% (77/242)
of all patients. The germline nature of the variation observed in
exon 2 was established as the variation observed in the tumor was
also identified in the adjacent normal tissue, blood and lymph node
of the same patient. The germline nature of the heterozygous
polymorphic variants at position 846 and 868 in normal breast
tissue was established as the variants were also observed in blood
and lymph node of the same patient.
The presence of BRCA2 mutations when compared with
various clinico-epidemiological attributes of sporadic breast
cancer patients demonstrated an association with age, menopausal
stage and advanced clinical stage of the disease, although this was
not statistically significant. However, there was no association
with early onset and late onset breast cancer cases, positive lymph
node status and the breast involved.
Discussion
Second only to lung carcinoma, breast cancer is a
leading cause of cancer mortality among females in the western
hemisphere. The lifetime risk of developing breast cancer and the
overall prognosis following diagnosis of the disease and the
likelihood of response to specific therapy is able to be determined
by exploiting the distinct characteristics of breast carcinoma. It
is quite evident that a myriad of factors, including steroid
hormones along with their receptors, peptide growth factors,
oncogenes and tumor suppressor genes are crucial in the
transformation of breast tissue (28). The contribution of the BRCA2 gene
to the incidence and prevalence of breast cancer is well
established worldwide (29,30,31,32).
However, how important BRCA2 is in pre-disposing Indians to breast
cancer is not well explored and to the best of our knowledge no
such study has been performed in the Kashmiri population. The aim
of the present study was to assess the mutational spectrum and
frequency of BRCA2 mutations as well as to explore the existence of
population specific mutations, if any, in Kashmiri breast cancer
patients. Direct nucleotide sequencing revealed somatic mutations
at codon positions 846 (rs11571654), 868 (novel), 991 (rs1799944)
and 1131 (rs1801406), and one germline variation in the UTR region
of exon 2 with rs1799943 in the BRCA2 gene.
Our studies revealed that a polymorphism at codon
868 lead to translation into either proline or threonine in 62% of
the normal breast tissue samples, whereas only proline was present
in 38% of the samples. The replacement of the heterozygous amino
acid pair proline/threonine with threonine/threonine at codon
position 868 is a novel change. This change was always linked with
the change at codon 846, where also a heterozygous set of codons
(TCC/TCA), although coding for the same amino acid serine (silent
mutation), was replaced by a homozygous pair of codons (TCA/TCA).
These changes were observed in the N-terminal region of exon 11 and
lie extremely close to the highly conserved BRCA1 repeat. This
amino acid change may be extremely significant as proline,
belonging to the non-polar aliphatic group, has been replaced by
threonine belonging to a group of polar and uncharged amino acids.
The proline reduces the structural flexibility of the protein at
that position as the secondary amino (imino) group of proline is
held in a rigid conformation, thus this substitution could be
important in altering the functional properties of the protein and
may contribute significantly in making Kashmiri population more
susceptible to breast cancer.. Although the genotypic change at
codon position 846 is a silent change, the strong linkage of the
changes at the two codon positions could be very important and
needs to be further elucidated. It may be possible that the duo
change is actually important in the transition from normal to
cancerous breast tissue.
The amino acid change at codon 991 observed in 24%
(58/242) of the patients is a missense change substituting
asparagine by aspartic acid. This amino acid lies in the BRCA1
repeat of exon 11, which constitutes one of the 4 repeats of the
BRCA2 protein that appear to be highly conserved in mammals
(33,34). These BRC repeats in the BRCA2
protein aid in directly binding BRCA2 to RAD51, a critical protein
for DNA recombination and double-stranded DNA repair (35,36,37).
The association of this missense mutation with breast cancer
susceptibility has been demonstrated in the Cyprist population
(38). The authors demonstrated a
significant association of this change with an increased risk of
breast cancer (P=0.01 and P=0.0076). A moderately strong
association of this BRCA2 polymorphism with malignant melanoma risk
has also been demonstrated (P=0.02 following Bonferroni correction)
(39). In silico prediction
methods also suggest that this is a non-tolerated amino acid
substitution within the limits of confidence in the alignments
(40). The role of this SNP in
breast cancer has also been investigated in the multi-ethnic cohort
study and no association has been demonstrated (41). The germ-line variation observed in
the UTR region of exon 2 was present in 32% (77/242) of total
patients. This variation observed is only 25 bases ahead of the
initiation codon in exon 2, thus there is a high probability that
it may be important in RNA processing. However, whether this
polymorphic variant represents a simple population variant or is
important in Kashmiri breast cancer patients needs to be elucidated
by further studies. No mutation of either germ-line or somatic
nature was observed in any other exons screened. The comparison of
various clinico-epidemiological attributes of sporadic breast
cancer patients to these somatic mutations demonstrated an
association with early onset and late onset breast cancer cases,
menopausal status and advanced clinical stage (III and IV) of the
disease. However, there was no significant association of BRCA2
mutations with positive lymph node status and the breast
involved.
Although no insertion or deletion was demonstrated
in any case, the missense variations observed in our population may
not be nullified as the prevailing polygenic model of breast cancer
risk suggests that a moderate number of genes, each conferring a
small amount of risk alone (relative risk 1.3–1.5), together
combines multiplicatively, resulting in modest susceptibility to
breast cancer (42). According to
this model, more than 100 genes may contribute to breast cancer
susceptibility. Each gene may have either common or rare variants
and females carrying more variant alleles may be at a greater risk
than those carrying fewer. Thus, there is a high possibility that
these somatic missense variants may be contributing to breast
cancer susceptibility along with variations in other low
penetrating genes in sporadic type of breast cancer in this
population cohort. Distinct characteristics of breast carcinoma may
be exploited for determining the lifetime risk of development of
the disease, the overall prognosis following a diagnosis of breast
carcinoma and the likelihood of response to specific therapy
(32). The findings from the
present study add to the body of knowledge concerning the
prevalence and nature of BRCA2 mutations in the ethnic Kashmiri
population and may inform strategies for genetic cancer risk
assessment (43). Additionally,
increased understanding of breast carcinoma pathways may enhance
our ability to produce targeted approaches for the prevention of
this disease.
Acknowledgements
This study was supported in part by the Council of
Scientific and Industrial Research (no. 9/251(12)/2004-EMR-1) and the Department of
Biotechnology, University of Kashmir (Srinagar, Jammu and Kashmir,
India). The authors gratefully acknowledge Dr Tariq Jan, Department
of Statistics, University of Kashmir (Srinagar, Jammu and Kashmir,
India) for his assistance with the statistical analysis of the
study.
Abbreviations:
|
IBC
|
inflammatory breast carcinoma
|
|
IDC
|
infiltrating ducta carcinoma
|
|
UTR
|
U-terminal region
|
References
|
1
|
Parkin DM: International variation.
Oncogene. 23:6329–6340. 2004. View Article : Google Scholar
|
|
2
|
Parkin DM, Whelan SL, Ferlay J, Raymond L
and Young J: Cancer Incidence in Five Continents. 8. IARC Press;
Lyon: 1997
|
|
3
|
McPherson K, Steel CM and Dixon JM: ABC of
breast diseases. Breast cancer-epidemiology, risk factors, and
genetics. BMJ. 321:624–628. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Indian Council of Medical Research.
National Cancer Registry programme: Consolidated report of the
population based cancer registries 1990–1996. Indian Council of
Medical Research; New Delhi: 2001
|
|
5
|
Mittra I, Badwe RA, Desai PB, Yeole BB and
Jussawalla DJ: Early detection of breast cancer in developing
countries. Lancet. 1:719–720. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rao DN and Ganesh B: Estimate of cancer
incidence in India in 1991. Indian J Cancer. 35:10–18.
1998.PubMed/NCBI
|
|
7
|
Magnusson C, Baron J, Persson I, Wolk A,
Bergström R, Trichopoulos D and Adami HO: Body size in different
periods of life and breast cancer risk in post-menopausal women.
Int J Cancer. 76:29–34. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hamajima N, Hirose K, Tajima K, et al:
Alcohol, tobacco and breast cancer-collaborative reanalysis of
individual data from 53 epidemiological studies, including 58,515
women with breast cancer and 95,067 women without the disease. Br J
Cancer. 87:1234–1245. 2002. View Article : Google Scholar
|
|
9
|
Gervais-Fagnou DD, Girouard C, Laperriere
N, Pintillie M and Goss PE: Breast cancer in women following
supradiaphragmatic irradiations for Hodgkin’s disease. Oncology.
57:224–231. 1999.
|
|
10
|
Calle EE, Rodriguez C, Walker-Thurmond K
and Thun MJ: Overweight, obesity, and mortality from cancer in a
prospectively studied cohort of U.S. adults. N Engl J Med.
348:1625–1638. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bray F, McCarron P and Parkin DM: The
changing global patterns of female breast cancer incidence and
mortality. Breast Cancer Res. 6:229–239. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Datta K and Biswas J: Influence of dietary
habits, physical activity and affluence factors on breast cancer in
East India: a case-control study. Asian Pac J Cancer Prev.
10:219–222. 2009.PubMed/NCBI
|
|
13
|
McMichael AJ and Giles GG: Cancer in
migrants to Australia: extending the descriptive epidemiological
data. Cancer Res. 48:751–756. 1988.PubMed/NCBI
|
|
14
|
Schulz M, Hoffmann K, Weikert C, Nöthlings
U, Schulze MB and Boeing H: Identification of a dietary pattern
characterized by high-fat food choices associated with increased
risk of breast cancer: the European Prospective Investigation into
Cancer and Nutrition (EPIC)-Potsdam Study. Br J Nutr. 100:942–946.
2008. View Article : Google Scholar
|
|
15
|
Collaborative Group on Hormonal Factors in
Breast Cancer. Breast cancer and breastfeeding: collaborative
reanalysis of individual data from 47 epidemiological studies in 30
countries, including 50302 women with breast cancer and 96973 women
without the disease. Lancet. 360:187–195. 2002. View Article : Google Scholar
|
|
16
|
Lord SJ, Bernstein L, Johnson KA, et al:
Breast cancer risk and hormone receptor status in older women by
parity, age of first birth, and breastfeeding: a case-control
study. Cancer Epidemiol Biomarkers Prev. 17:1723–1730. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Winer EP, Morrow M, Osborne CK and Harris
JR: Cancer of the breast. Section 2, Malignant Tumors of the
Breast. Cancer: Principles & Practice of Oncology. DeVita VT:
2. 6th edition. J.B. Lippincott; Philadelphia: pp. 1651–1659.
2000
|
|
18
|
Huo D, Adebamowo CA, Ogundiran TO, Akang
EE, Campbell O, Adenipekun A, Cummings S, Fackenthal J, Ademuyiwa
F, Ahsan H and Olopade OI: Parity and breastfeeding are protective
against breast cancer in Nigerian women. Br J Cancer. 98:992–996.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zeng Y, Xu MS, Tan SQ and Yin L: Analysis
of the risk factors of breast cancer. Nan Fang Y Yi Ke Da Xue Xue
Bao. 30:622–623. 2010.(In Chinese).
|
|
20
|
Beral V: Breast cancer and
hormone-replacement therapy in the Million Women Study. Lancet.
362:419–427. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moynahan ME, Pierce AJ and Jasin M: BRCA2
is required for homology-directed repair of chromosomal breaks. Mol
Cell. 7:263–272. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Davies AA, Masson JY, McIIwraith MJ,
Stasiak AZ, Stasiak A, Venkitaraman AR and West SC: Role of BRCA2
in control of the RAD51 recombination and DNA repair protein. Mol
Cell. 7:273–282. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pellegrini L, Yu DS, Lo T, Anand S, Lee M,
Blundell TL and Venkitaraman AR: Insights into DNA recombination
from the structure of a RAD51-BRCA2 complex. Nature. 420:287–293.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gayther SA, Mangion J, Russell P, Seal S,
Barfoot R, Ponder BA, Stratton MR and Easton D: Variation of risks
of breast and ovarian cancer associated with different germline
mutations of the BRCA2 gene. Nat Genet. 15:103–105. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thorlacius S, Olafsdottir G, Tryggvadottir
L, Neuhausen S, Jonasson JG, Tavtigian SV, Tulinius H,
Ogmundsdottir HM and Eyfjörd JE: A single BRCA2 mutation in male
and female breast-cancer families from Iceland with varied cancer
phenotypes. Nat Genet. 13:117–119. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Oddoux C, Struewing JP, Clayton CM, et al:
The carrier frequency of the BRCA2 6174delT mutation among
Ashkenazi Jewish individuals is approximately 1%. Nat Genet.
14:188–190. 1996.
|
|
27
|
Thorlacius S, Sigurdsson S, Bjarnadottir
H, Olafsdottir G, Jonasson JG, Tryggvadottir L, Tulinius H and
Eyfjord JE: Study of a single BRCA2 mutation with high carrier
frequency in a small population. Am J Hum Genet. 60:1079–1084.
1997.PubMed/NCBI
|
|
28
|
Phillips KA: Current perspectives on
BRCA1- and BRCA2-associated breast cancers. Inter Med J.
31:349–356. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Peto J, Collins N, Barfoot R, Seal S,
Warren W, Rahman N, Easton DF, Evans C, Deacon J and Stratton MR:
Prevalence of BRCA1 and BRCA2 mutations in patients with
early-onset breast cancer. J Natl Cancer Inst. 91:943–949. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hopper JL, Southey MC, Dite GS, Jolley DJ,
Giles GG, McCredie MR, Easton DF and Venter DJ: Population-based
estimate of the average age-specific cumulative risk of breast
cancer for a defined set of protein-truncating mutations in BRCA1
and BRCA2. Cancer Epidemiol Biomarkers Prev. 8:741–747.
1999.PubMed/NCBI
|
|
31
|
Thorlacius S, Struewing JP, Hartge P,
Olafsdottir GH, Sigvaldason H, Tryggvadottir L, Wacholder S,
Tulinius H and Eyfjörd JE: Population-based study of risk of breast
cancer in carriers of BRCA2 mutation. Lancet. 352:1337–1339. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Keen JC and Davidson NE: The biology of
breast carcinoma. Cancer. 97:825–833. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bignell G, Micklem G, Stratton MR,
Ashworth A and Wooster R: The BRC repeats are conserved in
mammalian BRCA2 proteins. Hum Mol Genet. 6:53–58. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Benson FE, Stasiak A and West SC:
Purification and characterization of the human Rad51 protein, an
analogue of E. coli RecA. EMBO J. 13:5764–5771. 1994.PubMed/NCBI
|
|
35
|
Donovan JW, Milne GT and Weaver DT:
Homotypic and heterotypic protein associations control Rad51
function in double-strand break repair. Genes Dev. 8:2552–2562.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hays SL, Firmenich AA and Berg P: Complex
formation in yeast double-strand break repair: participation of
Rad51, Rad52, Rad55, and Rad57 proteins. Proc Natl Acad Sci USA.
92:6925–6929. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Loizidou MA, Michael T, Neuhausen SL,
Newbold RF, Marcou Y, Kakouri E, Daniel M, Papadopoulos P, Malas S,
Hadjisavvas A and Kyriacou K: DNA-repair genetic polymorphisms and
risk of breast cancer in Cyprus. Breast Cancer Res Treat.
115:623–627. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Debniak T, Scott RJ, Górski B, et al:
Common variants of DNA repair genes and malignant melanoma. Eur J
Cancer. 44:110–114. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fackenthal JD, Sveen L, Gao Q, et al:
Complete allelic analysis of BRCA1 and BRCA2 variants in young
Nigerian breast cancer patients. J Med Genet. 42:276–281. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Freedman ML, Penney KL, Stram DO, Le
Marchand L, Hirschhorn JN, Kolonel LN, Altshuler D, Henderson BE
and Haiman CA: Common variation in BRCA2 and breast cancer risk: a
haplotype-based analysis in the Multiethnic Cohort. Hum Mol Genet.
13:2431–2441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Saxena S, Szabo CI, Chopin S, Barjhoux L,
Sinilnikova O, Lenoir G, Goldgar DE and Bhatanager D: BRCA1 and
BRCA2 in Indian breast cancer patients. Hum Mutat. 20:473–474.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hedau S, Jain N, Husain SA, Mandal AK, Ray
G, Shahid M, Kant R, Gupta V, Shukla NK, Deo SSV and Das BC: Novel
germline mutations in breast cancer susceptibility genes BRCA1,
BRCA2 and p53 gene in breast cancer patients from India. Breast
Cancer Res and Treat. 88:177–186. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Weitzel JN, Lagos V, Blazer KR, Nelson R,
Ricker C, Herzog J, McGuire C and Neuhausen S: Prevalence of BRCA
mutations and founder effect in high-risk Hispanic families. Cancer
Epidemiol Biomarkers Prev. 14:1666–1671. 2005. View Article : Google Scholar : PubMed/NCBI
|