Introduction
Large tumor suppressor (lats), which
encodes a putative serine/threonine kinase, has been identified as
a tumor suppressor gene in Drosophila (1,2).
Deterioration of the lats gene function results in promotion of
cell proliferation and tumor formation in Drosophila
(2). Two mammalian homologues of
the Drosophila lats, LATS1 and LATS2 have been
identified. LATS1-deficient mice developed soft tissue
sarcomas or ovarian stromal cell tumors, suggesting that
LATS1 is a tumor suppressor gene (3). Overexpression of LATS1 causes
G2-M arrest through the inhibition of CDC2 kinase activity in
vitro (4). Furthermore,
overexpression of LATS1 significantly suppresses
tumorigenicity in vivo by inducing apoptosis (4,5).
LATS2 overexpression results in cell cycle
arrest in the G2/M phase via inhibition of Cdc2-cyclin B kinase
activity leading eventually to apoptosis (6), inhibition of G1/S transition via
down-regulation of Cdk2-cyclin E kinase activity (7), or apoptosis via down-regulation of
Bcl-2 and Bcl-xL (8). LATS2
binds to Mdm2 and inhibits its E3 ubiquitin ligase activity,
resulting in the stabilization of p53 (9).
DNA methylation is an essential mechanism for the
regulation of genes which contain a defined CpG island, and
LATS2 hypermethylation has been recently associated with an
aggressive phenotype in breast cancers (10). Down-regulation of the LATS2
gene is associated with poor prognosis in acute lymphoblastic
leukemia (11). More recently,
LATS2 gene tumor-specific mutations and down-regulation have
been reported in non-small cell carcinoma (12). These findings have led us to analyze
the potential role of the promoter hypermethylation of the
LATS1 and 2 genes in non-small cell lung cancer
(NSCLC) patients. In this study, the methylation statuses of the
promoter regions of these genes were studied in Japanese lung
cancers. The methylation statuses of the promoter regions of
LATS1 and 2 were investigated by methylation-specific
PCR. The findings were compared to the clinicopathological features
of the lung cancer cases.
Patients and methods
Patients
The study group included lung cancer patients who
had undergone surgery at the Department of Surgery II, Nagoya City
University Medical School. The lung tumors were classified
according to the general rule for clinical and pathological
recording of lung cancer in Japan (13). All tumor samples were immediately
frozen and stored at −80°C until assayed. Since Strazisar et
al revealed that LATS2 mutations were predominantly
found in the squamous cell histotype of lung cancer while no
mutations were found in adenocarcinoma (12), we mainly focused on squamous cell
carcinoma for the LATS2 sequencing study. The clinical and
pathological characteristics of the 178 lung cancer patients for
the LATS2 sequencing analysis were as follows: 159 (89.3%)
were male and 19 were female. One hundred and sixteen (65.2%)
patients were diagnosed as squamous cell carcinomas, 42 were
adenocarcinomas and 17 were adenosquamous cell carcinomas. One
hundred and sixty-five (92.7%) were smokers and 13 were
non-smokers. The clinicopathological characteristics of the lung
cancer patients in the methylation analyses for LATS1 and
2 are listed in Tables I and
II, respectively. The samples from
these patients were previously sequenced for EGFR (13–16).
 | Table IClinicopathological data of 119 lung
cancer patients. |
Table I
Clinicopathological data of 119 lung
cancer patients.
| LATS1 gene
status | |
|---|
|
| |
|---|
| Factors | Methylated
patients | Unmethylated
patients | P-value |
|---|
| Mean age (years) | 64.5±8.9 | 66.3±13.3 | 0.2211 |
| Stage |
| I | 38 (40.0%) | 11 (45.8%) | 0.6470 |
| II–IV | 57 (60.0%) | 13 (54.2%) | |
| Lymph node
metastasis |
| N0 | 54 (56.8%) | 16 (66.7%) | 0.4880 |
| N+ | 41 (43.2%) | 8 (33.3%) | |
| Smoking |
| Never smoker | 22 (23.2%) | 11 (45.8%) | 0.0399 |
| Smoker | 73 (76.8%) | 13 (54.2%) | |
| EGFR mutation |
| Wild-type | 77 (81.1%) | 13 (54.2%) | 0.0143 |
| Mutation | 18 (18.9%) | 11 (45.8%) | |
| Pathological
subtypes |
| SCC | 36 (37.9%) | 3 (12.5%) | 0.0267 |
| Non-SCC | 59 (62.1%) | 21 (87.5%) | |
| Age |
| ≤65 | 39 (41.1%) | 9 (37.5%) | 0.8190 |
| >65 | 56 (58.9%) | 15 (62.5%) | |
| Gender |
| Male | 75 (78.9%) | 15 (62.5%) | 0.1872 |
| Female | 20 (21.1%) | 9 (37.5%) | |
 | Table IIClinicopathological data of 203 lung
cancer patients. |
Table II
Clinicopathological data of 203 lung
cancer patients.
| LATS2 gene
status | |
|---|
|
| |
|---|
| Factors | Methylated
patients | Unmethylated
patients | P-value |
|---|
| Mean age
(years) | 66.1±8.9 | 64.3±10.9 | 0.4076 |
| Stage |
| I | 70 (43.8%) | 22 (51.2%) | 0.3946 |
| II–IV | 90 (56.2%) | 21 (48.8%) | |
| Lymph node
metastasis |
| N0 | 95 (59.4%) | 27 (62.8%) | 0.7286 |
| N+ | 65 (40.6%) | 16 (37.2%) | |
| Smoking |
| Never smoker | 35 (21.9%) | 14 (32.6%) | 0.1622 |
| Smoker | 125 (78.1%) | 29 (67.4%) | |
| EGFR mutation |
| Wild-type | 126 (78.8%) | 35 (81.4%) | 0.8332 |
| Mutation | 34 (21.2%) | 8 (18.6%) | |
| Pathological
subtypes |
| SCC | 80 (50.0%) | 18 (41.9%) | 0.3920 |
| Non-SCC | 80 (50.0%) | 25 (58.1%) | |
| Age |
| ≤65 | 68 (42.5%) | 22 (51.2%) | 0.3876 |
| >65 | 92 (57.5%) | 21 (48.8%) | |
| Gender |
| Male | 123 (76.9%) | 31 (72.1%) | 0.5490 |
| Female | 37 (23.1%) | 12 (27.9%) | |
PCR assays for LATS2 mutations
Total RNA was extracted from lung cancer tissues
using the Isogen Kit (Nippon Gene, Tokyo, Japan), according to the
manufacturer’s instructions. The RNA concentration was determined
using a spectrophotometer and adjusted to a concentration of 200
ng/ml. Approximately 10 cases were excluded for each assay, since
the tumor cells were too few to sufficiently extract tumor RNA. RNA
(1 μg) was reverse transcribed by Superscript II enzyme (Gibco BRL,
Gaithersburg, MD, USA) with 0.5 μg oligo (dT)12–16
(Amersham Pharmacia Biotech Inc., Piscataway, NJ, USA). The
reaction mixture was incubated at 42°C for 50 min and then at 72°C
for 15 min. We then used 1 μl of each DNA for PCR analysis. The PCR
reactions were performed using the LA-Taq Kit (Takara Bio Inc.,
Shiga, Japan) in a 25-μl reaction volume. The primer sequences for
the LATS2 gene for exon 8 (including the S1073 region) were
as follows: forward primer 5-CGACCCCGTAGATGAAGAAA-3 and reverse
primer 5-AGCGATGCTGAGTCCTGTT-3 (454 bp, 3448–3901). The cycling
conditions were as follows: initial denaturation at 94°C for 5 min,
followed by 40 cycles at 94°C for 45 sec, 60°C for 45 sec and 72°C
for 45 sec. The products were purified using the Qiagen PCR
Purification Kit (Qiagen, Valencia, CA, USA). These samples were
sequenced by ABI PRISM 3100 analyzer (Applied Biosystems Japan
Ltd., Tokyo, Japan) and analyzed by BLAST and chromatograms by
manual review.
Methylation-specific polymerase chain
reaction analysis
DNA was prepared from tissue samples using the
standard methods, and bisulfite modification of genomic DNA was
performed using the MethylCode Bisulfite Conversion Kit
(Invitrogen). Briefly, 500 ng of genomic DNA was denatured by
incubation with CT Conversion Reagent for 10 min at 98°C, followed
by 2.5 h at 68°C and 4°C for several minutes. Modified DNA was
purified using a spin column and then eluted with dlution
buffer.
The primer sequences for the LATS1 gene for
methylated (M) sequences were as follows: forward primer 5-GGAGTT
CGTTTTGTC-3 and reverse primer 5-CGACGTAATAACG AACGCCTA-3. The
primer sequences for the LATS1 gene for unmethylated (U)
sequences were as follows: forward primer 5-TAGGTTGGAGTGTGGTGGT-3
and reverse primer 5-CCC AACATAATAACAAACACCT-3. The primer
sequences for the LATS2 gene for methylated (M) sequences
were as follows: forward primer 5-ATTTCGGTTTATTGTAATTTTC-3 and
reverse primer 5-AACCAACATAATAAAACCCCG-3. The primer sequences for
the LATS2 gene for unmethylated (U) sequences were as
follows: forward primer 5-TTTGTTTTTT GGGTTTAAGT-3 and reverse
primer 5-CCAACATAATA AAACCCCA-3. The cycling conditions were as
follows: initial denaturation at 94°C for 5 min, followed by 40
cycles at 94°C for 45 sec, 58°C (LATS1 and LATS2, M)
or 53°C (LATS1, U) or 50°C (LATS2, U) for 45 sec, and
72°C for 45 sec.
Statistical analysis
Statistical analyses were carried out using the
Mann-Whitney U test for unpaired samples and the Wilcoxon’s signed
rank test for paired samples. Linear relationships between
variables were determined by means of simple linear regression.
Correlation coefficients were determined by rank correlation using
the Spearman’s and χ2 tests. The overall survival of
lung cancer patients was examined by the Kaplan-Meier method, and
differences were examined by the log-rank test. Analysis was
carried out using the Stat-View software package (Abacus Concepts
Inc., Berkeley, CA, USA), and differences were considered
significant at a p-value <0.05.
Results
LATS2 gene mutation status in Japanese
lung cancer patients
We sequenced for the exon 8 of the LATS2 gene
in 178 NSCLC samples. No mutations were found in the 178 patients
from the direct sequencing using cDNA samples.
LATS gene methylation statuses in
Japanese lung cancer patients
Methylation-specific PCR showed that the
LATS1 promoter region was hypermethylated in 95 out of 119
(79.8%) lung cancers. The methylation status of LATS1 was
significantly associated with squamous histology (squamous cell
carcinoma, 92.3% vs. non-squamous cell carcinoma, 73.8%; p=0.0267)
and smoking status (never-smoker, 66.7% vs. smoker, 84.9%;
p=0.0399). However, LATS1 methylation status did not
correlate with gender (p=0.1872), age (p=0.8190), lymph node EGFR
metastasis (p=0.4880) and pathological stages (I vs. II–IV;
p=0.6470). LATS1 ummethylated patients harbored more EGFR
mutations (p=0.0143). LATS1 methylation status did not
correlate with patient survival (log-rank test; p=0.4109).
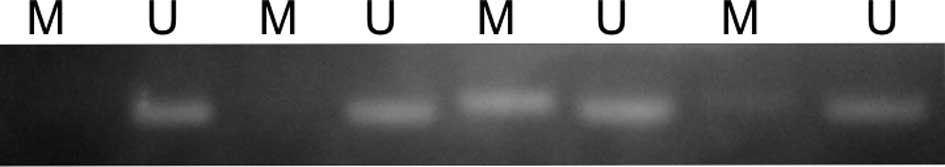
The LATS2 promoter region was hypermethylated
in 160 out of 203 (78.8%) lung cancers (Fig. 1). However, the methylation status
revealed no associations with the clinicopathologic characteristics
of the lung cancers. The LATS2 methylation status did not
correlate with patient survival (log-rank test; p=0.4598).
Discussion
We found that the LATS gene family was
hypermethylated in Japanese lung cancers. We did not find
correlations between the methylated statuses and gender,
pathological stages and survival in Japanese NSCLC. Although the
LATS1 methylation status was correlated with smoking status,
squamous histology and EGFR mutations, the methylation status of
the LATS genes was of limited value in Japanese lung
cancers. In addition, we did not find a LATS2 mutation at
exon 8, suggesting that an ethnic difference may exist.
The LATS tumor suppressor family has been
shown to play an important role in the control of tumor development
and the cell cycle (3–5,18,19).
Mechanistic studies concerning LATS1 revealed that it might
control tumorigenesis by negatively regulating the cell cycle.
Ectopic expression of LATS1 in human cancer cell lines leads
to the down-regulation of cyclin A and B at the protein level
(5), and/or inactivation of CDC2
kinase, thereby blocking cells at G2/M and preventing tumor
development in nude mice (4,5).
Ectopic expression of LATS1 in human tumor cell lines has
also been shown to induce apoptosis by up-regulating the level of
BAX protein (5) or up-regulating
caspase-3 activity (4), indicating
that LATS1 may also control tumorigenesis by inducing
apoptosis. Although it is unclear whether smoking induces the
methylation of LATS1, the methylation also occurred more
frequently in squamous cell carcinoma. Notably, a chromosomal
alteration was frequently noted at chromosome 6q24 (20) where the LATS1 gene is
localized (18).
LATS2, also known as KPM (21), is the second mammalian member of the
LATS tumor suppressor gene family (22). Human LATS2 has been mapped
onto human chromosome 13q11–12 (21), a hot spot (67%) for loss of
heterozygosity in NSCLC (23).
LATS2 encodes a putative Ser/Thr protein kinase. The
LATS2 protein shares 85% sequence identify to human
LATS1 proteins in the kinase domain (21,22).
LATS2 has a role in the maintenance of
mitotic fidelity and genomic stability, since
LATS−/− mutant embryonic cells exhibit an
increased frequency of cytokinetic defects, accumulation of
micronuclei, supernumerary centrosomes and aneuploidy (24,25).
LATS2 also functions as an inducer of apoptosis through
down-regulation of anti-apoptotic proteins of the Bcl-family
(8). More recent findings implicate
LATS2 as the key mediator of the G1 tetraploidy checkpoint,
while LATS2 translocates into the nucleus by mitotic
apparatus dysfunction and inactivates Mdm2 (9). Although down-regulation of the
LATS2 gene has been reported in several cancers (10,11)
including lung cancer (12), in our
analysis we did not find any correlation between LATS2
methylation and clinicopathological features. We did not find a
LATS2 mutation at exon 8. An ethnic difference between the
studies concerning mutant LATS2 may exist.
In conclusion, the LATS2 mutation in Japanese
lung cancers appears to be extremely rare, and the methylation
status of the LATS genes is of limited value in Japanese
lung cancers.
Acknowledgements
The authors would like to thank Mrs. Tomomi Shibata
for her excellent technical assistance. This study was supported by
a Grand-in-Aid for Research, Nagoya City University (2006) and
Grants-in-Aid for Scientific Research, Japan Society for the
Promotion of Science (JSPS) (nos. 21591820 and 21390394).
References
|
1
|
Justice RW, Zilian O, Woods DF, Noll M and
Bryant PJ: The Drosophila tumor suppressor gene warts
encodes a homolog of human myotonic dystrophy kinase and is
required for the control of cell shape and proliferation. Genes
Dev. 9:534–546. 1995.
|
|
2
|
Xu T, Wang W, Zhang S, Stewart RA and Yu
W: Identifying tumor suppressors in genetic mosaics: the
Drosophila lats gene encodes a putative protein kinase.
Development. 121:1053–1063. 1995.PubMed/NCBI
|
|
3
|
John MA, Tsao W, Fei X, et al: Mice
deficient of Lats1 develop soft-tissue sarcomas, ovarian tumors and
pituitary dysfunction. Nat Genet. 21:182–186. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang X, Li D, Chen W and Xu T: Human
homologue of the Drosophila lats, LATS1, negatively
regulates growth by inducing G2/M arrest or apoptosis. Oncogene.
20:6516–6523. 2001.
|
|
5
|
Xia H, Qi H, Li Y, et al: LATS1 tumor
suppressor regulates G2/M transition and apoptosis. Oncogene.
21:1233–1241. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kamikubo Y, Takaori-Kondo A, Uchiyama T
and Hori T: Inhibition of cell growth by conditional expression of
kpm, a human homologue of Drosophila warts/lats tumor
suppressor. J Biol Chem. 278:17609–17614. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Y, Pei J, Xia H, Ke H, Wang H and Tao
W: Lats2, a putative tumor suppressor, inhibits G1/S transition.
Oncogene. 22:4398–4405. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ke H, Pei J, Ni Z, et al: Putative tumor
suppressor Lats2 induces apoptosis through down-regulation of Bcl-2
and Bcl-x(L). Exp Cell Res. 298:329–338. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aylon Y, Michael D, Shumueli A, Yabuta N,
Nojima H and Oren M: A positive feedback loop between the p53 and
Lats2 tumor suppressors prevents tetraploidization. Genes Dev.
20:2687–2700. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takahashi Y, Miyoshi Y, Takahata C, et al:
Down-regulation of LATS1 and LATS2 mRNA expression by
hypermethylation and its association with biologically aggressive
phenotype in human breast cancers. Clin Cancer Res. 11:1380–1385.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jimenez-Velasco A, Roman-Gomez J, Agirre
X, et al: Down-regulation of the large tumor suppressor 2
(LATS/KPM) gene is associated with poor prognosis in acute
lymphoblastic leukemia. Leukemia. 19:2347–2350. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Strazisar M, Mlakar V and Glavac D: LATS2
tumor specific mutations and down-regulation of the gene in
non-small cell carcinoma. Lung Cancer. 64:257–262. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Japan Lung Cancer Society. General Rule
for Clinical and Pathological Record of Lung Cancer. 5th edition.
The Japan Lung Cancer Society; 5. pp. 1–177. 1999
|
|
14
|
Paez JG, Janne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Endo K, Konishi A, Sasaki H, et al:
Epidermal growth factor receptor gene mutation in non-small cell
lung cancer using highly sensitive and fast TaqMan PCR assay. Lung
Cancer. 50:375–384. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sasaki H, Shimizu S, Endo K, et al: EGFR
and erbB2 mutation status in Japanese lung cancer patients. Int J
Cancer. 118:180–184. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sasaki H, Endo K, Konishi A, et al: EGFR
mutation status in Japanese lung cancer patients: genotyping
analysis using LightCycler. Clin Cancer Res. 11:2924–2929. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nishiyama Y, Hirota T, Morisaki T, et al:
A human homolog of Drosophila warts tumor suppressor,
h-warts, localized to mitotic apparatus and specifically
phosphorylated during mitosis. FEBS Lett. 459:159–165.
1999.PubMed/NCBI
|
|
19
|
Tao W, Zhang S, Turenchalk GS, et al:
Human homologue of the Drosophila melanogaster lats tumor
suppressor modulates CDC2 activity. Proc Natl Acad Sci USA.
96:11335–11340. 1999.
|
|
20
|
Petersen S, Aninat-Meyer M, Schluns K,
Gellert K, Dietel M and Petersen I: Chromosomal alterations in the
clonal evolution to the metastatic stage of squamous cell carcinoma
of the lung. Br J Cancer. 82:65–73. 2000.PubMed/NCBI
|
|
21
|
Hori T, Takaori-Kondo A, Kamikubo Y and
Uchiyama T: Molecular cloning of a novel human protein kinase, kpm,
that is homologous to warts/lats a Drosophila tumor
supressor. Oncogene. 19:3101–3109. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yabuta N, Fujii T, Copeland NG, et al:
Structure, expression and chromosome mapping of LATS2, a mammalian
homologue of the Drosophila tumor suppressor gene
lats/warts. Genomics. 63:263–270. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Girard L, Zochbauer-Muller S, Virmani AK,
Gazdar AF and Minna JD: Genome-wide allelotyping of lung cancer
identifies new regions of allelic loss, differences between
small-cell lung cancer and non-small cell lung cancer, and loci
clustering. Cancer Res. 60:4894–4906. 2000.PubMed/NCBI
|
|
24
|
McPherson JP, Tamblyn L, Elia A, et al:
Lats2/Kpm is required for embryonic development, proliferation
control and genomic integrity. EMBO J. 23:3677–3688. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yabuta N, Okada N, Ito A, et al: Lats2 is
an essential mitotic regulator required for the coordination of
cell division. J Biol Chem. 282:19259–19271. 2007. View Article : Google Scholar : PubMed/NCBI
|