Introduction
Periosteal chondroma is a benign cartilage tumor
that accounts for <2% of chondromas. This type of tumor grows in
the periosteal region and frequently erodes the underlying bone
cortex. Periosteal chondroma can lead to sclerosis of underlying
bone, and the formation of a ‘dished’ area under the tumor with a
‘buttress’ or peripheral wall of reactive bone at its edge. The
bone cortex may be abnormal or thinned adjacent to the tumor.
Periosteal chondromas typically present in the second and third
decades of life with a male predilection (2:1), are usually
asymptomatic, and are commonly identified incidentally on
radiographs obtained for other reasons (1). Common sites of occurrence include the
proximal humerus, proximal and distal femur and the phalanges of
the hands and feet (1). A tender
swelling or the feeling of a mass may bring the lesion to clinical
attention. Periosteal chondroma may be confused with chondrosarcoma
or periosteal and parosteal osteosarcoma. Therefore, diagnosis
requires a thorough investigation that includes radiological
examination and biopsy. Simple excision of the tumor is often
curative.
Previous studies have described clonal cytogenetic
abnormalities in 7 periosteal chondromas, but no specific
aberration pattern has been detected (2–4). In other
studies, IDH1 mutations were observed in 6 out of the 8
periosteal chondromas examined (5,6). The
present study investigated cytogenetic and molecular genetic data
in 4 cases of periosteal chondroma.
Materials and methods
Ethics statement
The study was approved by the regional ethics
committee (Regional komité for medisinsk forskningsetikk Sør-Øst,
Norge, http://helseforskning.etikkom.no).
Patients
A total of 3 females and 1 male, between 8 and 47
years of age, were included in the study. Clinical data are
presented in Table I.
 | Table I.Cytogenetic and molecular analysis of
four periosteal chondromas. |
Table I.
Cytogenetic and molecular analysis of
four periosteal chondromas.
| Case | Gender | Age | Location | Tumor size
(cm) | Karyotype | IDH1 mutation | IDH2 R172
mutation | TERT promoter
-C228T/-C250T | MGMT promoter
methylation | CRBP1 promoter
methylation | S100A10 expression
(mean Cq) | HMGA2 expression
(mean Cq) |
|---|
| 1 | F | 8 | Humerus | 6.0 | 46,XX | R132C
(CGT>TGT) | No | No/No | No | No | Unknown | Unknown |
| 2 | F | 21 | Humerus | 2.5 | 46,XX | R132C
(CGT>TGT) | No | No/No | No | No | 28 | –/– |
| 3 | F | 22 | Femur | 4.3 | 46,XX,t(12;13)
(q13;p11) [10]/46,XX[3] | R132L
(CGT>CTT) | No | No/No | No | No | 34 | –/– |
| 4 | M | 47 | Humerus | 2.8 |
47,XY,+8[11]/45~87<2n>,
t(5;8)(q31;q12~13),t(9;10) (p13;q11),del(12)(q15q23), −13,add(22)(p11)[cp3]/46,XY[3] |
R132L(CGT>CTT) | No | No/No | No | No | 24 | –/– |
Chromosome banding analysis
Samples from the surgically resected tumors were
mechanically and enzymatically disaggregated and short-term
cultured as described previously (6).
The cultures were harvested and the chromosomes were G-banded using
Wright's stain (Sigma-Aldrich; St Louis, MO, USA) (7). The subsequent cytogenetic analysis and
karyotype description followed the recommendations of the
International System for Human Cytogenetic Nomenclature (8).
Polymerase chain reaction (PCR) for
IDH1 and IDH2 mutations
Genomic DNA was extracted using a Maxwell 16
Research Instrument system and a Maxwell 16 Tissue DNA Purification
Kit (Promega Corporation, Madison, WI, USA). For the detection of
possible IDH1 and IDH2 mutations, quantitative (q)PCR
with high resolution melt curve analysis (HRM) was performed
followed by Sanger sequencing to confirm positive HRM screens
(9). As positive and negative
controls, plasmids containing the wild-type IDH1R132 and
IDH2R172 and the mutated IDH1R132H and IDH2R172M were
used. The 20-µl PCR mixture contained 10 µl Precision Melt Supermix
(Bio-Rad Laboratories AB, Oslo, Norway), 0.2 µM of each of the
forward and reverse primers (custom-made; Invitrogen Life
Technologies, Carlsbad, CA, USA), and 20 ng of genomic DNA. To
identify possible IDH1R132 mutations, the following primers were
used: The forward primer was IDH1-F1, 5′-TCA GAG AAG CCA TTA TCT
GCA-3′ and the reverse was IDH1-R1, 5′-AAT CAC ATT ATT GCC AAC ATG
A-3′. To identify possible IDH1R172 mutations the following primers
were used: The forward was IDH2-F1New, 5′-TAG TCC CTG GCT
GGA CCA-3′ and the reverse was IDH2-R1New, 5′-TGC CCA GGT
CAG TGG ATC-3′. The PCRs were conducted on a CFX96 Touch Real-Time
PCR Detection System using the Bio-Rad CFX Manager 2.1 software
(Bio-Rad Laboratories AB). The PCR cycling and melt curve
conditions were as follows: An initial denaturation at 95°C for 2
min followed by 40 cycles of 10 sec at 95°C, 30 sec at 55°C (plus
plate read). Next, the melt curve started with denaturation at 95°C
for 30 sec and annealing at 60°C for 1 min. The melt curve program
then continued from 65°C to 95°C with increments of 0.2°C for 10
sec, plus a final plate read. The HRM of the data was made using
the Precision Melt Analysis software, version 1.2 (Bio-Rad
Laboratories AB). To confirm positive HRM screens, Sanger
sequencing was performed as follows: The PCR products were purified
using the NucleoSpin Gel and PCR Clean-up kit (Machery-Nagel GmbH,
Düren, Germany) and direct Sanger sequencing was performed using
the LIGHTrun Sequencing service (GATC Biotech AG, Konstanz,
Germany; www.gatc-biotech.com/en/products/sanger-services/lightrun-sequencing.html).
BLAST software (blast.ncbi.nlm.nih.gov/Blast.cgi) was used for
computer analysis of the sequence data.
PCR for -C228T and -C250T
mutations
In order to detect the possible -C228T and -C250T
mutations in the promoter region of telomerase reverse
transcriptase (TERT), which correspond to positions 124 and
146 bp, respectively, upstream of the TERT ATG start site
(10), PCR was conducted as follows:
The 25 µl PCR volume contained 1X PrimeSTAR GXL Buffer (Takara Bio
Europe SAS, Saint-Germain-en-Laye, France), 200 µM of each dNTP,
0.4 µM of each of the primers (custom-made; Invitrogen Life
Technologies), 1.25 units of PrimeSTAR GXL DNA polymerase and 20 ng
of genomic DNA. The primer sequences were as follows: TRETpromF2,
5′-GCC GGG CTC CCA GTG GAT TCG-3′ and the reverse primer
TRETpromR2, 5′-GGC TTC CCA CGT GCG CAG CAG-3′. The PCR was
conducted on a C-1000 Thermal Cycler (Bio-Rad Laboratories AB) with
an initial denaturation at 94°C for 30 sec, followed by 35 cycles
of 7 sec at 98°C, 90 sec at 68°C, and a final extension for 5 min
at 68°C. A total of 4 µl PCR products were stained with GelRed
Nucleic Acid Gel Stain (Biotium Inc., Hayward, CA, USA),
electrophoresed through a 1.0% agarose gel (certified molecular
biology agarose (Bio-Rad Laboratories AB) and images for analysis
were captured using a camera. The remaining amplified products were
purified using the NucleoSpin Gel and PCR Clean-up kit followed by
direct Sanger sequencing as above.
Methylation analysis
Methylation analysis of the
O-6-methylguanine-DNA methyltransferase (MGMT)
promoter was performed using the primers and PCR conditions
described by Esteller et al (11). Methylation analysis of the cellular
retinol binding protein 1 (CRBP1) promoter was performed
using the primers and PCR conditions described by Jerónimo et
al (11).
Total RNA and cDNA synthesis
Total RNA was extracted using an miRNeasy mini kit
and QIAcube (Qiagen AB, Sollentuna, Sweden) according to the
manufacturer's recommendations. Human Universal Reference Total RNA
was used as a control (Clontech Laboratories, Inc., Mountainview,
CA, USA); it is a mixture of total RNAs from male and female adult
human tissues selected to represent a broad range of expressed
genes. Reverse transcription (RT) was conducted as follows: 400–500
ng total RNA was reverse transcribed in a 20 µl reaction volume
using iScript Advanced cDNA Synthesis Kit for RT-qPCR according to
the manufacturer's instructions (Bio-Rad Laboratories AB). Next,
the cDNA was diluted to the equivalent of 10 ng/µl of RNA and 2 µl
was used as a template in subsequent PCR assays.
High-mobility group AT-hook 2 (HGMA2)
expression analysis
To assess HMGA2 expression in the periosteal
chondroma tissue samples by PCR, the 25 µl PCR volumes contained
12.5 µl Premix Taq polymerase (Takara Bio Europe SAS) 2 µl diluted
cDNA and 0.4 µM of each of the forward and reverse primers
(custom-made; Invitrogen Life Technologies). The PCRs were
conducted on a C-1000 Thermal Cycler. The PCR conditions were as
follows: An initial denaturation at 94°C for 30 sec followed by 35
cycles of 7 sec at 98°C, 120 sec at 68°C, and a final extension for
5 min at 68°C. The primer sequences used for the amplification of
transcripts of HMGA2 exons 1–3 were as follows:
HMGA2-846F1, 5′-CCA CTT CAG CCC AGG GAC AAC CT-3′ and
HMGA2-1021R, 5′-CCT CTT GGC CGT TTT TCT CCA GTG-3′. The
quality of the cDNA synthesis was assessed by amplification of a
cDNA fragment of the S100A10 gene, which is expressed in
chondrocytes (12). The following
primer sequences were used: S100A10-555F, 5′-TTC ACA AAT TCG CTG
GGG ATA AAG G-3′ and S100A10-840R, 5′-GAT TCC TTA AGC GAC CCT TTG
GGA C-3′. The PCR for the amplification of transcripts of
HMGA2 were repeated three times. A total of 3 µl PCR
products were stained with the GelRed Nucleic Acid Gel Stain,
electrophoresed through a 1.0% agarose gel and images for analysis
were captured using a camera.
qPCR was also conducted to determine the expression
level of HMGA2. The TaqMan Gene Expression Assays
Hs00171569_m1 (which probes HMGA2 exons 1–2), Hs00971725_m1 (which
probes HMGA2 exons 4–5) and the S100A10 gene (control) assay
Hs00237010_ml (Applied Biosystems Life Technologies, Foster City,
CA, USA) were used. Four replicates of each sample and endogenous
control were used to ensure reliability. The 20 µl PCR reaction
volume contained 1X TaqMan Universal Master Mix II with uracil
n-glycosylase, 1X TaqMan gene expression mix, and cDNA (equivalent
to 10 ng RNA). The PCR was run on the CFX96 Touch Real-Time PCR
Detection System. The thermal cycling included an initial step at
50°C for 2 min, followed by 10 min at 95°C, 40 cycles of 15 sec at
95°C and 1 min at 60°C. The Bio-Rad CFX Manager software, version
2.1, was used to analyze the data and to calculate the mean
quantification cycle (mean Cq).
Results
Clonal chromosomal aberrations
The clinical, cytogenetic and molecular analysis
data are presented in Table I. The
histologic examination of case 1 showing a nodular tumor of benign
chondrocytes covered by periosteum is presented in Fig 1A. Clonal chromosomal aberrations were
identified in two out of the four examined periosteal chondromas,
while the other two tumors presented with a normal karyotype.
Rearrangements of chromosome 12 q13-15 were observed in the two
tumors with chromosome abnormalities (Table I). Case 3 has a simple balanced
translocation between chromosomes 12 and 13, t(12;13)(q13;p11), as
the sole aberration. The tumor in case 4 had two cytogenetically
unrelated clones. The first clone displayed trisomy of chromosome
8, while the second clone had numerical and structural aberrations,
including a deletion in the long arm of chromosome 12, del(12)(q15q23) (Table
I, Fig. 1B).
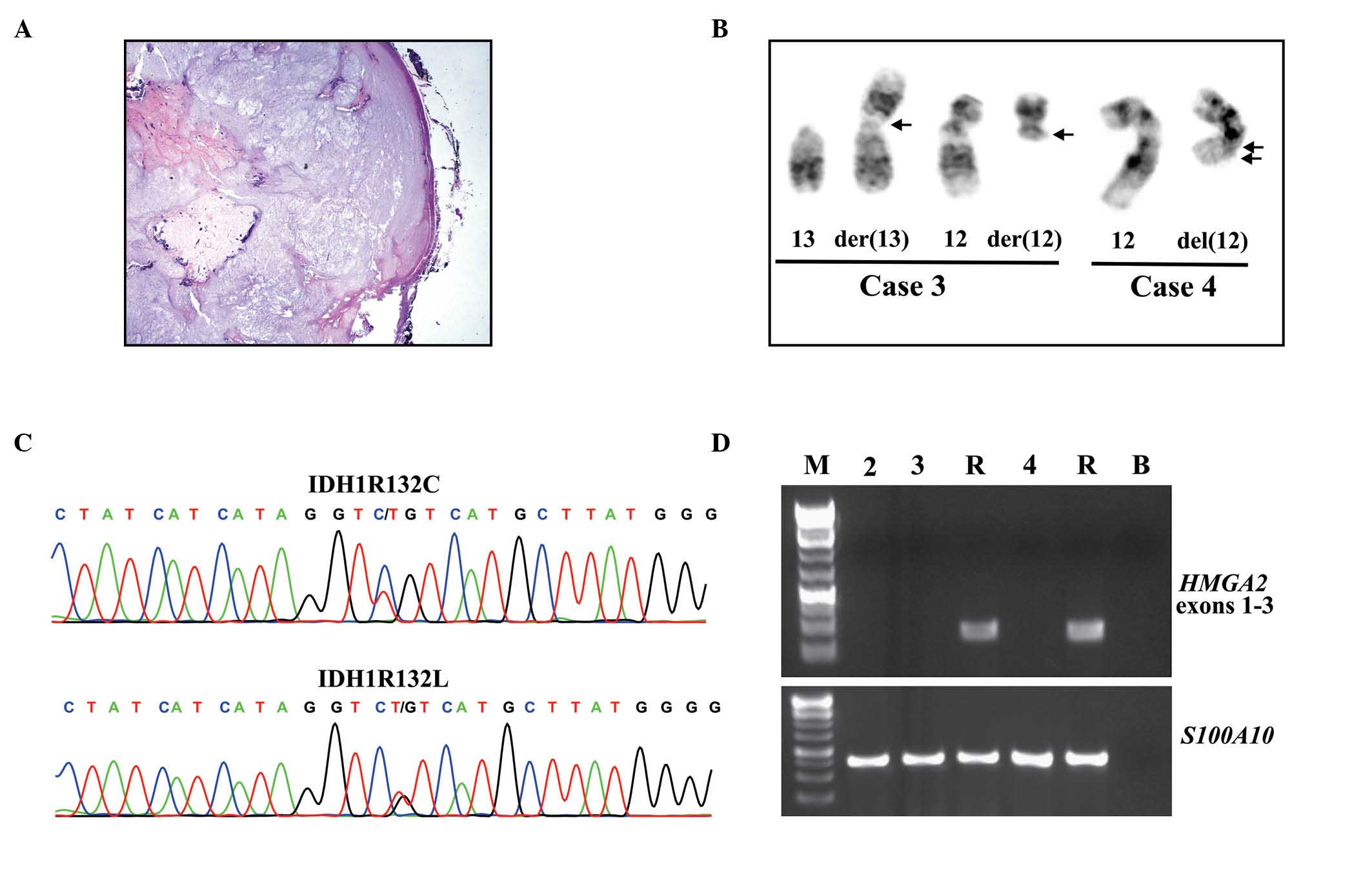 | Figure 1.Histology, cytogenetics, HMGA2
expression, and IDH1 mutations in periosteal chondromas. (A)
Histologic examination of case 1 exhibiting a nodular tumor of
benign chondrocytes covered by periosteum. (B) Partial karyotypes
presenting chromosome aberrations der(13)t(12;13)(q13;p11) and der(12)t(12;13)(q13;p11) in case 3, and
del(12)(q15q23) in case 4, with the
corresponding normal chromosome homologs; breakpoint positions are
indicated by arrows. (C) Partial sequence chromatogram displaying
the CGT>TGT (IDH1R132C; cases 1 and 2) and CGT>CTT in
(IDH1R132L; cases 3 and 4) IDH1. (D) Polymerase chain
reaction results demonstrating the amplification of HMGA2
exons 1–3 and S100A10. M, GeneRuler 1 Kb Plus DNA ladder
(Thermo Fisher Scientific, Waltham, MA, USA); R, cDNA synthesized
from Human Universal Reference Total RNA; B, no RNA in cDNA
synthesis. HMGA2, high-mobility group AT-hook 2; IDH,
isocitrate dehydrogenase. |
Molecular genetic analyses
All four periosteal chondromas had the R132 mutation
in IDH1, whereas none had mutations in codon 172 of
IDH2 (Table I, Fig. 1C). Two tumors carried an IDH1R132C
(CGT>TGT) mutation while the other two carried an IDH1R132L
(CGT>CTT) mutation. None of the periosteal chondromas had
methylated MGMT or CRBP1 promoters or mutations at
positions -C228T and -C250T in the promoter region of TERT
(Table I).
The total RNA was extracted from the three tumors
(cases 2–4) and a 310-bp S100A10 cDNA fragment was amplified
in all of them, indicating that the synthesized cDNA was of good
quality (Fig. 1D). RT-PCR with the
primer set HMGA2-846F1/HMGA2-1021R did not amplify any cDNA
fragments from the periosteal chondromas, whereas an amplified
HMGA2 cDNA fragment was observed in the positive control
(Fig. 1D). This result indicated that
the HMGA2 transcript was not expressed in the three examined
periosteal chondromas. Similar results were also obtained with
qPCR: For the control Human Universal Reference Total RNA, the mean
Cq was 25, 30 and 31 for S100A10, HMGA2 exons 1–2, and
HMGA2 exons 4–5, respectively (data not shown). The mean Cq
for S100A10 was 28, 34 and 24 for cases 2, 3 and 4,
respectively, indicating that the gene expression assay was
successful. However, there were no mean Cq values for HMGA2
exons 1–2/HMGA2 exons 4–5 since the TaqMan gene expression
assays did not amplify any product (Table
I). These results further indicate that HGMA2 was not
expressed in the periosteal chondromas examined in the present
study.
Discussion
To the best of our knowledge, prior to the present
study, karyotypic information on periosteal chondromas was
restricted to seven cases and no consistent abnormality was
recognized (4). Changes observed in
periosteal chondromas in the previous study included loss of
chromosome 6 and rearrangements of 2q37, 4q21–25, 11q13-15 and
12q13 (4). The present study
demonstrated that the q arm of chromosome 12 was involved in two
out of the two periosteal chondromas with an informative karyotype.
Taken together, the data from the present study and those
previously reported (3) demonstrate
that rearrangements of 12q13-15 may be recurrent in this type of
tumor. The involvement of the chromosome bands 12q13-15 in
periosteal chondromas may not be random, particularly as they are
frequently aberrant in benign connective tumors, such as lipoma and
leiomyoma (14). In addition, 12-q
rearrangements that result in the transcriptional activation of the
HMGA2 gene, were reported in mesenchymal chondromas by
Dahlén et al (15). They also
demonstrated that HMGA2 was expressed in four of six soft
tissue chondromas, of which three tumors possessed a truncated
(exons 1–3) transcript and one possessed the full-length (exons
1–5) transcript of HMGA2. In addition, Dahlén et al
(15) observed that HMGA2 was
expressed in two skeletal chondromas: One of the tumors, which
possessed a pericentric inversion of chromosome 12, expressed a
truncated transcript of HMGA2; whereas the other case, which
had no visible involvement of 12q by cytogenetic analysis,
expressed the full-length HMGA2 transcript (15). The description of these two tumors was
inconclusive as to whether they were enchondromas or periosteal
chondromas.
In the present study, neither conventional RT-PCR
nor qPCR demonstrated expression of HMGA2 in the examined
periosteal chondromas.
The mutation analysis of IDH1 and IDH2
revealed that the tumors carried heterozygous IDH1 mutations
at R132, which are in accordance with previous observations that a
majority of periosteal chondromas carry heterozygous mutations at
R132 of IDH1 (5,6). From the results of the present study it
can be hypothesized that the rearrangements and expression of
HMGA2 are mutually exclusive with IDH1 and
IDH2 mutations in periosteal chondromas.
The R132L mutation in IDH1 that was observed
in two of the periosteal chondromas in the current study has also
been identified previously by Amary et al (6), who observed this mutation in central
low-grade cartilaginous tumors (1/23), chondrosarcomas GII and GIII
(3/23) and dedifferentiated chondrosarcomas (2/13). Of the eight
periosteal chondromas previously analyzed for the presence of
mutations in IDH1, mutations were observed in six: Four
possessed IDH1R132C (CGT>TGT) and two possessed R132S
(CGT>AGT) (4,5). Viewing the present and previously
published data in concert (5,6), mutations in codon 12 of IDH1 have
been present in the majority (83%, 10/12) of examined periosteal
chondromas. Of those mutations in the IDH1, R132C comprises
60% (6/10), whereas R132L and R132S are observed in 20% (2 cases
each). However, R132H, the most common mutation in gliomas, has not
been reported thus far in periosteal chondromas. The exact role
that IDH1 mutations serve in neoplasia is not fully
understood, however it has been demonstrated that presence of a
heterozygous R132H mutation induces genome-wide alterations in DNA
methylation, leading to hypermethylation and hypomethylation
(16,17). IDH1 mutations are associated
with MGMT and CRBP1 promoter methylation in certain
brain tumors (18–21). However, this association was not
observed in the periosteal chondroma tissue specimens in the
present study, since none of them possessed methylated MGMT or
CRBP1 promoters, as determined using the MSP methodology (11,12).
Mutations at positions -C228T and -C250T in the
promoter region of TERT, which correspond to 124 and 146 bp
upstream of the TERT ATG start site, have been reported in
gliomas and other tumors (10) but
have yet to be studied in periosteal chondromas. TERT
promoter mutations have been demonstrated to be inversely
associated with IDH1/IDH2 mutations in tumors of the nervous
system (22,23). In the present study, none of the four
periosteal chondromas had -C228T or -C250T mutations. In
conclusion, the present study revealed that the involvement of
12q13-15 in structural chromosomal aberrations is a relatively
recurrent and common event in periosteal chondromas, that
HMGA2 is not frequently expressed, that the majority of
periosteal chondromas carry heterozygous IDH1R132 mutation, that
MGMT and CRBP1 promoters are not methylated, and that
neither -C228T nor -C250T is present in the promoter region of
TERT. Rearrangements of HMGA2 resulting in fusion
genes and expression of HMGA2-fusion transcripts appear to
be mutually exclusive with IDH1 and 2 mutations.
Acknowledgements
The authors thank Ms. Hege Brandt Gehrken for
technical help. The present study was supported by grants from the
Norwegian Cancer Society and the Norwegian Radium Hospital
Foundation.
References
|
1
|
Lichtenstein L and Hall JE: Periosteal
chondroma; a distinctive benign cartilage tumor. J Bone Joint Surg
Am. 34:691–697. 1952.
|
|
2
|
Buddingh EP, Naumann S, Nelson M, Neffa
JR, Birch N and Bridge JA: Cytogenetic findings in benign
cartilaginous neoplasms. Cancer Genet Cytogenet. 141:164–168. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mandahl N, Willén H, Rydholm A, Heim S and
Mitelman F: Rearrangement of band q13 on both chromosomes 12 in a
periosteal chondroma. Genes Chromosomes Cancer. 6:121–123. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sakai Junior N, Abe KT, Formigli LM, et
al: Cytogenetic findings in 14 benign cartilaginous neoplasms.
Cancer Genet. 204:180–186. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Damato S, Alorjani M, Bonar F, et al: IDH1
mutations are not found in cartilaginous tumours other than central
and periosteal chondrosarcomas and enchondromas. Histopathology.
60:363–365. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Amary MF, Bacsi K, Maggiani F, et al: IDH1
and IDH2 mutations are frequent events in central chondrosarcoma
and central and periosteal chondromas but not in other mesenchymal
tumours. J Pathol. 224:334–343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mandahl N: Methods in solid tumour
cytogeneticsHuman cytogenetics: malignancy and acquired
abnormalities. Rooney DE: Oxford University Press; New York, NY:
pp. 165–203. 2001
|
|
8
|
Schaffer LG, Slovak ML and Campbell LJ:
ISCN 2009: an International System for Human Cytogenetic
Nomenclature. Karger; Basel: 2009
|
|
9
|
Patel KP, Barkoh BA, Chen Z, et al:
Diagnostic testing for IDH1 and IDH2 variants in acute myeloid
leukemia an algorithmic approach using high-resolution melting
curve analysis. J Mol Diagn. 13:678–686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Killela PJ, Reitman ZJ, Jiao Y, et al:
TERT promoter mutations occur frequently in gliomas and a subset of
tumors derived from cells with low rates of self-renewal. Proc Natl
Acad Sci USA. 110:6021–6026. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Esteller M, Hamilton SR, Burger PC, Baylin
SB and Herman JG: Inactivation of the DNA repair gene
O6-methylguanine-DNA methyltransferase by promoter hypermethylation
is a common event in primary human neoplasia. Cancer Res.
59:793–797. 1999.PubMed/NCBI
|
|
12
|
Jerónimo C, Henrique R, Oliveira J, et al:
Aberrant cellular retinol binding protein 1 (CRBP1) gene expression
and promoter methylation in prostate cancer. J Clin Pathol.
57:872–876. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song C, Zhou X, Dong Q, et al: Regulation
of inflammatory response in human chondrocytes by lentiviral
mediated RNA interference against S100A10. Inflamm Res.
61:1219–1227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Heim S and Mitelman F: Cancer
Cytogenetics: Chromosomal and Molecular Genetic Abberations of
Tumor Cells. 3rd. Wiley-Blackwell; Oxford, UK: 2009
|
|
15
|
Dahlén A, Mertens F, Rydholm A, et al:
Fusion, disruption, and expression of HMGA2 in bone and soft tissue
chondromas. Mod Pathol. 16:1132–1140. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Duncan CG, Barwick BG, Jin G, et al: A
heterozygous IDH1R132H/WT mutation induces genome-wide alterations
in DNA methylation. Genome Res. 22:2339–2355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Turcan S, Rohle D, Goenka A, et al: IDH1
mutation is sufficient to establish the glioma hypermethylator
phenotype. Nature. 483:479–483. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chou AP, Chowdhury R, Li S, et al:
Identification of retinol binding protein 1 promoter
hypermethylation in isocitrate dehydrogenase 1 and 2 mutant
gliomas. J Natl Cancer Inst. 104:1458–1469. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Leu S, von Felten S, Frank S, et al:
IDH/MGMT-driven molecular classification of low-grade glioma is a
strong predictor for long-term survival. Neuro Oncol. 15:469–479.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mellai M, Monzeglio O, Piazzi A, et al:
MGMT promoter hypermethylation and its associations with genetic
alterations in a series of 350 brain tumors. J Neurooncol.
107:617–631. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Noushmehr H, Weisenberger DJ, Diefes K, et
al: Cancer Genome Atlas Research Network: Identification of a CpG
island methylator phenotype that defines a distinct subgroup of
glioma. Cancer Cell. 17:510–522. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Koelsche C, Sahm F, Capper D, et al:
Distribution of TERT promoter mutations in pediatric and adult
tumors of the nervous system. Acta Neuropathol. 126:907–915. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nonoguchi N, Ohta T, Oh JE, Kim YH,
Kleihues P and Ohgaki H: TERT promoter mutations in primary and
secondary glioblastomas. Acta Neuropathol. 126:931–937. 2013.
View Article : Google Scholar : PubMed/NCBI
|















