Introduction
Breast cancer is the most common malignancy in
females worldwide, and the disease is often fatal due to metastasis
dissemination (1). The disease
develops from breast tissue with symptoms including a lump in the
breast, dimpling of the skin, a red scaly patch of skin, changes in
breast shape or fluid coming from the nipple (2). Generally, the first noticeable symptom
of breast cancer is a lump that feels different from the rest of
the breast tissue. Several factors, including smoking tobacco, lack
of physical exercise, ionizing radiation, obesity, and having
children late in life or not at all, may induce breast cancer
(3). The survival rates of breast
cancer in developing countries are poor, with 1.68 million cases
and 522,000 mortalities in 2012 (4).
Therefore, numerous researchers and physicians are focusing on the
pathogenesis of the disease with the aim of improving current
therapies and identifying new treatments.
microRNAs (miRNAs) are small non-coding RNA
molecules, which participate in post-transcriptional gene
regulation in a sequence-specific manner. It has been suggested
that they are involved in the establishment and progression of
human tumors, appearing to be critical biomarkers of cancer
(5). For instance, miR-148b is a
major coordinator of breast cancer progression in a
relapse-associated miRNA signature by targeting integrin α5,
Rho-associated coiled-coil containing protein kinase 1
(ROCK1), and phosphatidylinositol-4,5-bisphosphate 3-kinase,
catalytic subunit α (PIK3CA) (5). miR-335 was observed to inhibit
proliferation, cell cycle progression, colony formation and
invasion by targeting paired box 6 (PAX6) in breast cancer
cells (6). In addition, miR-185
suppresses tumor proliferation by directly targeting E2F
transcription factor 6 (E2F6) and indirectly upregulating
breast cancer 1, early onset (BRCA1) in triple-negative
breast cancer (7). Although the
pathogenesis of breast cancer has been investigated, the mechanism
is not fully understood. More crucial miRNAs that might play
significant roles in breast cancerogenesis need to be
identified.
Therefore, in our present study, the underlying
mechanisms of breast cancer were further explored and predicted by
bioinformatics approaches, including screening breast
cancer-related miRNAs, predicting target genes of the miRNAs,
performing functional analysis for target genes, and constructing a
protein-protein interaction network. The findings may provide new
insights into breast cancer pathogenesis and bring about novel
targets for cancer therapy.
Materials and methods
Microarray data
The Gene Expression Omnibus database (GEO,
http://www.ncbi.nlm.nih.gov/geo/) in the
National Center for Biotechnology Information (NCBI) is currently
the largest fully public gene expression resource, and includes
214,268 samples and 4,500 platforms (8). In the present study, microarray data set
GSE26659 (5) was downloaded from GEO,
which included 77 gene chips from ductal breast carcinoma biopsies
and 17 from mammoplasties. The 77 frozen tumor specimens were
selected from the Tumor Bank of the Department of Obstetrics and
Gynecology at the University of Turin, Italy. They were obtained
from patients who were diagnosed with invasive breast cancer at T
and N stages and underwent primary surgical treatment between 1988
and 2001 at a median age of 54 years. The 17 frozen mammoplasty
samples were from the École polytechnique fédérale (Swiss Federal
Institute of Technology), Lausanne, Switzerland, and were included
in the study as normal breast controls. Raw data were collected
based on the platform GPL8227 Agilent-019118 Human miRNA Microarray
2.0 G4470B (miRNA ID version; Affymetrix Inc., Santa Clara, CA,
USA).
Data preprocessing and differential
analysis
The original data and annotation files were
downloaded and normalized. Then according to the SOFT formatted
family files, the normalized data series matrix files were mapped
to their corresponding miRNA names. The expression values of
multiple probes for a given miRNA were reduced to a single value by
taking the average expression value. The Affy package (9) in R (https://www.r-project.org/) was selected for
background correction, normalization and calculation of expression
values. After that, the linear models for microarray analysis
(Limma) package (10) in R was used
to calculate the probability of miRNAs being differentially
expressed between cases and controls. The fold change (FC) and its
logarithm value (log FC) were also determined. A false discovery
rate (FDR) <0.01 and |log FC| >1 were set as the cut-offs to
screen differentially expressed miRNAs.
Identification of breast
cancer-related differentially expressed miRNAs
The selected differentially expressed miRNAs were
mapped into the human miRNA disease database (HMDD; http://cmbi.bjmu.edu.cn/hmdd and http://202.38.126.151/hmdd/tools/hmdd2.html) to
further select the differentially expressed miRNAs related to
breast cancer. As a database for experimentally supported human
miRNA and disease associations, HMDD serves as a valuable resource
for investigating the roles of miRNAs in human disease (11).
Target gene prediction
The target genes of breast cancer-related
differentially expressed miRNAs were predicted by five miRNA
databases, namely miRanda (http://microrna.sanger.ac.uk) (12), MirTarget2 (http://nar.oxfordjournals.org/cgi/content/abstract/34/5/1646)
(13), PicTar (http://pictar.bio.nyu.edu) (14), PITA (http://genie.weizmann.ac.il/pubs/mir07) (15) and TargetScan (http://targetscan.org) (16). In order to obtain more solid results,
target genes that occurred in not less than three databases were
regarded as the target genes of breast cancer-related
differentially expressed miRNAs. In addition, the published
oncogenes and suppressors of breast cancer were selected from
TSGene (http://bioinfo.mc.vanderbilt.edu/TSGene/) (17) and Tumor Associated Gene (TAG;
http://www.binfo.ncku.edu.tw/TAG/)
databases (18).
Functional enrichment analysis
The Database for Annotation, Visualization and
Integrated Discovery (DAVID) bioinformatics resource consists of an
integrated biological knowledge base and analytic tools aimed at
systematically extracting biological meaning from large gene or
protein lists (19). In the present
study, DAVID was applied to conduct Kyoto encyclopedia of genes and
genomes (KEGG) pathway and gene ontology (GO) enrichment analyses
for the identified target genes. KEGG is a knowledge base for
systematic analysis of gene functions (20). GO analysis predicts the function of
the target genes in three aspects, including biological processes,
cellular components and molecular function (21). P<0.05 and FDR<0.05 were set as
thresholds.
miRNA-target gene network
construction
Based on the identified breast cancer-related
differentially expressed miRNAs and their target genes, the
miRNA-target gene network was constructed and visualized using
Cytoscape (http://www.cytoscape.org/) (22), which is open source software used for
visualizing biological network and integrating data.
Results
Differentially expressed miRNAs
Among the 77 breast cancer samples and 17 controls,
a total of 34 differentially expressed miRNAs were screened out,
which included 19 upregulated and 15 downregulated miRNAs in breast
cancer patients (Table I). By mapping
them to HMDD, 13 of these were selected as breast cancer-related
differentially expressed miRNAs; namely hsa-let-7d, hsa-let-7e,
hsa-let-7g, hsa-miR-100, hsa-miR-125b, hsa-miR-143, hsa-miR-145,
hsa-miR-155, hsa-miR-197, hsa-miR-21, hsa-miR-223, hsa-miR-425 and
hsa-miR-497.
 | Table I.List of differentially expressed genes
with |log FC| >1 and FDR <0.01. |
Table I.
List of differentially expressed genes
with |log FC| >1 and FDR <0.01.
| Category | miRNA ID | Log FC | P-value | FDR |
|---|
| Upregulated | hsa-miR-21 | 3.068525 | 5.10E-31 | 6.12E-29 |
|
| hsa-miR-142-3p | 2.128498 | 5.73E-10 | 2.46E-09 |
|
| hsa-miR-155 | 2.087345 | 1.35E-14 | 2.32E-13 |
|
| hsa-miR-15b | 1.991570 | 4.72E-22 | 2.83E-20 |
|
| hsa-miR-425 | 1.950364 | 3.00E-13 | 2.77E-12 |
|
| hsa-miR-342-3p | 1.807487 | 1.77E-09 | 6.84E-09 |
|
| hsa-miR-331-3p | 1.672587 | 8.82E-20 | 2.65E-18 |
|
|
hsa-miR-125a-5p | 1.616122 | 4.28E-12 | 2.85E-11 |
|
| hsa-miR-130b | 1.581679 | 5.43E-12 | 3.43E-11 |
|
| hsa-miR-98 | 1.521842 | 1.72E-12 | 1.37E-11 |
|
| hsa-let-7e | 1.510875 | 3.49E-14 | 5.20E-13 |
|
| hsa-miR-197 | 1.447683 | 1.51E-11 | 8.64E-11 |
|
| hsa-miR-223 | 1.385350 | 3.56E-08 | 1.19E-07 |
|
| hsa-miR-455-3p | 1.256636 | 2.83E-06 | 7.37E-06 |
|
| hsa-let-7d | 1.210423 | 1.21E-14 | 2.32E-13 |
|
| hsa-let-7g | 1.161949 | 2.53E-13 | 2.53E-12 |
|
| hsa-miR-150 | 1.106677 | 0.000447 | 0.000851 |
|
| hsa-let-7f | 1.061395 | 1.83E-12 | 1.37E-11 |
|
| hsa-miR-16 | 1.040802 | 2.48E-13 | 2.53E-12 |
| Downregulated | hsa-miR-130a | −1.025820 | 4.14E-08 | 1.34E-07 |
|
| hsa-miR-939 | −1.078370 | 8.47E-10 | 3.39E-09 |
|
| hsa-miR-143 | −1.089560 | 3.02E-06 | 7.70E-06 |
|
| kshv-miR-K12-3 | −1.127510 | 2.79E-09 | 9.85E-09 |
|
| hsa-miR-768-3p | −1.277500 | 6.53E-13 | 5.60E-12 |
|
| hsa-miR-101 | −1.295530 | 9.80E-14 | 1.18E-12 |
|
| hsa-miR-188-5p | −1.342530 | 2.76E-09 | 9.85E-09 |
|
| hsa-miR-497 | −1.385440 | 3.74E-12 | 2.64E-11 |
|
| hsa-miR-26a | −1.408450 | 3.90E-14 | 5.20E-13 |
|
| hsa-miR-125b | −1.419680 | 5.30E-10 | 2.35E-09 |
|
| hsa-miR-100 | −1.432230 | 4.55E-10 | 2.10E-09 |
|
| hsa-miR-99a | −1.560150 | 6.26E-09 | 2.15E-08 |
|
| hsa-miR-140-3p | −1.822440 | 2.00E-21 | 7.99E-20 |
|
| hsa-miR-923 | −2.289380 | 2.18E-10 | 1.09E-09 |
|
| hsa-miR-145 | −2.552090 | 2.89E-15 | 6.94E-14 |
Target genes of breast cancer-related
miRNAs
The target genes of the 13 miRNAs were identified
from the Miranda, MirTarget2, PicTar, PITA and TargetScan
databases. Finally, 3,086 target genes of these 13 miRNAs were
screened from not less than three databases. Among these genes, 47
oncogenes [including zinc finger protein 217 (ZNF217),
Wolf-Hirschhorn syndrome candidate 1 (WHSC1) and ubiquitin
specific peptidase 6 (USP6)] and 123 tumor suppressor genes
(including ZIC family member 1 (ZIC1), zinc finger homeobox
3 (ZFHX3) and wingless-type MMTV integration site family,
member 5A (WNT5A)] were further filtered by combining the
TSGene and TAG databases (Table
II).
 | Table II.Oncogenes and tumor suppressor genes
among target genes of breast cancer-related differentially
expressed microRNAs. |
Table II.
Oncogenes and tumor suppressor genes
among target genes of breast cancer-related differentially
expressed microRNAs.
| Category | Genes |
|---|
| Oncogenes | BCL11A, BCL2L2,
CBL, CCNA2, CCND1, CCND2, CCNE1, CHKA, CRK, CRKL, CSF1R, DEK,
EIF5A2, ELK1, ERBB3, ERG, FGF2, FGFR1, FGFR3, FOS, GNAS, HMGA2,
HOXA10, KRAS, LMO2, MAP3K8, MCF2, MYBL1, MYCL1, NET1, NRAS, PDGFRA,
PIM1, PTPN11, RAF1, RALA, RUNX1T1, SALL4, SERTAD2, SET, SKI, TAF15,
TAL1, TRIM32, USP6, WHSC1, ZNF217 |
| Tumor suppressor
genes | ADAMTS18, AKAP12,
APAF1, APC, ARHGAP20, ARHGAP35, ARHGEF12, ARID1A, ARID2, ARID3B,
AXIN2, BACH2, BHLHE41, BLCAP, BTG2, CADM1, CADM4, CAMTA1, CBFA2T3,
CDC73, CDKN1B, CDKN2A, CHEK1, CREBL2, CSMD1, CTDSPL, CTNND1, CUL2,
CUL5, CYGB, CYLD, DAB2, DAPK1, DDX3X, DIRAS1, DKK3, EGLN1, EGR1,
ENC1, EPB41L3, ERF, FOXO1, FOXO3, FOXP1, HBP1, IGF2R, IGFBP3,
IGFBP5, ITGB3, JUP, KIF1B, KLF5, KRIT1, LATS2, LIN9, LOX, LRIG3,
LRP1B, MFHAS1, MTAP, MTSS1, MTUS1, NF2, NR4A3, NRCAM, OPCML,
PAFAH1B1, PDCD4, PDS5B, PHF6, PHLDA3, PHLPP2, PPAP2A, PPP1CA,
PPP2R1B, PRDM1, PRDM4, PRKAR1A, PTCH1, PTPN2, PTPN3, PTPRD, RARB,
RASL10B, RASSF5, RBM6, RCN2, RECK, REV3L, RINT1, RND3, RSRC2,
SASH1, SERPINB5, SIAH1, SLC39A1, SMAD2, SMAD3, SMARCA4, SOCS1,
SOX11, SPRY2, ST5, STARD13, TCF4, TGFBI, TGFBR2, TGFBR3, TIMP3,
TOPORS, TP53, TP53INP1, TSC1, TUSC2, UBE4B, UHRF2, UNC5A, VCL,
WHSC1L1, WIF1, WNT5A, ZFHX3, ZIC1 |
Functional analysis for target
genes
The top five pathways and GO terms are listed in
Tables III and IV, respectively. In Table III, the most significant KEGG
pathway was hsa05200, pathways in cancer (P=3.33E-11) and 76 target
genes, including E2F2, fibroblast growth factor 9
(FGF9) and WNT3A, were enriched in this pathway.
Moreover, 60, 36, 26 and 23 differentially expressed miRNAs
affected the MAPK signaling pathway (P=2.02E-08), neurotrophin
signaling pathway (P=3.42E-08), colorectal cancer (P=1.10E-06) and
chronic myeloid leukemia (P=6.37E-06), respectively (Table III). Additionally, the most
significant GO term was GO:0045449, regulation of transcription
(P=7.43E-13; Table IV). A total of
360 target genes, including myocyte enhancer factor 2C
(MEF2C), MEF2A and growth differentiation factor 6
(GDF6) were enriched in this GO term. Enzyme-linked receptor
protein signaling pathway (P=7.45E-11), nucleoplasm (P=1.18E-10),
positive regulation of transcription (P=1.96E-10) and nuclear lumen
(P=2.22E-10) were the next four most significant GO terms after
regulation of transcription (Table
IV).
 | Table III.Top five significant pathways
regulated by target genes of breast cancer-related differentially
expressed microRNAs. |
Table III.
Top five significant pathways
regulated by target genes of breast cancer-related differentially
expressed microRNAs.
| Category | Term | Count | P-value | FDR |
|---|
| KEGG_PATHWAY | hsa05200:Pathways
in cancer | 76 | 3.33E-11 | 4.05E-08 |
| KEGG_PATHWAY | hsa04010:MAPK
signaling pathway | 60 | 2.02E-08 | 2.46E-05 |
| KEGG_PATHWAY |
hsa04722:Neurotrophin signaling
pathway | 36 | 3.42E-08 | 4.15E-05 |
| KEGG_PATHWAY | hsa05210:Colorectal
cancer | 26 | 1.10E-06 | 0.001339 |
| KEGG_PATHWAY | hsa05220:Chronic
myeloid leukemia | 23 | 6.37E-06 | 0.007737 |
 | Table IV.Top five significant gene ontology
terms enriched by target genes of breast cancer-related
differentially expressed microRNAs. |
Table IV.
Top five significant gene ontology
terms enriched by target genes of breast cancer-related
differentially expressed microRNAs.
| Category | Term | Count | P-value | FDR |
|---|
| GOTERM_BP_FAT |
GO:0045449~regulation of
transcription | 380 | 7.43E-13 | 1.37E-09 |
| GOTERM_BP_FAT |
GO:0007167~enzyme-linked receptor protein
signaling pathway | 78 | 7.45E-11 | 1.37E-07 |
| GOTERM_CC_FAT |
GO:0005654~nucleoplasm | 143 | 1.18E-10 | 1.74E-07 |
| GOTERM_BP_FAT | GO:0045941~positive
regulation of transcription | 110 | 1.96E-10 | 3.62E-07 |
| GOTERM_CC_FAT | GO:0031981~nuclear
lumen | 209 | 2.22E-10 | 3.26E-07 |
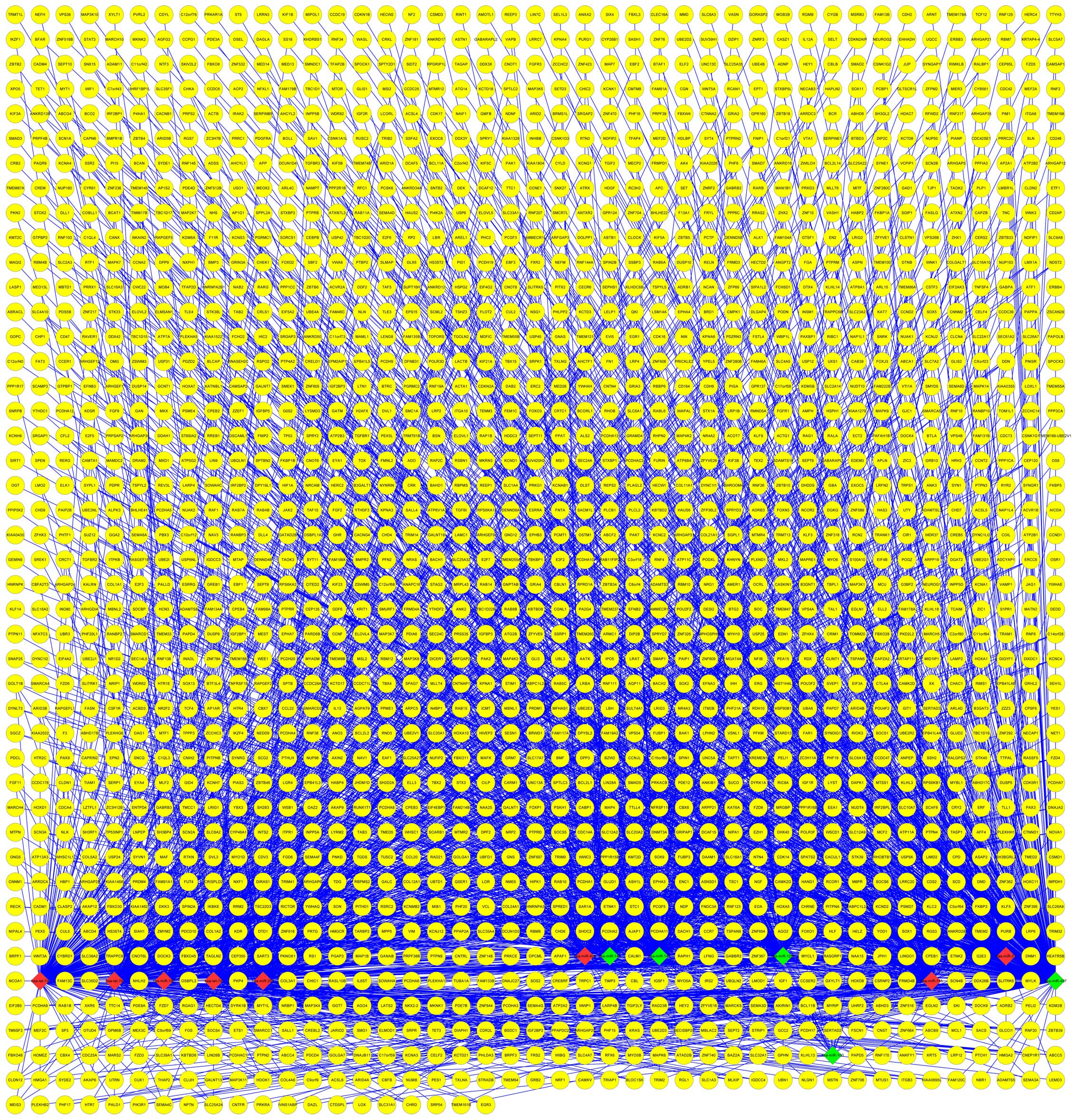
Construction of miRNA-target gene
network
With the correlations between the breast
cancer-related differentially expressed miRNAs and their target
genes, a miRNA-target gene network was constructed using Cytoscape.
In Fig. 1, interactions between eight
upregulated miRNAs (including hsa-miR-425) and five downregulated
miRNAs (including hsa-miR-143, hsa-miR-145 and hsa-miR-125b) and
their various target genes are shown.
Discussion
Like other cancers, breast cancer occurs due to
interactions between an environmental factor and a genetically
susceptible host. In the present study, the underlying mechanism of
the disease was analyzed by using a series of bioinformatics
approaches. Gene expression profile including 77 breast cancer
samples and 17 controls was used in our study, and 34
differentially expressed miRNAs were identified. Furthermore,
breast cancer-related miRNAs that may play roles in breast cancer
development and progression were selected from HMDD.
A total of 13 breast cancer-related differentially
expressed miRNAs, including let-7d, miR-100 and miR-125b, were
identified. Moreover, the target genes of these 13 miRNAs were
predicted from five databases, and the miRNA-target gene network
containing eight upregulated miRNAs (including miR-21, miR-197,
let-7g, let-7e and let-7d) and five downregulated miRNAs (including
miR-143, miR-145 and miR-100) was visualized. The let-7 family is a
tumor-suppressor gene family, which is often inactivated in human
cancers, including breast cancer. As one of the target genes of the
let-7 family, lin-28 homolog A (LIN28) has been reported to
promote tumorigenic activity by suppressing let-7 miRNA maturation
in breast cancer cells (23). miR-100
has been reported to inhibit tumorigenesis in oral squamous cell
carcinoma, ovarian cancer and breast cancer by interfering with
proliferation and survival signaling (24–26).
Additionally, miR-143 and miR-145 have been observed to
synergistically regulate v-erb-b2 avian erythroblastic leukemia
viral oncogene homolog 3 to suppress cell proliferation and
invasion in breast cancer (27).
Moreover, miR-197 and miR-21 were noted to be prominently
upregulated in male breast cancer (28). The deregulation of these miRNAs and
target genes is associated with breast tumorigenesis, and our
results are consistent with these studies, indicating that these
miRNAs and their target genes might be potential therapeutic
targets for breast cancer treatment.
Furthermore, following functional analysis, target
genes were identified to be notably enriched in regulation of
transcription (P=7.43E-13) and pathways related to cancer
(P=3.33E-11). The biological process associated with regulation of
transcription means any process that modulates the frequency, rate
or extent of cellular DNA-templated transcription. A total of 380
target genes, including MEF2C, MEF2A and GDF6,
were enriched in this GO term. MEF2C is involved in cardiac
morphogenesis, myogenesis and vascular development. MEF2C,
which is regulated by let-7 g, has been reported as the main
regulator in primary breast cancer (29). Mutations and deletions of this gene
have been associated with severe mental retardation, stereotypic
movement, epilepsy and cerebral malformation (30). MEF2A, regulated by miR-155, is
involved in vertebrate skeletal muscle development and
differentiation (31). Another target
gene, GDF6, is associated with growth and differentiation of
developing embryos, and is involved in early regulation of cell
growth and development (32). In the
miRNA-target gene network, GDF6 was regulated by several
miRNAs, including let-7d. Taken together, these results might be
useful in developing breast cancer treatments.
Additionally, 76 target genes, including
E2F2, FGF9 and WNT3A, regulated pathways
related to cancer. As a target of the transcription proteins of
small DNA tumor viruses, transcription factor E2F2 controls the
cell cycle and the action of tumor suppressor proteins. In
particular, miR-155 has been reported to regulate tumor development
and metastasis in a mouse model of metastatic breast cancer by
targeting E2F2 (33).
FGF9, regulated by let-7e, is associated with
epithelial-to-mesenchymal transition and invasion by inducing
vascular endothelial growth factor expression (34). WNT3A has been implicated in
oncogenesis, adipogenesis, regulation of cell fate and patterning
during embryogenesis. However, the roles of FGF9 and
WNT3A in breast cancer have not been investigated. In breast
tumorigenesis, the up- or downregulated expression of these miRNAs
and genes may affect cancer pathways, and then induce the
disease.
Our results provide new insights into the
pathogenesis and treatment of breast cancer. The disturbed
cancer-related pathways and regulation of transcription may play
significant roles in breast cancer development. Moreover,
MEF2A, GDF6, FGF9 and WNT3A may be new
targets for breast cancer therapy. These findings may be useful to
researchers and physicians in future studies. However, due to the
limitations of bioinformatics approaches, the results in our study
are predicted, and the findings are not confirmed by traditional
bio-molecular methods. Therefore, further genetic and experimental
studies with a larger sample size are necessary to verify these
results.
References
|
1
|
Liu S, Clouthier SG and Wicha MS: Role of
microRNAs in the regulation of breast cancer stem cells. J Mammary
Gland Biol Neoplasia. 17:15–21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang F, Li H, Tan PH, Chua ET, Yeo RM, Lim
FL, Kim SW, Tan DY and Wong FY: Validation of a nomogram in the
prediction of local recurrence risks after conserving surgery for
Asian women with ductal carcinoma in situ of the breast. Clin Oncol
(R Coll Radiol). 26:684–691. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fortune-Greeley AK, Wheeler SB, Meyer AM,
Reeder-Hayes KE, Biddle AK, Muss HB and Carpenter WR: Preoperative
breast MRI and surgical outcomes in elderly women with invasive
ductal and lobular carcinoma: a population-based study. Breast
Cancer Res Treat. 143:203–212. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hansen BT, Nygård M, Falk RS and Hofvind
S: Breast cancer and ductal carcinoma in situ among women with
prior squamous or glandular precancer in the cervix: a
register-based study. Br J Cancer. 107:1451–1453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cimino D, De Pittà C, Orso F, Zampini M,
Casara S, Penna E, Quaglino E, Forni M, Damasco C, Pinatel E, et
al: miR148b is a major coordinator of breast cancer progression in
a relapse-associated microRNA signature by targeting ITGA5, ROCK1,
PIK3CA, NRAS and CSF1. FASEB J. 27:1223–1235. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meng Y, Zou Q, Liu T, Cai X, Huang Y and
Pan J: microRNA-335 inhibits proliferation, cell-cycle progression,
colony formation, and invasion via targeting PAX6 in breast cancer
cells. Mol Med Rep. 11:379–385. 2015.PubMed/NCBI
|
|
7
|
Tang H, Liu P, Yang L and Xie X, Ye F, Wu
M, Liu X, Chen B, Zhang L and Xie X: miR-185 suppresses tumor
proliferation by directly targeting E2F6 and DNMT1 and indirectly
up-regulating BRCA1 in triple negative breast cancer. Mol Cancer
Ther. 13:3185–3197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: archive for functional
genomics data sets-update. Nucleic Acids Res. 41(Database Issue):
D991–D995. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wilson CL and Miller CJ: Simpleaffy: a
BioConductor package for Affymetrix Quality Control and data
analysis. Bioinformatics. 21:3683–3685. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy - analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T
and Cui Q: HMDD v2.0: a database for experimentally supported human
microRNA and disease associations. Nucleic Acids Res. 42(Database
Issue): D1070–D1074. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila.
Genome Biol. 5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang X and El Naqa IM: Prediction of both
conserved and nonconserved microRNA targets in animals.
Bioinformatics. 24:325–332. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nature
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nature Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao M, Sun J and Zhao Z: TSGene: A web
resource for tumor suppressor genes. Nucleic Acids Res. 41(Database
Issue): D970–D976. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen JS, Hung WS, Chan HH, Tsai SJ and Sun
HS: In silico identification of oncogenic potential of fyn-related
kinase in hepatocellular carcinoma. Bioinformatics. 29:420–427.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord G, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: a novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tran NT and Huang CH: Gene expression and
gene ontology enrichment analysis for H3K4me3 and H3K4me1 in mouse
liver and mouse embryonic stem cell using ChIP-Seq and RNA-Seq.
Gene Regul Syst Bio. 8:33–43. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sakurai M, Miki Y, Masuda M, Hata S,
Shibahara Y, Hirakawa H, Suzuki T and Sasano H: LIN28: a regulator
of tumor-suppressing activity of let-7 microRNA in human breast
cancer. J Steroid Biochem Mol Biol. 131:101–106. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gebeshuber CA and Martinez J: miR-100
suppresses IGF2 and inhibits breast tumorigenesis by interfering
with proliferation and survival signaling. Oncogene. 32:3306–3310.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Henson BJ, Bhattacharjee S, O'Dee DM,
Feingold E and Gollin SM: Decreased expression of miR-125b and
miR-100 in oral cancer cells contributes to malignancy. Genes
Chromosomes Cancer. 48:569–582. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nagaraja AK, Creighton CJ, Yu Z, Zhu H,
Gunaratne PH, Reid JG, Olokpa E, Itamochi H, Ueno NT, Hawkins SM,
et al: A link between mir-100 and FRAP1/mTOR in clear cell ovarian
cancer. Mol Endocrinol. 24:447–463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan X, Chen X, Liang H, Deng T, Chen W,
Zhang S, Liu M, Gao X, Liu Y, Zhao C, et al: miR-143 and miR-145
synergistically regulate ERBB3 to suppress cell proliferation and
invasion in breast cancer. Mol Cancer. 13:2202014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lehmann U, Streichert T, Otto B, Albat C,
Hasemeier B, Christgen H, Schipper E, Hille U, Kreipe HH and Länger
F: Identification of differentially expressed microRNAs in human
male breast cancer. BMC Cancer. 10:1092010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baca-López K, Mayorga M, Hidalgo-Miranda
A, Gutiérrez-Nájera N and Hernández-Lemus E: The role of master
regulators in the metabolic/transcriptional coupling in breast
carcinomas. PLoS One. 7:e426782012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sturtzel C, Testori J, Schweighofer B,
Bilban M and Hofer E: The transcription factor MEF2C negatively
controls angiogenic sprouting of endothelial cells depending on
oxygen. PloS One. 9:e1015212014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou Y, Liu Y, Jiang X, Du H, Li X and Zhu
Q: Polymorphism of chicken myocyte-specific enhancer-binding factor
2A gene and its association with chicken carcass traits. Mol Biol
Rep. 37:587–594. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Banka S, Cain SA, Carim S, Daly SB,
Urquhart JE, Erdem G, Harris J, Bottomley M, Donnai D, Kerr B, et
al: Leri's pleonosteosis, a congenital rheumatic disease, results
from microduplication at 8q22.1 encompassing GDF6 and SDC2 and
provides insight into systemic sclerosis pathogenesis. Ann Rheum
Dis. 74:1249–1256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bollig-Fischer A, Marchetti L, Mitrea C,
Wu J, Kruger A, Manca V and Drăghici S: Modeling time-dependent
transcription effects of HER2 oncogene and discovery of a role for
E2F2 in breast cancer cell-matrix adhesion. Bioinformatics.
30:3036–3043. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Teishima J, Yano S, Shoji K, Hayashi T,
Goto K, Kitano H, Oka K, Nagamatsu H and Matsubara A: Accumulation
of FGF9 in prostate cancer correlates with
epithelial-to-mesenchymal transition and induction of VEGF-A
expression. Anticancer Res. 34:695–700. 2014.PubMed/NCBI
|