Introduction
microRNAs (miRs) are short, non-coding RNAs that
canonically function to repress translation and/or initiate
degradation of target messenger RNAs (mRNAs) (1). The members of the miR-183 family, which
are transcribed as the polycistronic miR-183-96-182 cluster
(1), are consistently expressed at
high levels in a number of human cancer types, including
hormone-dependent breast and prostate cancer (1–8). When
these miRs are overexpressed in cell cultures they behave primarily
as oncogenes or ‘oncomiRs’, and alter phenotypes consistent with
transformation and oncogenesis, including cell proliferation,
migration, invasion and xenograft growth (2–8).
In addition to their intracellular role, mature miRs
are present in secretory vesicles, which may facilitate
cell-to-cell transfer of miRs (9–12).
Exosomes are a subclass of such vesicles, which are 50–150 nm in
size, and are released from the majority of cell types (12,13).
Exosomes were discovered in the 1980s in mammalian reticulocytes
and were originally thought to be a mechanism for the cell to rid
itself of waste (14). However,
exosomes are now known to contain valuable cellular material
including RNA, DNA and proteins (10,15,16). Cells
may take up exosomes by fusion (17)
or by internalization, and thus exosomes are mediators of
cell-to-cell communication (18).
miRs within exosomes are functional in the receiving cells
(9,10,16) and,
in the context of cancer, may act on nearby cells to modify the
local microenvironment during carcinogenesis (16,19,20).
The majority of circulating miRs are within exosomes
(21); thus, exosomes are potentially
a cancer-specific source of miR biomarkers (21–23).
Circulating miRs have been isolated and examined as biomarkers for
breast and prostate cancer (24,25), but,
to the best of our knowledge, few studies have focused specifically
on miRs within exosomes.
Given the well-documented oncogenic role of the
miR-183 family in breast and prostate cancer and the emerging role
of exosomes in miR trafficking, the present study examined the
levels of these miRs in exosomes isolated from cultured breast and
prostate cancer cells. In addition, the present study examined
miR-182 levels in exosomes from cells with stable miR-182
overexpression, and investigated cell-to-cell transfer of miR-182
via exosomes. The expression of the miR-183 family was also
determined in exosomes isolated from fresh serum.
Materials and methods
Cell culture and patient samples
Prostate and breast cell lines were maintained as
recommended by the supplier [American Type Culture Collection
(ATCC), Manassas, Virginia, USA]. Prostate cell lines, RWPE-1 and
RWPE-2, were cultured in keratinocyte serum-free media (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with bovine
pituitary extract (BPE) (Thermo Fisher Scientific, Inc.) and
epidermal growth factor (EGF) (Thermo Fisher Scientific, Inc.), as
recommended by the ATCC. The DU145 prostate cell line was cultured
in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (FBS)
(Sigma-Aldrich, St. Louis, MO, USA). The PC3 prostate cell line was
cultured in RPMI-1640 media (Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS. Primary prostate epithelial cells (PrE)
were derived at University of Illinois at Chicago (Chicago, IL,
USA; University of Illinois at Chicago Cancer Center IRB-approved
protocol) as previously described (6,26,27) and cultured in prostate epithelial cell
growth medium (PrEGM; Lonza Group, Basel, Switzerland) supplemented
with BPE, cholera toxin (Sigma-Aldrich) and EGF. For 48 h prior to
exosome collection, the PrEGM was not supplemented with BPE, and
all cell lines that require FBS were supplemented with exosome-free
FBS. All cells were harvested or used for exosome isolation at 70%
cell density.
A total of three sets of pooled de-identified serum
were collected under an IRB-approved protocol from male and female
patients at University of Illinois at Chicago Hospital (Chicago,
IL, USA). A total of 10 ml each from five patients of fresh, never
frozen sera was combined to provide 50 ml total for exosome
isolation.
Overexpression of miR-182 in
MDA-MB-231 cells
MDA-MB-231 cells were transfected with pCMV-MIR
(scrambled or miR-182; OriGene Technologies, Inc., Rockville, MD,
USA) using Lipofectamine (Thermo Fisher Scientific, Inc.) and
selected with Geneticin (G418; Sigma-Aldrich) according to the
manufacturer's protocol. Following transfection, subpopulations
were clonally selected with G418 and measured for miR expression by
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR). One negative scramble miR control (miR-NEG) and two
miR-182 overexpressing clones (182-1 and 182-7) were used in the
experiments.
Exosome isolation
Prior to isolation, exosome-free FBS was prepared by
two sequential 100,000 × g centrifugations at 4°C of 70 min each.
For PrE and RWPE cells, BPE was omitted from culture medium 48 h
prior to collection for exosome isolation. To collect cell-derived
exosomes, cells were plated on five 100 mm dishes. When cells
reached 70% density, they were washed twice with 1X HEPES-buffered
saline and the media was replaced with exosome-depleted media.
Following 48 h of incubation, the media was harvested and exosomes
isolated by differential centrifugation as previously described
(28). Briefly, cells were removed by
a spin at 300 × g at 4°C for 10 min. The supernatant containing the
exosomes was removed and centrifuged at 2,000 × g at 4°C for 20
min. Supernatant was subsequently placed in polycarbonate bottles
and ultracentrifuged at 10,000 × g at 4°C for 30 min, and the
resulting supernatant was put into a clean polycarbonate bottle and
centrifuged at 100,000 × g at 4°C for 70 min. The supernatant was
discarded and exosomes were washed in 1X phosphate-buffered saline
(PBS), and subsequently repelleted at 100,000 × g at 4°C for 70
min. The supernatant was discarded and exosomes were harvested in
100 µl 1X PBS. Collected exosomes were subjected to RNase A
treatment for 1 h at 37°C prior to RNA analysis.
Human serum exosomes were isolated by an identical
procedure starting with 50 ml of serum diluted with 1X PBS to
completely fill the polycarbonate ultracentrifuge bottles.
Transmission electron microscopy
A total of 20 µl of isolated exosomes were placed on
copper coated grids and allowed to settle for 5 min. Grids were
subsequently stained with 20 µl of uranyl acetate (Sigma-Aldrich)
for 1 min. Grids were allowed to dry overnight prior to viewing and
were analyzed on a JEM-1220 transmission electron microscope (JEOL,
Ltd., Tokyo, Japan).
Exosome quantification
Exosome number was quantified using enzyme-linked
immunosorbent assay (ELISA) for cluster of differentiation (CD)81
[breast cells (29)] and CD9
[prostate cells (30)]. ELISAs were
performed according to the manufacturer's protocol (System
Biosciences, Palo Alto, CA, USA) on freshly harvested exosomes.
Prior to cell treatment with isolated exosomes, the cell number was
determined for the exosome-donating cells, and the exosome number
was normalized to the cell number between treatment and controls
for each cell line.
RT-qPCR
RNA was extracted using the TRIzol reagent (Thermo
Fisher Scientific, Inc.) according to manufacturer's protocol for
preservation of small RNAs. RNA for miR detection was reverse
transcribed using the Universal cDNA Synthesis kit (Exiqon, Inc.,
Woburn, MA, USA) with 100 ng RNA, under the following conditions:
60 min at 42°C, followed by 5 min at 95°C and hold at 4°C. qPCR was
performed on the cDNA using the ExiLENT SYBR Green Master mix
(Exiqon, Inc.) according to the manufacturer's protocol. The
StepOne Plus Real-Time PCR System (Thermo Fisher Scientific, Inc.)
was used for detection and quantitation. The following cycling
conditions were employed for qPCR: 95°C for 10 min, followed by 40
cycles of 95°C for 10 sec and 60°C for 1 min. Primers for the
following were used: small nucleolar RNA U66 (snoR-U66) (product
no. 203905), miR-96 (product no. 204417), miR-182 (product no.
206070) and miR-183 (product no. 206030; Exiqon, Inc.). Relative
quantity was calculated by the ΔΔCq method (31) and normalized to snoR-U66.
Luciferase reporter for miR-182
activity
LightSwitch Synthetic miR Target GoClone Reporter
for miR-182 and the empty vector were purchased from SwitchGear
Genomics (Menlo Park, CA, USA). The LightSwitch vector contains a
Renilla luciferase gene with a synthetic 3′ untranslated
region (UTR) with repeats of the miR-182 binding site. A total of
50 ng of LightSwitch and 50 ng of pGL4-empty vector (Promega
Corporation, Madison, WI, USA) were co-transfected into MDA-231-MB
cells with Dharmafect (Dharmacon; GE Healthcare Life Sciences,
Chalfont, UK). Luciferase activity was measured 24 h later with the
Dual-Luciferase® Assay (Promega Corporation).
Lightswitch Renilla activity was normalized to pGL4 to
assess transfection efficiency.
Co-culture
Naïve MDA-MB-231 cells were seeded into a 12-well
plate and 182-1, 182-7 or NEG control cells were seeded in inserts.
After 24 h of incubation at 37°C in 5% CO2, the inserts
were placed into the wells containing the naïve cells with and
without 10 µM of microvesicle release inhibitor hydrochloride
hydrate (GW4869; Sigma-Aldrich; dimethyl sulfoxide as a control)
and were co-cultured for 5 days at 37°C in 5% CO2. The
medium and GW4869 were replaced after 72 h. Cells were collected
following five days of co-culture and miR-182 levels were measured
by RT-qPCR in the naïve cells.
Statistical analysis
Statistical analysis was performed by Student's
t-test in GraphPad Prism 5 software (GraphPad Software, Inc., La
Jolla, CA, USA). It was noted that when the repeat experiments had
values of zero, the t-test was not valid. P<0.05 was considered
to indicate a statistically significant difference.
Results
Exosomes are produced by breast and
prostate cells
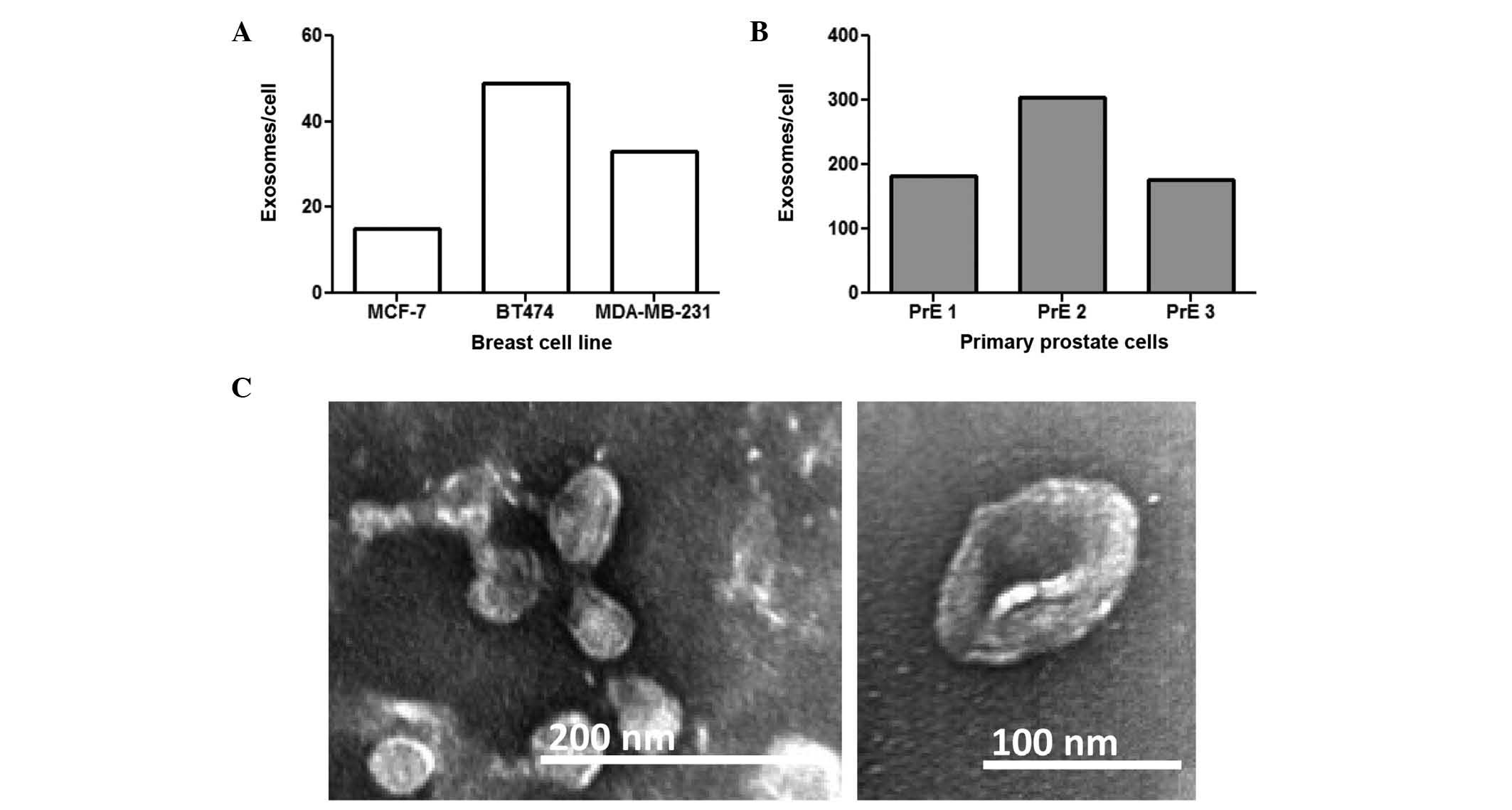
The present study examined exosome production from
several prostate and breast cell lines. Exosomes were isolated by
ultracentrifugation and quantified by ELISA. CD81 was used for
breast cancer cells (29) and CD9 was
used for prostate cells (30) based
on previous reports. All of the breast cancer cell lines examined
(MCF-7, BT474, and MDA-MB-231) released exosomes (Fig. 1A). A total of three distinct
patient-derived primary prostate cell (PrE) populations also
secreted exosomes (Fig. 1B).
Transmission electron micrographs (TEM) confirmed
that the isolated vesicles were exosomes and were the correct shape
and size in the range of 50–150 nM. Representative TEM images of
exosomes from MDA-MB-231 cells are presented in Fig. 1C.
miR-182 is present in exosomes from
breast and prostate cell lines and human serum
Intracellular and exosomal levels of the miR-183
family members were compared in five prostate and three breast cell
lines (Fig. 2A-D). Although miRs-183,
−96 and −182 were all present intracellularly, only miR-182 was
robustly detected in the exosomes (Fig.
2C and D). Notably, RWPE-1 benign immortalized prostate
epithelial cell line and DU145 prostate cancer cell line cells
expressed miR-182 at high levels, but did not have detectable
miR-182 in their exosomes. In the breast cancer cell lines,
MDA-MB-231 cells had the lowest amount of miR-182 in the exosomes
compared with MCF-7 and BT474 cells.
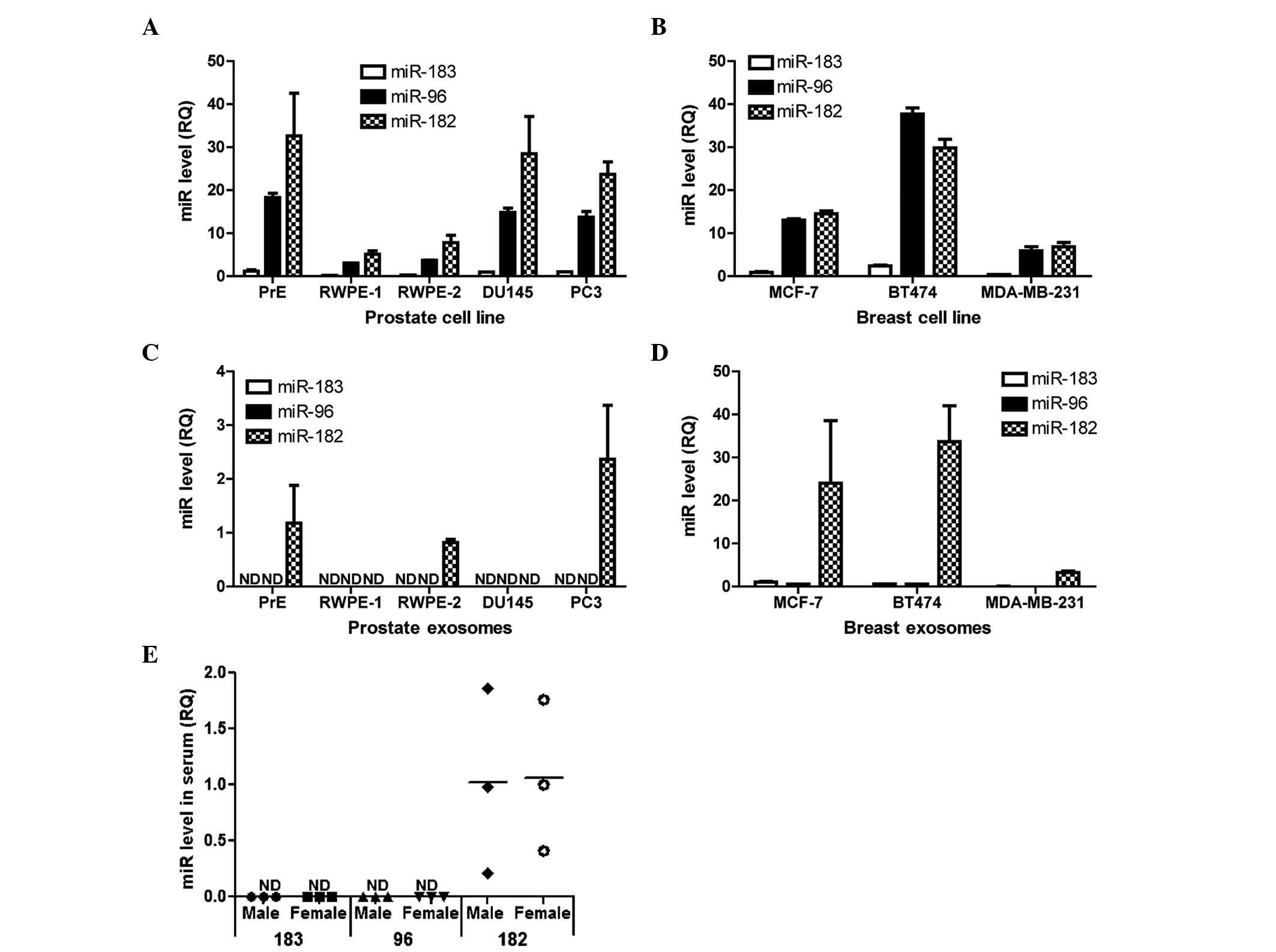 | Figure 2.miR-182 is present in exosomes from
breast cells, prostate cells and human serum. RT-qPCR analyses of
the miR-183 family members (miR-183, miR-96 and miR-182) in (A)
prostate cells (PrE, RWPE-1. RWPE-2, DU145 and PC3), (B) breast
cells (MCF7, BT474 and MDA-MB-231), (C) exosomes secreted by
prostate cells and (D) exosomes secreted by breast cells. Bar
graphs are representative of at least 3 experiments. Data are
presented as the mean ± standard deviation. (E) RT-qPCR analyses of
miR-183, −96 and −182 in pooled serum from men and women. The graph
shows three independent pooled serum samples from male and female
donors. RQ is shown normalized to small nucleolar RNA U66. miR,
microRNA; RT-qPCR, reverse transcription-polymerase chain reaction;
RQ, relative quantity; ND, not detected. |
Exosomes were isolated from fresh human serum to
determine the presence of miR-183 family members. A total of three
independent sets of pooled male and female serum were used for
exosome isolation. miR-182 (Fig. 2E),
but not miRs-183 and 96, was detected in exosomes from males and
females. These data indicate that miR-182 is preferentially
packaged into exosomes in all the cell lines examined, as well as
present in human patient sera.
Overexpression of miR-182 in
MDA-MB-231 breast cancer cells dose-dependently increases exosome
levels of miR-182
To determine if an increase in intracellular miR-182
would alter miR-182 levels in exosomes, miR-182 was stably
overexpressed in the MDA-MB-231 breast cancer cell line. MDA-MB-231
cells were used for these experiments as they had the lowest level
of miR-182 in the exosome experiments. miR-182 was overexpressed
using a hairpin premiR expression plasmid, resulting in a 10- to
50-fold increase in miR-182, as compared with the miR-NEG scrambled
hairpin control (Fig. 3A). Exosomes
isolated from the media of the MDA-182 cells dose-dependently
increased miR-182 10- to 50-fold compared with the miR-NEG control
(Fig. 3B). To adjust for potential
differences in cell density during exosome collection, RNA input
was normalized to the cell number. Activity of the overexpressed
miR-182 was confirmed via the reduction in luciferase activity of a
synthetic 3′UTR reporter (Fig.
3C).
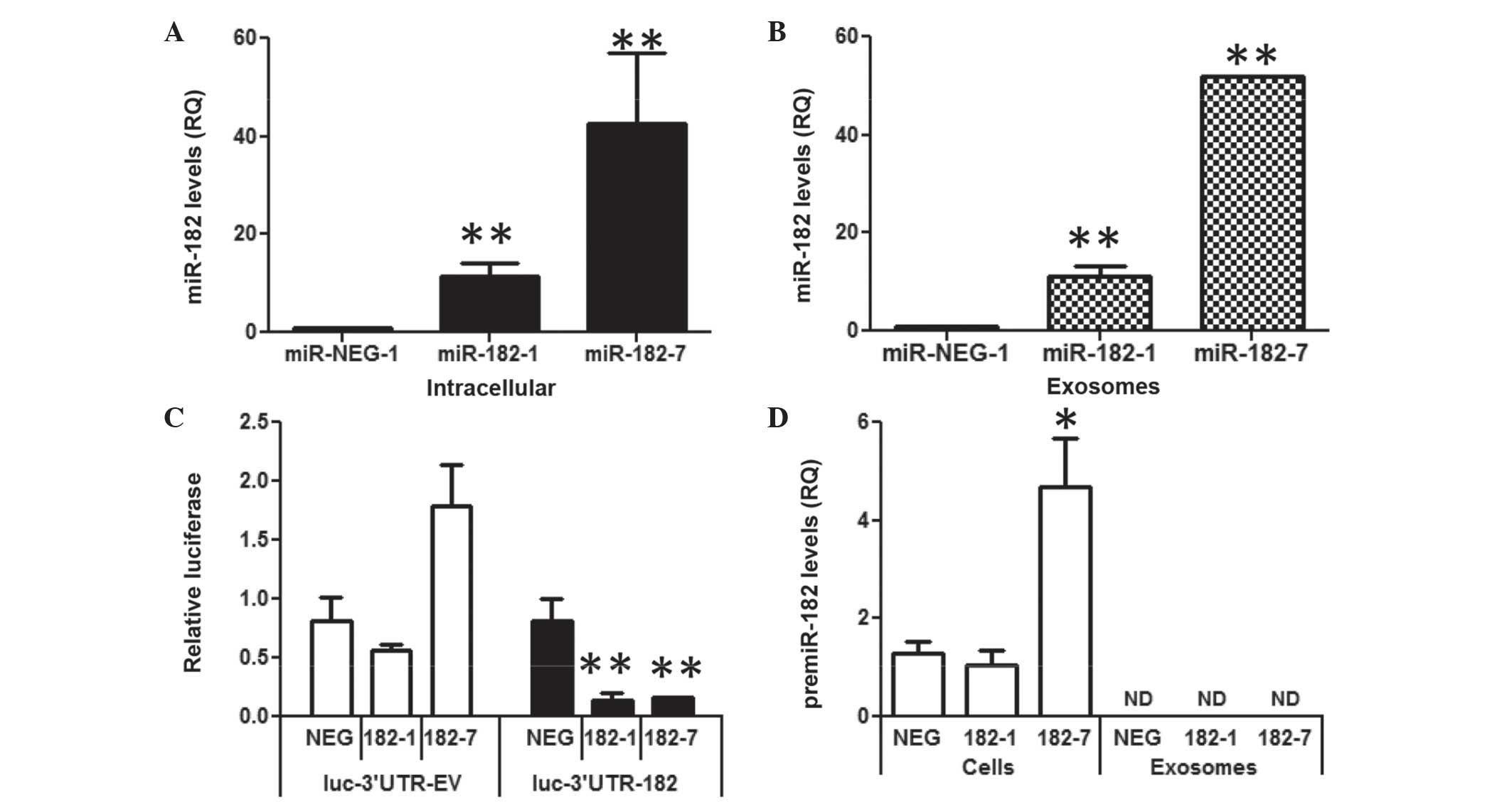 | Figure 3.Overexpression of miR-182 in
MDA-231-MB cells increases intracellular and exosomal mature
miR-182. (A) Intracellular miR-182 levels in 2 clones of MDA-MB-231
cells stably overexpressed miR-182, as compared to an miR-NEG-1
clone, assessed by RT-qPCR. (B) Levels of miR-182 in exosomes
isolated from two clones of MDA-MB-231 cells stably overexpressing
miR-182, as compared to an miR-NEG-1 clone, assessed by RT-qPCR.
(C) Luciferase activity of a reporter with a 3′ untranslated region
containing miR-182 binding sites or EV. (D) premiR-182 expression
in MDA-NEG, MDA-182-1 and MDA-182-7 cells and exosomes, assessed by
RT-qPCR. Data are presented as the mean ± standard error of three
replicate experiments. **P≤0.01 and *P≤0.05 compared with the
negative control, assessed by Student's t-test. miR, microRNA;
RT-qPCR, reverse transcription-polymerase chain reaction; EV, empty
vector; NEG, negative control; RQ, relative quantity; ND, not
detected. |
It has been reported that miR-182 is only present in
its mature form in exosomes (16) in
contrast to other miRs that are present as premiRs and mature miRs
in exosomes (16). In the MDA-182
cells the premiR-182 was only detectable intracellularly and not in
the exosomes (Fig. 3D). Furthermore,
levels of premiR-182 were not significantly different in the
MDA-182 clones compared with MDA-NEG, suggesting a short half-life
for premiR-182 RNA. These data show that in these cells increased
miR-182 expression resulted in increased levels of the mature miR
in the exosomes.
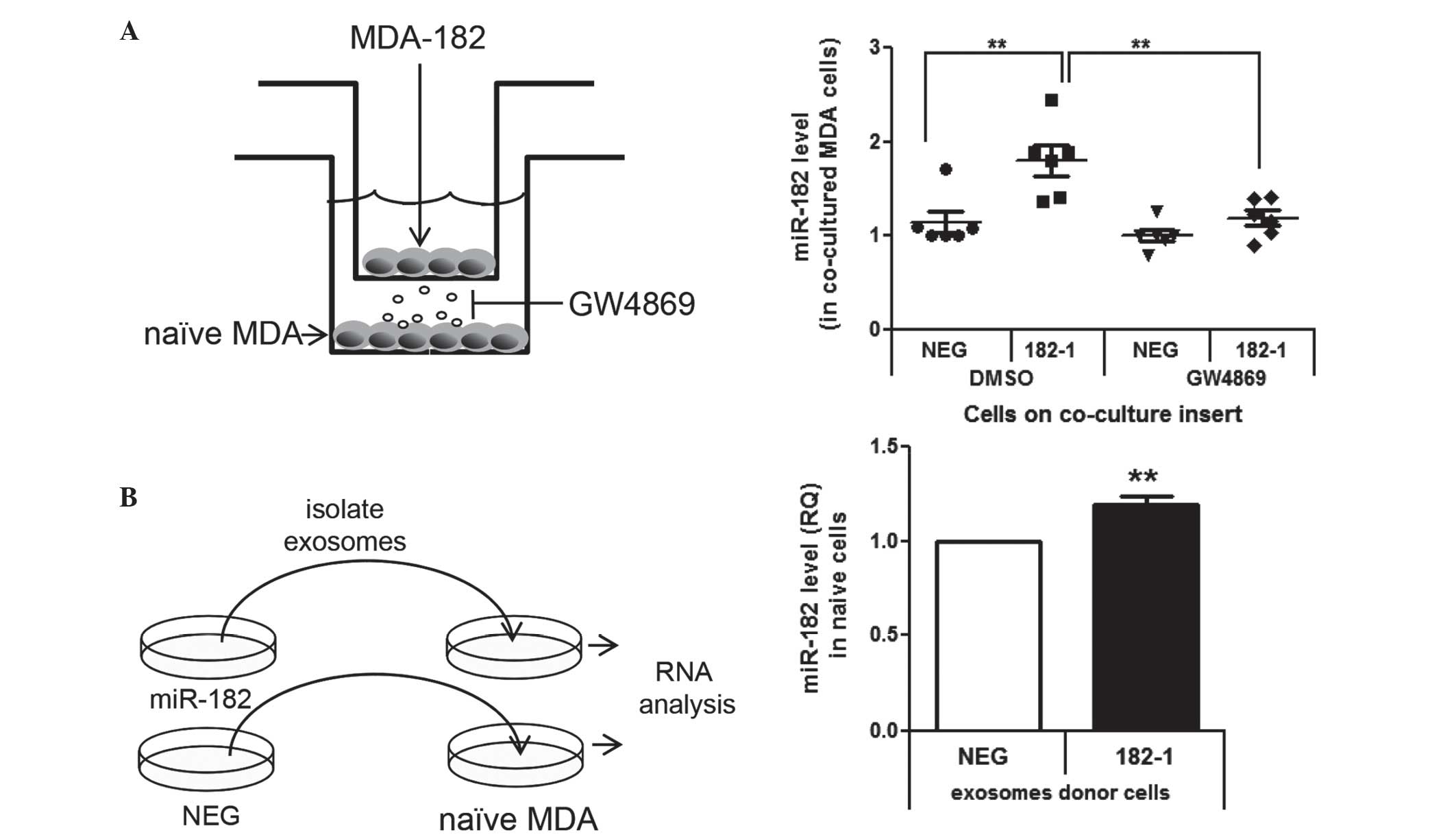
Cell-to-cell transfer of miR-182 in
MDA-MB-231 cells by exosomes
Experiments were designed to determine if miR-182
can be transferred between cells via exosome-mediated mechanisms.
This was achieved by two methods: Co-culture of cells and direct
transfer of isolated exosomes. MDA-182-1 cells were selected for
the following experiments as this clone had an elevated expression
of miR-182 without alterations in cell proliferation. MDA-182-1
cells were co-cultured with naïve MDA-MB-231 cells (Fig. 4A), resulting in an increase in miR-182
in the naïve cells (Fig. 4A).
Addition of GW4869, an inhibitor of microvesicle release (32), blocked this increase, demonstrating
that the transfer of miR-182 between cells was dependent on
microvesicle transfer (Fig. 4A).
Furthermore, direct transfer of isolated exosomes from MDA-182-1
cells to the naïve MDA-MB-231 cells significantly increased levels
of miR-182 in the naïve cells (Fig.
4B).
Discussion
The miR content of cancer cell-derived exosomes
varies based on aggressiveness of phenotype (33,34), and
cancer cells secrete increased exosomal miR compared with normal
cells (16). Consistent with these
previous findings, the present study demonstrated that prostate and
breast cancer cells, as well as noncancerous prostate cells,
secrete exosomes. Furthermore, the present study demonstrated that
miR-182 expression and secretion changes with breast cancer cell
line aggressiveness.
The present study has two significant findings that
have not been previously shown to the best of our knowledge: i)
miR-182 is the only member of the miR-183 family detected in
exosomes from human serum and multiple breast/prostate cell types,
and ii) overexpression of miR-182 dose-dependently increased
miR-182 in exosomes. miR-183, miR-96 and miR-182 have all been
identified to be overexpressed in prostate and breast cancer
tissues (35); however, the present
study observed miR-182 to be present at significantly increased
levels in exosomes compared to miR-96 and miR-183. miR-182 may be
selectively packaged into exosomes. Alternatively, as miR-182 is
expressed at the highest level of the three miRs in the cluster, it
may be passively taken up into the vesicles. Regardless of the
mechanism for preferential miR-182 packaging, the results of the
present study suggest that miR-182, more than the other miR-183
family members, may have significance in cell-cell communication
via microvesicle transfer.
The present study complements a previous study by
Melo et al (16), who
demonstrated that miR-182 could be transferred between cells and
that miR-182 is secreted exclusively as a mature miR, unlike other
exosomal miRs that are secreted as premiRs and processed to the
mature form within exosomes (16).
The present study additionally did not detect premiR-182 in any of
the exosomes, which was consistent with the data from Melo et
al (16). The present study also
observed that exosomes facilitated transfer of miR-182 in breast
cancer cells, as exosomes derived from miR-182 overexpressing
breast cancer cells were able to increase the cellular
concentration of miR-182 in receiving cells. Melo et al
(16) additionally demonstrated that
miR-182 from MDA-231-MB exosomes was functional and led to
decreased mRNA target levels and altered the phenotype of receiving
cells. Therefore, miR-182, not miRs-96 or −183, is likely to
contribute to the cancer microenvironment.
In addition to their known functions within cells,
miRs have been reported to be present at varying levels in the
circulation of cancer patients, suggesting that they may have
utility as biomarkers for diagnosis or prognosis (36–38). miRs
are stable in the serum (36) and
thus are attractive candidates for biomarkers. The miR-183 cluster
members are consistently overexpressed in breast cancer tissues
(39). miR-182 specifically has been
observed to be present at higher levels in the serum of breast
cancer patients and may be used to distinguish between estrogen
receptor and progesterone receptor positive breast cancers
(35,39). However, it remains to be elucidated
whether the serum miR-182 is within exosomes and, if it is, where
these exosomes originate. The present study observed that miR-182,
and not the remaining miR-183 family members, was detected within
exosomes from human male and female serum. However, the present
study did not examine differential levels of miR-182 in cancer
patients. The results of the present study suggest that utility of
serum miR-182 as a biomarker may be improved if exosomes are
examined.
The results of the present study support and
complement existing findings to demonstrate that the miR content of
cancer cell-derived exosomes varies based on the aggressiveness of
the cell (33,34). Furthermore, it was demonstrated that
miR-182 is the miR-183 family member that is trafficked by exosomes
in the investigated cell types and in human serum. Given the
oncogenic role of miR-182, increased exosomal levels of miR-182 in
breast cancer may not only contribute to disease aggressiveness,
but may additionally be exploited as a biomarker. Future studies
are required to elucidate whether miR-182 in exosomes is
functionally relevant to carcinogenesis and/or a biomarker of
disease prognosis.
Acknowledgements
The authors would like to thank Dr. Linda Juarez
(University of Illinois at Chicago) for assistance with
transmission electron microscopy experiments. The present research
was supported by the National Institutes of Health (grant no.
R01-CA166588) and University of Illinois in Chicago Center for
Clinical and Translational Sciences Pre-doctoral Education for
Clinical and Translational Scientists Fellowship.
References
|
1
|
Dambal S, Shah M, Mihelich B and Nonn L:
The microRNA-183 cluster: The family that plays together stays
together. Nucleic Acids Res. 43:7173–7188. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hannafon BN, Sebastiani P, de las Morenas
A, Lu J and Rosenberg CL: Expression of microRNA and their gene
targets are dysregulated in preinvasive breast cancer. Breast
Cancer Res. 13:R242011. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hirata H, Ueno K, Shahryari V, Deng G,
Tanaka Y, Tabatabai ZL, Hinoda Y and Dahiya R: MicroRNA-182-5p
promotes cell invasion and proliferation by down regulating FOXF2,
RECK and MTSS1 genes in human prostate cancer. PLoS One.
8:e555022013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu
C, Li J, Wang X and Song L: Unregulated miR-96 induces cell
proliferation in human breast cancer by downregulating
transcriptional factor FOXO3a. PLoS One. 5:e157972010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lowery AJ, Miller N, Dwyer RM and Kerin
MJ: Dysregulated miR-183 inhibits migration in breast cancer cells.
BMC Cancer. 10:5022010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mihelich BL, Khramtsova EA, Arva N,
Vaishnav A, Johnson DN, Giangreco AA, Martens-Uzunova E, Bagasra O,
Kajdacsy-Balla A and Nonn L: miR-183-96-182 cluster is
overexpressed in prostate tissue and regulates zinc homeostasis in
prostate cells. J Biol Chem. 286:44503–44511. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schaefer A, Jung M, Mollenkopf HJ, Wagner
I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G and Jung
K: Diagnostic and prognostic implications of microRNA profiling in
prostate carcinoma. Int J Cancer. 126:1166–1176. 2010.PubMed/NCBI
|
|
8
|
Ueno K, Hirata H, Shahryari V, Deng G,
Tanaka Y, Tabatabai ZL, Hinoda Y and Dahiya R: microRNA-183 is an
oncogene targeting Dkk-3 and SMAD4 in prostate cancer. Br J Cancer.
108:1659–1667. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Montecalvo A, Larregina AT, Shufesky WJ,
Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G,
Wang Z, et al: Mechanism of transfer of functional microRNAs
between mouse dendritic cells via exosomes. Blood. 119:756–766.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chevillet JR, Kang Q, Ruf IK, Briggs HA,
Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK,
Gallichotte EN, et al: Quantitative and stoichiometric analysis of
the microRNA content of exosomes. Proc Natl Acad Sci USA.
111:14888–14893. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Koga K, Matsumoto K, Akiyoshi T, Kubo M,
Yamanaka N, Tasaki A, Nakashima H, Nakamura M, Kuroki S, Tanaka M
and Katano M: Purification, characterization and biological
significance of tumor-derived exosomes. Anticancer Res.
25:3703–3707. 2005.PubMed/NCBI
|
|
13
|
Dragovic RA, Gardiner C, Brooks AS,
Tannetta DS, Ferguson DJ, Hole P, Carr B, Redman CW, Harris AL,
Dobson PJ, et al: Sizing and phenotyping of cellular vesicles using
nanoparticle tracking analysis. Nanomedicine. 7:780–788.
2011.PubMed/NCBI
|
|
14
|
Trams EG, Lauter CJ, Salem N Jr and Heine
U: Exfoliation of membrane ecto-enzymes in the form of
micro-vesicles. Biochim Biophys Acta. 645:63–70. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Johnstone RM, Adam M, Hammond JR, Orr L
and Turbide C: Vesicle formation during reticulocyte maturation.
Association of plasma membrane activities with released vesicles
(exosomes). J Biol Chem. 262:9412–9420. 1987.PubMed/NCBI
|
|
16
|
Melo SA, Sugimoto H, O'Connell JT, Kato N,
Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, et
al: Cancer exosomes perform cell-independent microRNA biogenesis
and promote tumorigenesis. Cancer Cell. 26:707–721. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Parolini I, Federici C, Raggi C, Lugini L,
Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A,
et al: Microenvironmental pH is a key factor for exosome traffic in
tumor cells. J Biol Chem. 284:34211–34222. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Keller S, König AK, Marmé F, Runz S,
Wolterink S, Koensgen D, Mustea A, Sehouli J and Altevogt P:
Systemic presence and tumor-growth promoting effect of ovarian
carcinoma released exosomes. Cancer Lett. 278:73–81. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jang JY, Lee JK, Jeon YK and Kim CW:
Exosome derived from epigallocatechin gallate treated breast cancer
cells suppresses tumor growth by inhibiting tumor-associated
macrophage infiltration and M2 polarization. BMC Cancer.
13:4212013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Umezu T, Ohyashiki K, Kuroda M and
Ohyashiki JH: Leukemia cell to endothelial cell communication via
exosomal miRNAs. Oncogene. 32:2747–2755. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gallo A, Tandon M, Alevizos I and Illei
GG: The majority of microRNAs detectable in serum and saliva is
concentrated in exosomes. PLoS One. 7:e306792012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng L, Sharples RA, Scicluna BJ and Hill
AF: Exosomes provide a protective and enriched source of miRNA for
biomarker profiling compared to intracellular and cell-free blood.
J Extracell Vesicles. 3:2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Taylor DD and Gercel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Oncol. 110:13–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mihelich BL, Maranville JC, Nolley R,
Peehl DM and Nonn L: Elevated serum microRNA levels associate with
absence of high-grade prostate cancer in a retrospective cohort.
PLoS One. 10:e01242452015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang F, Zheng Z, Guo J and Ding X:
Correlation and quantitation of microRNA aberrant expression in
tissues and sera from patients with breast tumor. Gynecol Oncol.
119:586–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peehl DM: Are primary cultures realistic
models of prostate cancer? J Cell Biochem. 91:185–195. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Peehl DM: Primary cell cultures as models
of prostate cancer development. Endocr Relat Cancer. 12:19–47.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Théry C, Amigorena S, Raposo G and Clayton
A: Isolation and characterization of exosomes from cell culture
supernatants and biological fluids. Curr Protoc Cell Biol Chapter.
3:Unit 3.22. 2006. View Article : Google Scholar
|
|
29
|
Luga V, Zhang L, Viloria-Petit AM,
Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M and
Wrana JL: Exosomes mediate stromal mobilization of autocrine
Wnt-PCP signaling in breast cancer cell migration. Cell.
151:1542–1556. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jansen FH, Krijgsveld J, van Rijswijk A,
van den Bemd GJ, van den Berg MS, van Weerden WM, Willemsen R,
Dekker LJ, Luider TM and Jenster G: Exosomal secretion of
cytoplasmic prostate cancer xenograft-derived proteins. Mol Cell
Proteomics. 8:1192–1205. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak and Schmittgen: Analysis of relative
gene expression data using real-time quantitative PCR and the
2-ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Trajkovic K, Hsu C, Chiantia S, Rajendran
L, Wenzel D, Wieland F, Schwille P, Brügger B and Simons M:
Ceramide triggers budding of exosome vesicles into multivesicular
endosomes. Science. 319:1244–1247. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Llorente A, Skotland T, Sylvänne T,
Kauhanen D, Róg T, Orłowski A, Vattulainen I, Ekroos K and Sandvig
K: Molecular lipidomics of exosomes released by PC-3 prostate
cancer cells. Biochim Biophys Acta. 1831:1302–1309. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ohshima K, Inoue K, Fujiwara A, Hatakeyama
K, Kanto K, Watanabe Y, Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K
and Mochizuki T: Let-7 microRNA family is selectively secreted into
the extracellular environment via exosomes in a metastatic gastric
cancer cell line. PLoS One. 5:e132472010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang QH, Sun HM, Zheng RZ, Li YC, Zhang
Q, Cheng P, Tang ZH and Huang F: Meta-analysis of microRNA-183
family expression in human cancer studies comparing cancer tissues
with noncancerous tissues. Gene. 527:26–32. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Heneghan HM, Miller N, Lowery AJ, Sweeney
KJ, Newell J and Kerin MJ: Circulating microRNAs as novel minimally
invasive biomarkers for breast cancer. Ann Surg. 251:499–505. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang PY, Gong HT, Li BF, Lv CL, Wang HT,
Zhou HH, Li XX, Xie SY and Jiang BF: Higher expression of
circulating miR-182 as a novel biomarker for breast cancer. Oncol
Lett. 6:1681–1686. 2013.PubMed/NCBI
|