Introduction
Renal cell carcinoma (RCC) accounts for ~3% of
malignant tumors in adults and >90% of neoplasms originating
from the kidney (1,2). It has been reported that RCC is the
ninth most common cancer worldwide, with approximately 337,860 new
cases diagnosed in 2012 (3). The
incidence of RCC differs geographically and the highest incidence
occurs in developed countries (1). In
addition, it possesses the highest mortality rate of the
genitourinary cancers and up to 30% of patients exhibit metastases
at the time of initial diagnosis (4).
The increasing incidence and mortality predicts that RCC will
continue to be a significant health burden in the future (5). Although a minority of RCC patients have
a high risk of recurrence, RCC is largely curable by surgery if
detected early (6). However,
detection of cancerous cells at early stages is challenging due to
the lack of early symptoms and incorrect distinction between benign
and malignant masses through imaging or needle biopsies (7). Therefore, investigation of innovative
noninvasive approaches allowing for early detection of RCC has been
performed (8).
Previously, promoter hypermethylation involving DNA
methylation of CpG islands has been considered to be a critical
mechanism during cancer development (9–11).
Aberrant DNA methylation in the regulatory region of
cancer-associated genes has now been established as an alternative
mechanism to heritably silence gene transcription (12,13).
Transcription factor 21 (TCF21) is a validated target of
aberrant promoter hypermethylation in cancer, and is crucial for
the differentiation of epithelial cells adjacent to the mesenchyme
(14). TCF21 is a member of
the basic helix-loop-helix transcription factor family and has a
significant role in the regulation of cell differentiation and cell
fate decisions during development of the lung, kidney and spleen
(15). Furthermore, it has been
considered to be a candidate tumor suppressor at 6q23-q24 that is
epigenetically inactivated in several types of human cancer
(16–18). A previous study reported that the
methylation level of TCF21 was markedly increased in
patients with clear cell RCC (ccRCC) and was additionally an
independent prognostic factor for poor survival (16). Costa et al (19) identified that TCF21 was part of
an innovative panel of biomarkers for simultaneous detection of
bladder cancer, RCC and prostate cancer; however, to the best of
our knoweldege, correlations between TCF21 methylation
levels and clinical parameters have rarely been reported since
then.
In the present study, the clinical potential of
TCF21 methylation in the diagnosis of RCC was investigated
and the correlations between TCF21 methylation levels and
certain clinical parameters in renal tissues and urine samples were
analyzed. The results of the present study indicate that detection
of TCF21 methylation may provide an effective novel method
for diagnosing RCC.
Materials and methods
Patient and tumor sample
collection
The present study was approved by the ethics
committee of First Hospital of Quanzhou Affiliated Fujian Medical
University (Quanzhou, China)(batch number, 20131016) and written
informed consent was obtained from all participants. Between
February 2011 and December 2013, 55 consecutive patients with RCC,
including 30 men and 25 women, who had received treatment at the
First Hospital of Quanzhou Affiliated Fujian Medical University
were enrolled in the present study. Tumor samples were obtained
from these patients subsequent to resection. Samples were instantly
snap-frozen, stored at −80°C in liquid nitrogen and cut in a
cryostat (Reichert Jung Cryocut 1800; Leica Microsystems, Inc.,
Buffalo Grove, IL, USA) for subsequent DNA extraction. The normal
renal cell tissue samples from 22 RCC-free individuals were used as
controls.
Urine sample collection and
processing
First morning voided urine samples (1 sample per
patient; 20–50 ml) were collected from 33 patients (28 males and 5
females) with RCC, who had been diagnosed and treated between
February 2011 and December 2013 in the Department of Urology of
First Hospital of Quanzhou Affiliated Fujian Medical University.
Healthy individuals with no history of occupational exposure to
carcinogens or personal/familial history of cancer were selected as
the control group (n=15; 10 males and 5 females). Written informed
consent was obtained from all subjects prior to enrolling into the
study. The storage and processing procedures for urine samples were
standardized. Briefly, samples were centrifuged at 1,776 × g for 10
min, and the pelleted urine sediment was rinsed with
phosphate-buffered saline (Shanghai Qifa Biotechnology Co., Ltd.,
Shanghai, China) 2 times for 10 min each and stored at −80°C until
required.
Isolation of nucleic acids and
bisulfite treatment
DNA was extracted from the frozen tissue and urine
samples using the AllPrep DNA Mini kit (Qiagen, Inc., Valencia, CA,
USA) according to the manufacturer's protocol. The concentration
and purity of DNA were assessed by determining the
OD260/OD280 ratio using a Nanodrop 1000
spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA,
USA). DNA was treated by bisulfite using an EZ-96 DNA
Methylation-Gold™ kit (Zymo Research, Irvine, CA, USA) according to
the manufacturer's protocol, as well as the previously described
protocol (20), and stored at −80°C
until use.
Quantitative pyrosequencing
methylation analysis
In order to detect the methylation level of the
TCF21 gene in urine and tissue samples, a total of 22 CpG
loci of 3 fragments in the TCF21 gene were selected for
methylation. Polymerase chain reaction (PCR) and TCF21
sequencing primers were designed with the PyroMark Assay Design 2.0
software (Qiagen, Inc.). The primers were as follows: Forward,
TTAGTTAGGAGGGGAAGTAGGTTT; reverse, ACACCCAAAACAAAATAATCTTAAATCT;
and sequencing, GGGGAAGTAGGTTTAG for TCF21 1; forward,
AGTGTTTTAGGGGTTGTAGTTGTAGTTTA; reverse, CACACCCCCACTCCCAAC; and
sequencing, GTTGTAGTTGTAGTTTAGG TCF21 2; and forward,
GGTGGAAGGTTTAGAAAGAGTTA; reverse, ACCACCTTCTCCCAACTA; and
sequencing, GGAAGGTTTAGAAAGAGTTAA TCF21 3. PCR was performed
with the HotStarTaq Master Mix kit (Qiagen, Inc.) according to the
manufacturer's protocol, with 1 µl of bisulfate-converted DNA. PCR
amplification was performed as follows: Denaturation at 95°C for 5
min, followed by 45 cycles at 95°C for 1 min, annealing at 36°C for
1 min and a final extension at 72°C for 5 min. Following PCR
amplification, pyrosequencing was performed using a PyroMark Gold
Q96 SQA Reagents kit (Qiagen, Inc.) and a PSQ96 HS DNA analyzing
system (Qiagen, Inc.) according to the manufacturer's protocol.
Pyro Q-CpG software version 1.0.9 (Qiagen, Inc.) was used to
calculate the percentage of methylation w, as follows: Methylation
(%) = methylated cytosine / total cytosine × 100.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Statistical analyses were performed using SPSS for Windows, version
20 (IBM SPSS, Armonk, NY, USA). Pairwise comparison utilized the
Mann-Whitney U test. Spearman's rank correlation was employed in
order to examine the association between multiple clinical
parameters. A receiver operating characteristic (ROC) curve was
constructed in order to evaluate the accuracy of the predictive
model. P<0.05 was considered to indicate a statistically
significant difference.
Results
Methylation level of the TCF21
gene
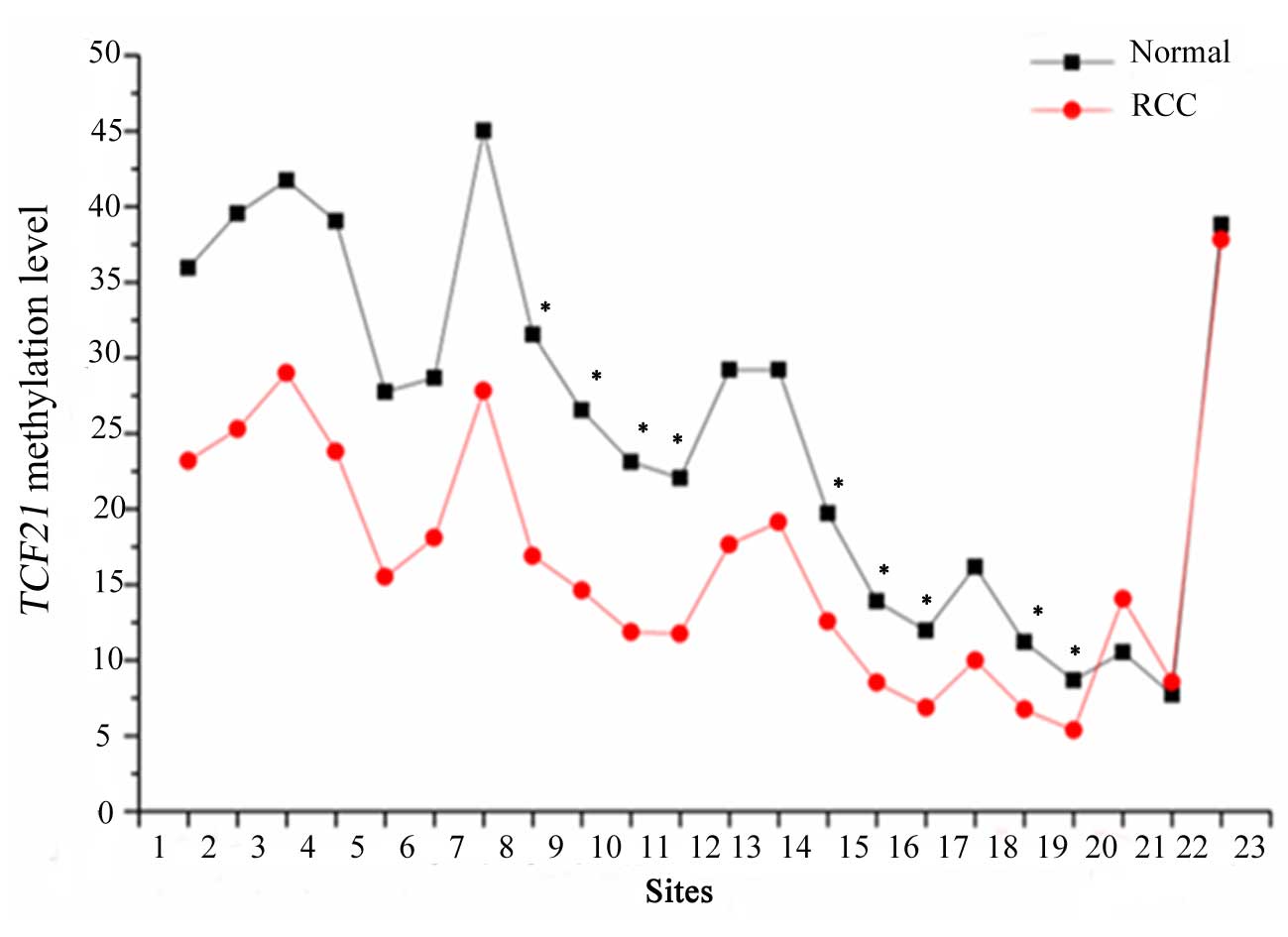
The data analysis of the first fragment of
TCF21 is presented in Fig. 1.
A total of 22 CpG sites of 3 fragments in the TCF21 gene
promoter in each sample were detected. The results revealed that
there were significant differences in the methylation level of the
first fragment in the cancer and normal tissues at 9 sites (sites
8–11, 14–16, 18–19). In addition, TCF21 methylation levels
were significantly increased in cancerous renal tissue and urine
samples compared with normal renal tissue and urine samples.
Clinicopathological analysis of TCF21
methylation level in RCC tissue
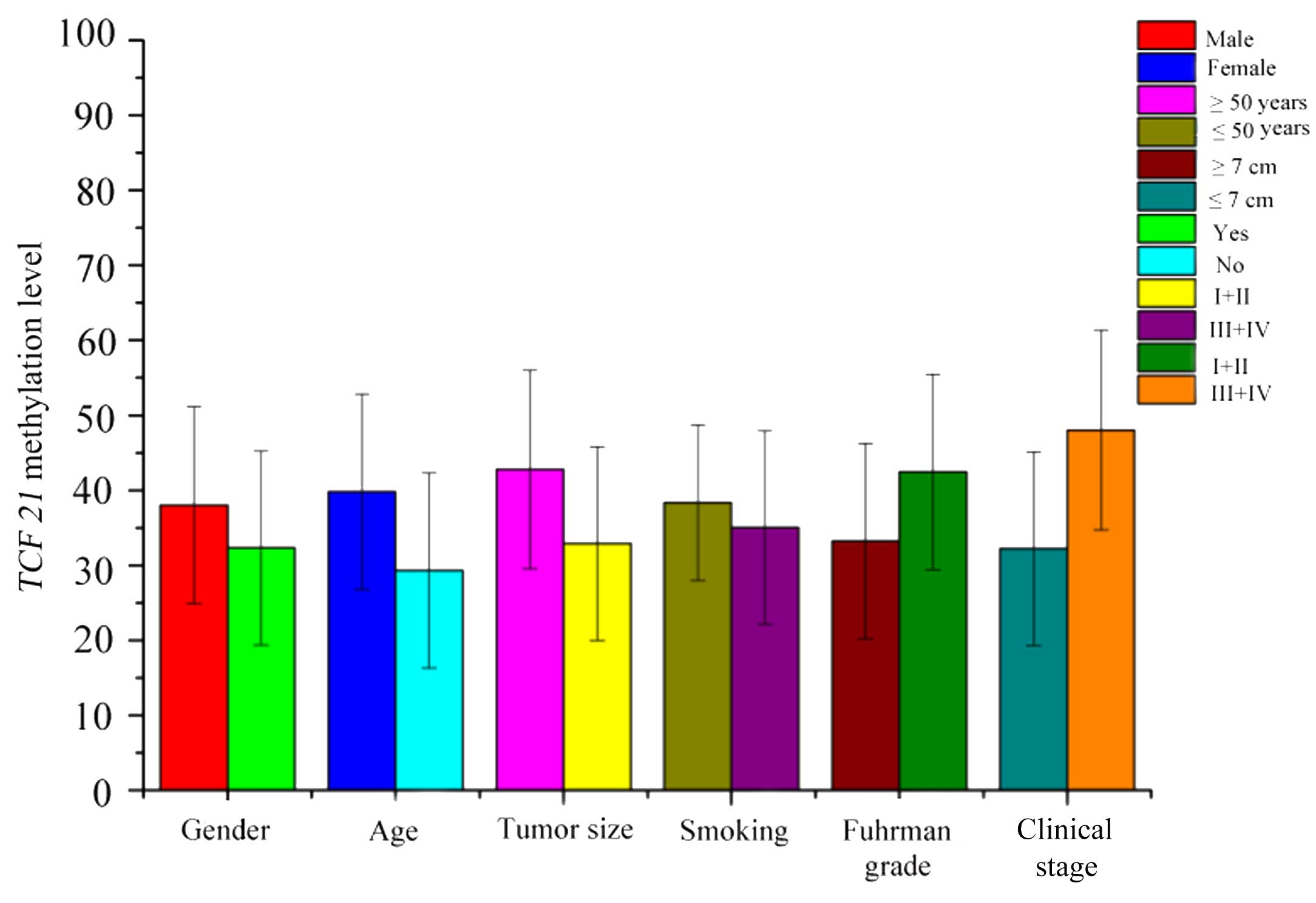
An analysis of TCF21 methylation levels in
RCC tissues in relation to various clinical parameters is presented
in Fig. 2. The results of the
analysis demonstrated that TCF21 methylation levels were
increased in male patients, patients aged ≥50 years, smokers,
patients with tumor size ≥7 cm and clinical stage III or IV
(21) compared with female patients,
patients aged <50 years, nonsmokers, patients with tumor size
<7 cm and Fuhrman grade I or II (22). The Spearman's correlation coefficient
between TCF21 methylation levels and clinical parameters is
summarized in Table I. The results
demonstrated that TCF21 methylation level was positively
associated with age (r=0.403; P=0.002), smoking history (r=0.321;
P=0.017) and Fuhrman grade (r=0.271; P=0.045). However, there were
no significant differences for gender (r=0.166; P=0.227), tumor
size (r=0.110; P=0.423) and clinical stage (r=0.258; P=0.057).
 | Table I.Correlation between transcription
factor 21 methylation levels and clinical parameters in renal cell
carcinoma tissue. |
Table I.
Correlation between transcription
factor 21 methylation levels and clinical parameters in renal cell
carcinoma tissue.
| Clinical
parameter | P-value | r |
|---|
| Gender | 0.227 | 0.166 |
| Age | 0.002 | 0.403 |
| Tumor size | 0.423 | 0.11 |
| Smoking | 0.017 | 0.321 |
| Fuhrman grade | 0.045 | 0.271 |
| Clinical stage | 0.057 | 0.258 |
Clinicopathological analysis of TCF21
methylation level in urine samples with RCC
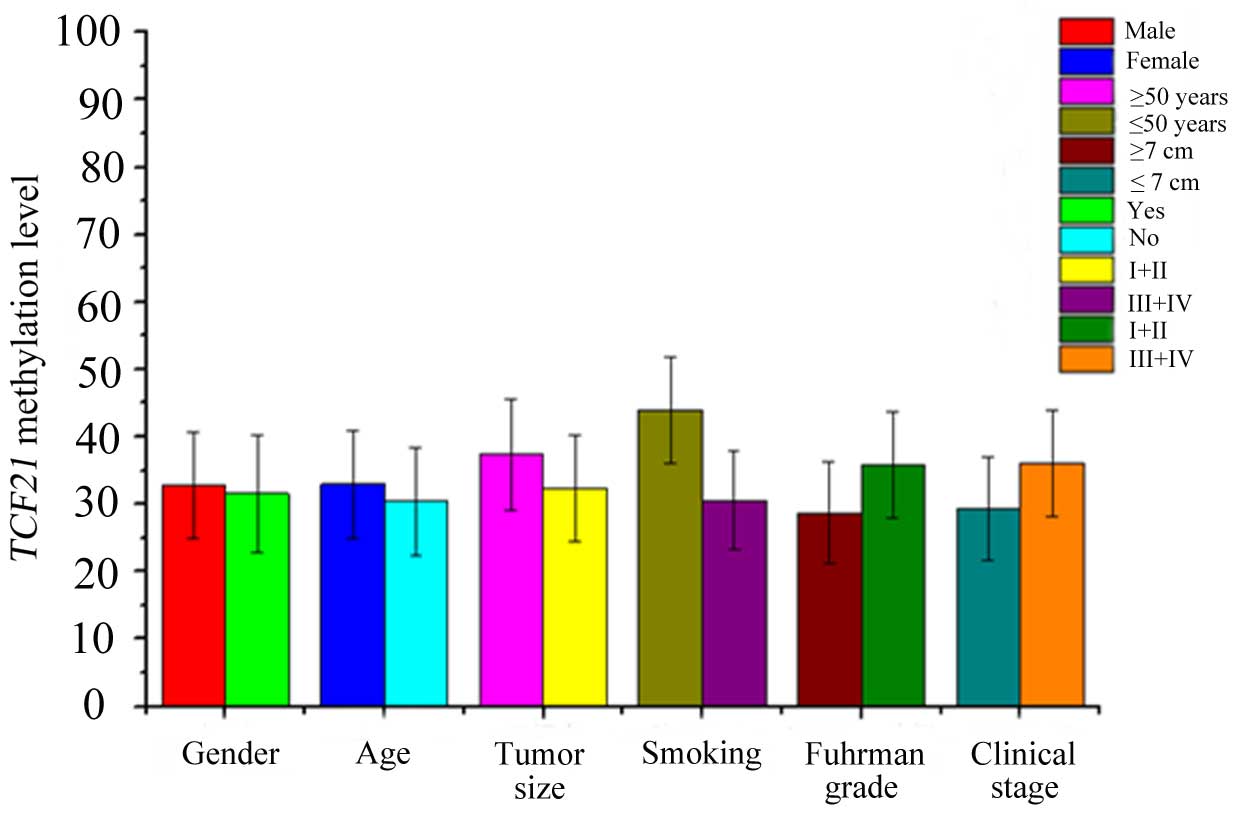
A comparison between TCF21 methylation levels
in urine samples with RCC and various clinical parameters is
presented in Fig. 3. Similarly,
TCF21 methylation levels of male patients, patients aged ≥50
years, smokers, patients with tumor size ≥7 cm and clinical stage
III or IV were increased compared with female patients, patients
aged <50 years, nonsmokers, patients with tumor size <7 cm
and Fuhrman grade I or II. Spearman's correlation analysis revealed
that the TCF21 methylation level was positively associated
with tumor size (r=0.622; P<0.001), Fuhrman grade (r=0.411;
P=0.017) and clinical stage (r=0.411; P=0.017). However, there were
no significant differences for gender (r=0.142; P=0.430), age
(r=0.067; P=0.713) and smoking history (r=0.441; P=0.010; Table II).
 | Table II.Correlation between transcription
factor 21 methylation levels and clinical parameters in urine
samples with renal cell carcinoma. |
Table II.
Correlation between transcription
factor 21 methylation levels and clinical parameters in urine
samples with renal cell carcinoma.
| Clinical
parameter | P-value | r |
|---|
| Gender | 0.43 | 0.142 |
| Age | 0.713 | 0.067 |
| Tumor size | 0.000 | 0.622 |
| Smoking | 0.204 | 0.227 |
| Fuhrman grade | 0.010 | 0.441 |
| Clinical stage | 0.017 | 0.411 |
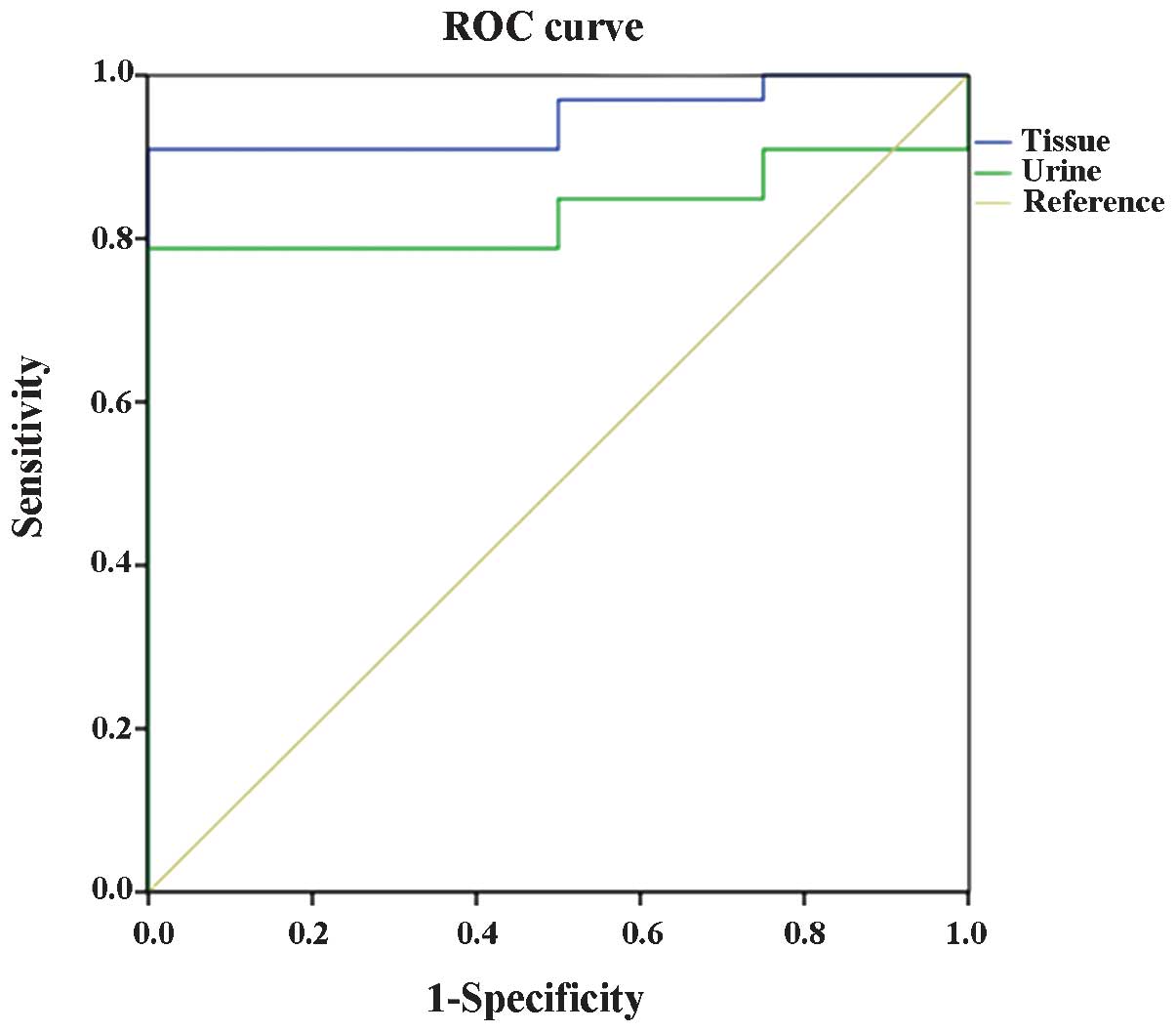
ROC curve of TCF21 methylation level
in RCC tissue and urine samples
The ROC curve of TCF21 methylation level in
RCC tissue and urine samples is presented in Fig. 4. In the ROC curve analysis of
TCF21 methylation level in RCC tissue, the cut-off value was
23.61% and the sensitivity and specificity were 89.00 and 61.90%,
respectively for predicting RCC. The ROC curve analysis of
TCF21 methylation level in urine samples with RCC
demonstrated a cut-off value of 26.84% and the sensitivity and
specificity were 79.00 and 100.00%, respectively for predicting
RCC. Furthermore, there were significant differences in the area
under the curve between the tissue and urine samples (P=0.004),
which may indicate that the diagnostic validity of the TCF21
methylation level in RCC tissue was increased compared with urine
samples with RCC.
Discussion
In the present study, the clinical potential of the
observation of TCF21 methylation for the diagnosis of RCC
was investigated in tissue and urine samples. The results of the
present study demonstrated that TCF21 methylation levels
were significantly increased in RCC tissue and urine samples
compared with normal tissue and urine samples. Furthermore,
TCF21 methylation was additionally positively associated
with age, smoking history and Fuhrman grade in RCC tissues and
tumor size, Fuhrman grade and clinical stage in urine samples. In
addition, the sensitivity was significantly increased in RCC tissue
samples compared with urine samples. It was concluded that
TCF21 may be a useful biomarker for diagnosing RCC, and
TCF21 methylation levels in urine samples may be a novel
method of diagnosing RCC.
It is a challenging task to screen for the early
stages of RCC. RCC tends to be clinically asymptomatic during its
earliest stages; therefore, approximately 20–30% of cases are
diagnosed at a locally advanced stage or with metastasis (23). Although noninvasive imaging (including
computed tomography and ultrasonography) has been widely used in
the detection of RCC, diagnostic challenges are arising due to
problems with accurate discrimination of benign from malignant
masses and accurate categorization (24). Thus, the development of more efficient
and accurate detection of RCC at early stages is imperative. The
role of epigenetic alterations and gene promoter hypermethylation
in the early diagnosis of cancer is receiving increasing attention
(7,25–28). An
emerging class of cancer biomarkers, including
prostaglandin-endoperoxide synthase 2, Ras association domain
family 1 isoform A, retinoic acid receptor β and Kelch-like family
member 35, has been previously reported to be methylated in RCC,
with a sensitivity of 31–77% and specificity of 75–91% (29–32). In
addition, Costa et al (19)
demonstrated that protocadherin-17 and TCF21 aberrant
promoter methylation may have a critical role in urological
cancers. Furthermore, Ye et al (16) indicated that TCF21 may be a
predictive biomarker of poor prognosis in patients with ccRCC.
However, correlations between TCF21 methylation levels and
clinical parameters were not reported in this previous study
(16).
TCF21 is a newly recognized target of
aberrant promoter hypermethylation in malignancies, which encodes a
basic helix-loop-helix transcription factor (33,34). It is
involved in mesenchymal-epithelial cell transition and controls
cell fate determination and tissue differentiation in the embryo
(35). The present study investigated
the clinical potential of TCF21 methylation in the diagnosis
of RCC, and additionally evaluated the correlations between
TCF21 methylation levels and certain clinical parameters in
renal tissues and urine samples. The results of the present study
revealed that TCF21 methylation levels were significantly
increased in RCC samples compared with those of normal renal
tissues or urine samples, which was in line with the findings of
previous studies (16,19). The Spearman's correlation coefficient
revealed that the TCF21 methylation level was positively
associated with age, smoking history and Fuhrman grade in RCC
tissues and associated with tumor size, Fuhrman grade and clinical
stage in urine samples. The above results indicated that the
TCF21 methylation level was associated with the degree of
malignancy and differentiation of RCC; the higher the level of
TCF21 methylation, the higher the degree of RCC malignancy.
In addition, age and smoking history were associated with RCC;
however, the mechanisms underlying these associations remain to be
elucidated. The present study hypothesized that aging and smoking
may act on the aberrant methylation of TCF21.
ROC curves revealed the cut-off value, sensitivity
and specificity for predicting RCC were 23.61, 89.00 and 61.90%,
respectively in tissue samples and were 26.84, 79.00 and 100.00%,
respectively in urine samples. Significant differences were
identified in the area under the curve between the tissue and urine
samples. These results are similar to a previous finding which
demonstrated a significant decrease in sensitivity in urine sample
(36). One of the primary reasons for
these observed results may be due to the varying amount of
neoplastic cells in tissues and urine samples. Therefore, the
results might be improved by increase the volume of tested urine
samples, because it seems to increase the content of DNA from RCC
tumors. Despite these marked differences in the area under the
curve, TCF21 methylation levels in urine samples may be
considered to be a novel and useful method of diagnosing RCC.
In conclusion, the results of the present study
demonstrated that TCF21 methylation levels were
significantly increased in RCC tissue and urine samples compared
with normal tissue and urine samples. TCF21 may be utilized
as a biomarker for the diagnosis of RCC and the detection of
methylation levels in RCC urine samples may be a novel and
efficient method of diagnosing RCC, due to the noninvasive nature
of this method
Acknowledgements
The present study was supported by Quanzhou Science
and Technology Project (grant no. 2013Z52; Quanzhou, China) and
Quanzhou Medical College Science and Technology Project (grant no.
XJ1305; Quanzhou, China).
References
|
1
|
Ljungberg B, Campbell SC, Choi HY, Jacqmin
D, Lee JE, Weikert S and Kiemeney LA: The epidemiology of renal
cell carcinoma. Eur Urol. 60:615–621. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sachdeva K, Curti B and Bagi JRP: Renal
cell carcinoma. http://emedicine.medscape.com/article/281340-overviewAccessed.
May 17–2016
|
|
3
|
Maher ER: Genomics and epigenomics of
renal cell carcinoma. Semin Cancer Biol. 23:10–17. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Janzen NK, Kim HL, Figlin RA and
Belldegrun AS: Surveillance after radical or partial nephrectomy
for localized renal cell carcinoma and management of recurrent
disease. Urol Clin North Am. 30:843–852. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Smith SJ, Bosniak MA, Megibow AJ, Hulnick
DH, Horii SC and Raghavendra BN: Renal cell carcinoma: Earlier
discovery and increased detection. Radiology. 170:699–703. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cairns P: Gene methylation and early
detection of genitourinary cancer: The road ahead. Nat Rev Cancer.
7:531–543. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Scélo G and Brennan P: The epidemiology of
bladder and kidney cancer. Nat Clin Pract Urol. 4:205–217. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park SY, Kwon HJ, Lee HE, Ryu HS, Kim SW,
Kim JH, Kim IA, Jung N, Cho NY and Kang GH: Promoter CpG island
hypermethylation during breast cancer progression. Virchows Arch.
458:73–84. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park JY: Promoter hypermethylation in
prostate cancer. Cancer Control. 17:245–255. 2010.PubMed/NCBI
|
|
11
|
Akhavan-Niaki H and Samadani AA: DNA
methylation and cancer development: Molecular mechanism. Cell
Biochem Biophys. 67:501–513. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vucic EA, Brown CJ and Lam WL: Epigenetics
of cancer progression. Pharmacogenomics. 9:215–234. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Duffy M, Napieralski R, Martens JW, Span
PN, Spyratos F, Sweep FC, Brunner N, Foekens JA and Schmitt M:
EORTC PathoBiology Group: Methylated genes as new cancer
biomarkers. Eur J Cancer. 45:335–346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Quaggin SE, Schwartz L, Cui S, Igarashi P,
Deimling J, Post M and Rossant J: The basic-helix-loop-helix
protein pod1 is critically important for kidney and lung
organogenesis. Development. 126:5771–5783. 1999.PubMed/NCBI
|
|
16
|
Ye YW, Jiang ZM, Li WH, Li ZS, Han YH, Sun
L, Wang Y, Xie J, Liu YC, Zhao J, et al: Down-regulation of TCF21
is associated with poor survival in clear cell renal cell
carcinoma. Neoplasma. 59:599–605. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Arab K, Smith LT, Gast A, Weichenhan D,
Huang JP, Claus R, Hielscher T, Espinosa AV, Ringel MD, Morrison
CD, et al: Epigenetic deregulation of TCF21 inhibits metastasis
suppressor KISS1 in metastatic melanoma. Carcinogenesis.
32:1467–1473. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Richards KL, Zhang B, Sun M, Dong W,
Churchill J, Bachinski LL, Wilson CD, Baggerly KA, Yin G, Hayes DN,
et al: Methylation of the candidate biomarker TCF21 is very
frequent across a spectrum of early-stage nonsmall cell lung
cancers. Cancer. 117:606–617. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Costa VL, Henrique R, Danielsen SA, Eknaes
M, Patrício P, Morais A, Oliveira J, Lothe RA, Teixeira MR, Lind GE
and Jerónimo C: TCF21 and PCDH17 methylation: An innovative panel
of biomarkers for a simultaneous detection of urological cancers.
Epigenetics. 6:1120–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shaw RJ, Liloglou T, Rogers SN, Brown JS,
Vaughan ED, Lowe D, Field JK and Risk JM: Promoter methylation of
P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer:
Quantitative evaluation using pyrosequencing. Br J Cancer.
94:561–568. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zisman A, Pantuck AJ, Dorey F, Said JW,
Shvarts O, Quintana D, Gitlitz BJ, deKernion JB, Figlin RA and
Belldegrun AS: Improved prognostication of renal cell carcinoma
using an integrated staging system. J Clin Oncol. 19:1649–1657.
2001.PubMed/NCBI
|
|
22
|
Lang H, Lindner V, de Fromont M, Molinié
V, Letourneux H, Meyer N, Martin M and Jacqmin D: Multicenter
determination of optimal interobserver agreement using the Fuhrman
grading system for renal cell carcinoma. Cancer. 103:625–629. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lam JS, Leppert JT, Belldegrun AS and
Figlin RA: Novel approaches in the therapy of metastatic renal cell
carcinoma. World J Urol. 23:202–212. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang XJ, Sugimura J, Schafernak KT,
Tretiakova MS, Han M, Vogelzang NJ, Furge K and Teh BT:
Classification of renal neoplasms based on molecular signatures. J
Urol. 175:2302–2306. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jerónimo C, Henrique R and Sidransky D:
Uses of DNA methylation in cancer diagnosis and risk assessment.
DNA Methylation: Approaches, Methods and Applications. Esteller M:
(1st). CRC Press. (Boca Raton, FL). 11–26. 2004. View Article : Google Scholar
|
|
26
|
Henrique R, Luís AS and Jerónimo C: The
epigenetics of renal cell tumors: From biology to biomarkers. Front
Genet. 3:942012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jerónimo C and Henrique R: Epigenetic
biomarkers in urological tumors: A systematic review. Cancer Lett.
342:264–274. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Morris MR and Maher ER: Epigenetics of
renal cell carcinoma: The path towards new diagnostics and
therapeutics. Genome Med. 2:592010. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Costa VL, Henrique R, Ribeiro FR, Pinto M,
Oliveira J, Lobo F, Teixeira MR and Jerónimo C: Quantitative
promoter methylation analysis of multiple cancer-related genes in
renal cell tumors. BMC Cancer. 7:1332007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Morris MR, Ricketts C, Gentle D,
Abdulrahman M, Clarke N, Brown M, Kishida T, Yao M, Latif F and
Maher ER: Identification of candidate tumour suppressor genes
frequently methylated in renal cell carcinoma. Oncogene.
29:2104–2117. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hoque MO, Begum S, Topaloglu O, Jeronimo
C, Mambo E, Westra WH, Califano JA and Sidransky D: Quantitative
detection of promoter hypermethylation of multiple genes in the
tumor, urine and serum DNA of patients with renal cancer. Cancer
Res. 64:5511–5517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Morris MR, Ricketts CJ, Gentle D, McRonald
F, Carli N, Khalili H, Brown M, Kishida T, Yao M, Banks RE, et al:
Genome-wide methylation analysis identifies epigenetically
inactivated candidate tumour suppressor genes in renal cell
carcinoma. Oncogene. 30:1390–1401. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Smith LT, Lin M, Brena RM, Lang JC,
Schuller DE, Otterson GA, Morrison CD, Smiraglia DJ and Plass C:
Epigenetic regulation of the tumor suppressor gene TCF21 on
6q23-q24 in lung and head and neck cancer. Proc Natl Acad Sci USA.
103:982–987. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Quaggin SE, Vanden Heuvel GB and Igarashi
P: Pod-1, a mesoderm-specific basic-helix-loop-helix protein
expressed in mesenchymal and glomerular epithelial cells in the
developing kidney. Mech Dev. 71:37–48. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Quaggin SE, Schwartz L, Cui S, Igarashi P,
Deimling J, Post M and Rossant J: The basic-helix-loop-helix
protein pod1 is critically important for kidney and lung
organogenesis. Development. 126:5771–5783. 1999.PubMed/NCBI
|
|
36
|
Costa VL, Henrique R, Danielsen SA, Eknaes
M, Patricio P, Morais A, Olieira J, Lothe RA, Teixeira MR, Lind GE
and Jerónimo C: TCF21 and PCDH17 methylation: An innovative panel
of biomarkers for a simultaneous detection of urological cancers.
Epigenetics. 6:1120–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|