Introduction
Glioblastoma multiforme accounts for up to 60% of
all malignant primary brain tumors in adults (1). The average patient survival remains poor
despite treatments including surgery, radiotherapy and chemotherapy
(1). Numerous studies have focused on
glioblastoma ‘stem cells’ or ‘initiating cells’, which constitute a
small population of the tumor, but which possess significant
tumorigenicity and resistance to conventional radiotherapy and
chemotherapy (2).
Due to the marked heterogeneity of glioblastoma
tissue, methods for selecting the stem cell population specifically
and effectively remain challenging (3). One simple and popular method to isolate
cancer stem cells is based on the use of a specific stem cell
marker, for example, the widely used cluster of differentiation
(CD) 133, a pentaspan transmembrane molecule (4). However, the application of this molecule
as a stem cell marker remains debatable. In glioma, CD133-negative
tumor cells are able to produce CD133-positive cells; furthermore,
CD133-negative cells demonstrate tumorigenicity similar to
CD133-positive cells (5,6). The glial progenitor marker A2B5 and the
neural stem cell marker CD15 have been used to isolate tumor stem
cells, which may also be negative for CD133 (7,8).
Additionally, CD133-positive cells are not necessarily positive for
A2B5 or CD15 (7,8). Therefore, a more specific cell surface
marker for glioma stem cells is required.
One way to circumvent the limitations of the current
stem cell markers is to isolate stem cells based on their inherent
features, which are distinct from their progenitor or
differentiated cells (9–11). Normal stem cells are characterized by
dormancy or slow cycling (12,13).
During dormancy, no DNA synthesis-associated mutations occur, thus
maintaining the homeostasis of the stem cell pool. Upon stress or
injury, dormant cells enter into the active proliferation stage and
reconstitute the tissue, thus maintaining the homeostasis of the
body (12,13). Such a small group of dormant cells
exists, for instance, among well-characterized mouse hematopoietic
stem cells (12). These cells divide
~5 times in a lifetime, and the division cycle may last as long as
145 days. Upon bone marrow injury or growth factor stimulus
treatment, for example granulocyte-colony stimulating factor or
interferon α, the dormant cells are activated into fast-cycling
cells to reconstitute the blood system (12). Among intestinal stem cells, a similar
dormant cell pool has a role similar to that of the cells in the
hematopoietic system (13).
Therefore, dormant stem cells reside at the top of the cellular
hierarchy, and screening dormant cells may be a rational way to
enrich stem cells.
Fluorescence label-retaining assays are able to
effectively discriminate dormant or slow-cycling cells from
fast-cycling cells (14). Following
the initial uniform fluorescence labeling of a whole cell
population, the fluorescence intensity in cycling cells decreases
by half due to cell division (14).
Following approximately 6 or 7 rounds of division, the fluorescence
intensity is so weak that no label is observable in fast-cycling
cells, whereas the fluorescence is retained in dormant or
slow-cycling cells; thus, the two cell populations can be
effectively discriminated based on their varying proliferative
abilities (14).
In the stem cell-permissive culture condition of
serum-free medium, suspended glioblastoma spheres are considered
rich in glioma stem cells (15).
However, the majority of cells in the spheres are progenitors with
a distinct proliferative potential compared with stem cells
(15). The present study hypothesized
that the label-retaining assay may further enrich stem cells in
glioblastoma spheres. To address this hypothesis, glioblastoma
sphere cells were fluorescently labeled, and the alterations in
fluorescence intensity during cultivation were monitored, screened,
and used to phenotypically characterize the label-retaining
cells.
Materials and methods
Cell lines and cell culture
The NCH421k, NCH441 and NCH644 glioblastoma sphere
cell lines were kindly provided by Professor Christel Herold-Mende
from the Department of Neurosurgery at Heidelberg University
(Heidelberg, Germany) and were cultivated in serum-free medium,
which consisted of Dulbecco's modified Eagle's medium (DMEM)/F-12
medium (Sigma-Aldrich, St. Louis, MO, USA) containing 20% BIT
serum-free supplement (STEMCELL Technologies, Inc., Vancouver, BC,
Canada), 20 ng/ml basic fibroblast growth factor (provitro GmbH,
Berlin, Germany) and 20 ng/ml epidermal growth factor (provitro
GmbH). To induce differentiation, glioblastoma spheres were grown
in DMEM containing 10% fetal calf serum (Biochrom, Ltd., Cambridge,
UK).
DiI staining of glioblastoma
spheres
Glioblastoma spheres were trypsinized (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) into single cell suspensions.
DiI cell-labeling solution (5 µl/ml; Thermo Fisher Scientific,
Inc.) was added to cell suspensions at a density of
1×106/ml. Following incubation for 15 min at 37°C, the
labeled cell suspensions were washed twice with phosphate-buffered
saline (PBS) and subsequently suspended in the aforementioned
serum-free medium. Following incubation for 10 min at 37°C, the
cells were uniformly stained with DiI.
Fluorescence-activated cell sorting
(FACS) analysis
Following staining with DiI and culturing in the
serum-free medium in a humidified 5% CO2/95% air
incubator at 37°C for 2 weeks, the glioblastoma spheres were
trypsinized into single cell suspensions. The cells
(2×106) were subjected to FACS analysis (FACSAria; BD
Biosciences, Franklin Lakes, New Jersey, USA) to separate
DiI-retaining and DiI-negative cells. Single cell suspensions
without DiI staining were used as the isotype control. The sorted
DiI-retaining and DiI-negative cells were expanded in the
serum-free mediumn in a humidified 5% CO2/95% air
incubator at 37°C for 2–4 weeks. When cell growth reached the
exponential phase and clear tumor spheres were observed, the
spheres were used for the subsequent experiments.
Cell proliferation assay
Dissociated spheres (100 µl) containing ~5,000 cells
were seeded in 5 replicates in a 96-well plate (Thermo Fisher
Scientific, Inc.). The proliferative ability of these cells was
measured using a cell counting kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) according to the
manufacturer's protocol.
Clonogenic assay
The glioblastoma spheres were trypsinized into
single cell suspensions and plated in 96-well plates in 0.2 ml
serum-free medium at a density of 1,000 cells/well, 500 cells/well
or 200 cells/well in each row. The cultures were fed 0.02 ml
serum-free medium/well every 3 days, until day 14. The glioma
spheres with dimensions >75 µm were counted. The sphere-forming
frequency was obtained by dividing the number of observed spheres
by the number of initially plated cells. For the serial clonogenic
assay, the spheres were harvested and cultivated in the serum-free
mediumn in a humidified 5% CO2/95% air incubator at 37°C
for approximately 1–2 weeks. Subsequently, the spheres were
trypsinized into single cell suspensions and replated in 96-well
plates as described above. Finally, the sphere-forming frequency
was recalculated.
Flow cytometric analysis
The glioblastoma spheres were trypsinized into
single cell suspensions for the flow cytometric analysis of
CD133-positive cells. Single cell suspensions containing
~5×105 cells were incubated with mouse monoclonal
anti-human CD133/1 primary antibody (dilution, 1:10; catalog no.,
130-090-422; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) for
30 min at room temperature; mouse monoclonal anti-human
immunoglobulin G1 (dilution, 1:10; catalog no., AM01157PU-N; Acris
Antibodies GmbH, Herford, Germany) was used as the isotype control.
Following washing 3 times with Flow Cytometry buffer (eBioscience,
Inc., San Diego, CA, USA), the single cell suspensions of the
NCH421k, NCH441 and NCH644 glioblastoma spheres were incubated with
fluorescein isothiocyanate-conjugated goat polyclonal anti-mouse
secondary antibody (dilution, 1:64; catalog no., BA1101; Boster
Systems, Inc., Pleasanton, CA, USA) in the dark for ~20 min.
Following washing 3 times with Flow Cytometry buffer (eBioscience,
Inc.), the aforementioned NCH421k, NCH441 and NCH644 glioblastoma
sphere single cell suspensions were subjected to flow cytometric
analysis (FACSCalibur™; BD Biosciences). Only cells with staining
intensities above the maximal level of the isotype control were
defined as positive.
Western blot analysis
Following differentiation for 1 week, DiI-retaining
and DiI-negative cells were harvested and lysed in
radioimmunoprecipitation assay buffer [50 mM Tris (pH 7.4), 0.15 M
NaCl, 1% Triton X-100, 1% sodium deoxycholate and 0.1% sodium
dodecyl sulfate; Sigma-Aldrich]. The supernatants were recovered,
and the total protein concentrations were determined using the
Bradford assay (16). In total, 75 µg
of protein per well were loaded onto the gel. The proteins were
separated by 8% sodium dodecyl sulfate polyacrylamide gel
electrophoresis and electrophoretically transferred onto a
polyvinylidene difluoride membrane at 4°C overnight. Subsequently,
the membrane was blocked at room temperature for 1 h in TST buffer
(1.211 g Tris, 8.766 g sodium chloride and 5 ml Tween-20 in 1 l
double distilled water at pH 7.4; G-Biosciences, St. Louis, MO,
USA) containing 5% non-fat milk (Sigma-Aldrich). The polyvinylidene
difluoride membrane was washed with TST buffer (G-Biosciences) and
subsequently incubated with the following primary antibodies at 4°C
overnight: Mouse monoclonal anti-human glial fibrillary acidic
protein (GFAP; dilution, 1:10; catalog no., MA1045; Boster Systems,
Inc.), rabbit polyclonal anti-human platelet-derived growth factor
receptor α (PDGFRα; dilution, 1:10; catalog no., PA1678; Boster
Systems, Inc.) or mouse monoclonal anti-human βIII-tubulin
(dilution, 1:10; catalog no., MA1112; Boster Systems, Inc.). The
polyvinylidene difluoride membrane was washed again with TST buffer
(G-Biosciences) and subsequently incubated with horseradish
peroxidase (HRP)-conjugated anti-mouse or anti-rabbit polyclonal
secondary antibody (dilution, 1:10,000; catalog nos., NA931 and
NA934, respectively; GE Healthcare Life Sciences, Chalfont, UK) and
HRP-conjugated mouse monoclonal anti-human β-actin antibody
(dilution, 1:25,000; catalog no., ab49900; Abcam, Cambridge, UK) at
room temperature for 1 h. Finally, the target proteins on the
membrane were detected using an ECL Plus kit (GE Healthcare Life
Sciences) according to the manufacturer's protocol. The band
intensities of the target proteins were normalized to β-actin using
ImageJ 1.46r software (imagej.nih.gov/ij/).
Irradiation
Glioblastoma spheres were trypsinized into single
cell suspensions and plated at a density of 2×104/ml in
96-well plates. The cells were irradiated at room temperature with
a single exposure to photon irradiation at a dose of 10 Gy
delivered by a linear accelerator (Elekta Synergy®;
Elekta AB, Stockholm, Sweden). Following 72 h of irradiation, the
proliferation of the irradiated cells was measured using a CCK-8
according to the manufacturer's protocol.
Chemotherapy
Glioblastoma spheres were trypsinized into single
cell suspensions and plated at a density of 2×104/ml in
96-well plates. The cells were treated with 500 µmol/l temozolomide
(Sigma-Aldrich). Following 72 h of treatment, the proliferation of
the treated cells was measured using a CCK-8 according to the
manufacturer's protocol.
In vivo tumorigenicity
In total, 40 female non-obese diabetic/severe
combined immunodeficiency (NOD/SCID) mice (6–8 weeks old; Charles
River Laboratories, Wilmington, MA, USA) were housed under specific
pathogen-free conditions at the temperature of 25±5°C, with 12 h
light per day. The mice had free access to sterilized food and
water. All the animal experiments were performed according to the
China Animal Protection Law following institutional guidelines for
animal welfare and experimental conduct. Ethical approval was
obtained from the Ethical Committee of Tonji Medical University,
Tongji Medical College, Huazhong University of Science and
Technology (Wuhan, China). In total, 20 mice were randomly selected
to be implanted with the DiI-retaining cells and the remaining mice
were implanted with the DiI-negative cells. Cells were implanted
stereotactically into the right hemispheres of the NOD/SCID mice.
To avoid reflux of cells along the needle tract, small carrier
volumes (5 µl) were injected 4 mm parasagittal along the coronary
suture at an adequate depth of 3.5 mm. Following pausing for 10 min
to allow for diffusion of the carrier fluid into the parenchyma,
the injection needle was slowly extracted. Subsequent to the
injection, no macroscopic reflux was observed in any of the
animals, and the needle tract was sealed with biodegradable bone
wax (Ethicon, Inc., Somerville, NJ, USA). The survival and general
performance of the mice were monitored daily for 6 months following
tumor cell implantation. At this point, the remaining live mice
were sacrificed by cervical dislocation, and all the brains were
harvested.
Immunohistochemistry
Mouse brains were fixed in 10% neutral-buffered
formalin (Sigma-Aldrich) and embedded in paraffin (Leica
Biosystems, Wetzlar, Germany). For immunohistochemistry, the
dewaxed and hydrated sections were incubated in a warm water bath
at 95°C for 20 min in neutral (pH 7.0) antigen retrieval solution
(Dako, Glostrup, Denmark). Subsequently, the sections were
incubated with the following primary antibodies overnight at 4°C:
rabbit anti-human Nestin (dilution, 1:10; Boster Systems, Inc.),
mouse anti-human GFAP (dilution, 1:10; Boster Systems, Inc.), mouse
anti-human Ki67 (dilution, 1:10; Boster Systems, Inc.) or mouse
anti-human CD31 (dilution, 1:10; Boster Systems, Inc.). The
subsequent procedures were performed according to the
manufacturer's protocol for a mouse- or rabbit-specific HRP/AEC
Polymer Detection Immunohistochemistry kit (Abcam). A total of 4
images were randomly acquired under magnification, ×200 using a
microscope (Olympus BX52; Olympus, Tokyo, Japan) to calculate the
percentages of Nestin-, GFAP- or Ki67-positive cells and the tumor
microvessel density according to the method previously reported by
Weidner et al (17).
Statistical analysis
Quantitative data are presented as the mean ±
standard deviation. The data were analyzed for statistical
significance using a two-sided Student's t-test with Excel software
2010 (Microsoft Corporation, Redmond, WA, USA). The survival
analysis was performed using a log-rank test. The survival data are
presented as a Kaplan-Meier plot using SPSS version 19.0 (IBM SPSS,
Armonk, NY, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Cell populations of DiI
fluorescence-retaining cells and DiI-negative cells with varying
proliferative potentials may be differentiated within glioblastoma
spheres at 2 weeks subsequent to initial DiI labeling
The strong photostable fluorescence, excellent
cellular retention and minimal cytotoxicity of DiI make it
particularly suitable for long-term labeling and tracking of cells
(18). Following the initial uniform
staining of DiI in the dissociated glioblastoma sphere cell lines
NCH421k, NCH441 and NCH644, the fluorescence intensity was
monitored every 2–3 days. With cell proliferation and sphere
reformation, the fluorescence intensity of DiI in certain cells
decayed in a sustained manner, whereas the fluorescence intensity
of DiI in other cells remained strong and constant (Fig. 1A and B). At 2 weeks, two cell
populations could be clearly distinguished based on fluorescence
intensity, namely, a group of DiI-retaining cells and a group of
DiI-negative cells (Fig. 1B). Flow
cytometric analysis revealed that DiI-retaining cells accounted for
a small population within the glioblastoma spheres at 2 weeks. In
the NCH421k cell line, the proportion of DiI-retaining cells was
~8±2%, in NCH441 cells it was ~6±1%, and in NCH644 cells it was
~5±1% (Fig. 1C). FACS analysis was
performed to isolate DiI-retaining and DiI-negative cells. The
CCK-8 cell proliferation assay revealed that DiI-retaining cells
proliferated significantly more slowly compared with DiI-negative
cells (P=0.011, P=0.035 and P=0.023 in the NCH421k, NCH441 and
NCH644 cell lines, respectively; Fig.
1D). Therefore, based on the fluorescence intensity of DiI in
the cells, a group of fast-cycling DiI-negative cells and a group
of slow-cycling DiI-retaining cells may be successfully
differentiated at 2 weeks subsequent to initial DiI labeling.
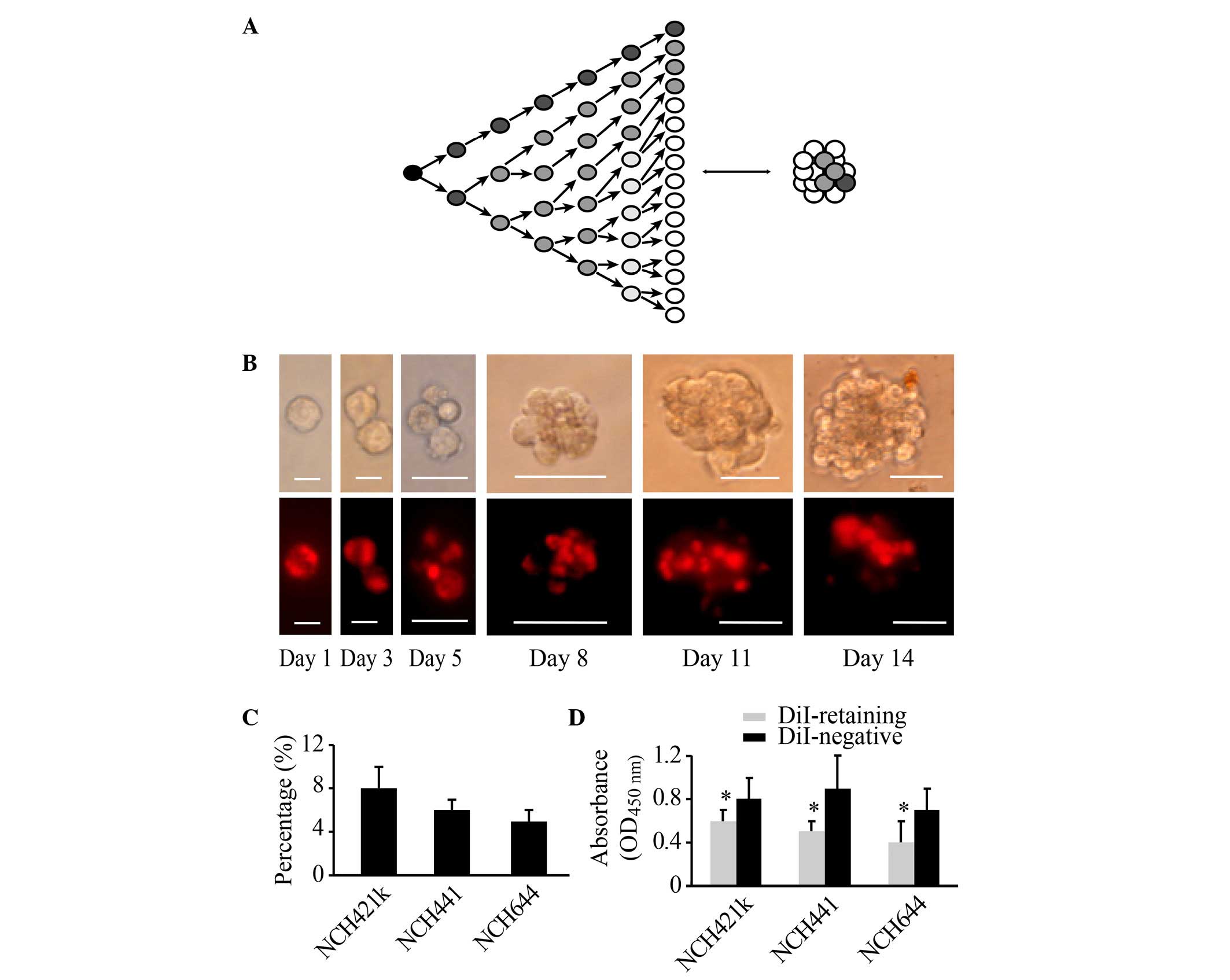 | Figure 1.Populations of DiI-retaining cells
and DiI-negative cells with varying proliferative potentials may be
differentiated within glioblastoma spheres at 2 weeks subsequent to
initial DiI labeling. (A) Diagram illustrating that the change in
the cellular fluorescence intensity in initially labeled cells
reflects the proliferative state. (B) Representative images showing
the dynamic changes in and distribution of DiI fluorescence (lower
panel) during cell proliferation and sphere reformation (upper
panel) in the NCH421k cell line at the indicated times (Day 1, Day
3, Day 5, Day 8, Day 11 and Day 14). Scale bars: Day 1 and day 3,
10 µm; day 5, 20 µm; day 8, day 11 and day 14, 50 µm. The cells
were initially stained with DiI fluorescence through incubation
with DiI cell-labeling solution, as indicated in the Materials and
methods. (C) Percentage of DiI-retaining cells in the NCH421k,
NCH441 and NCH644 cell lines as analyzed by flow cytometry at 2
weeks subsequent to initial DiI labeling. (D) Proliferative
potential of DiI-retaining and DiI-negative cells in the NCH421k,
NCH441 and NCH644 cell lines as analyzed by the cell counting kit-8
assay. The data are presented as the mean ± standard deviation from
three independent experiments. *P<0.05 compared with the
DiI-negative group. OD, optical density. |
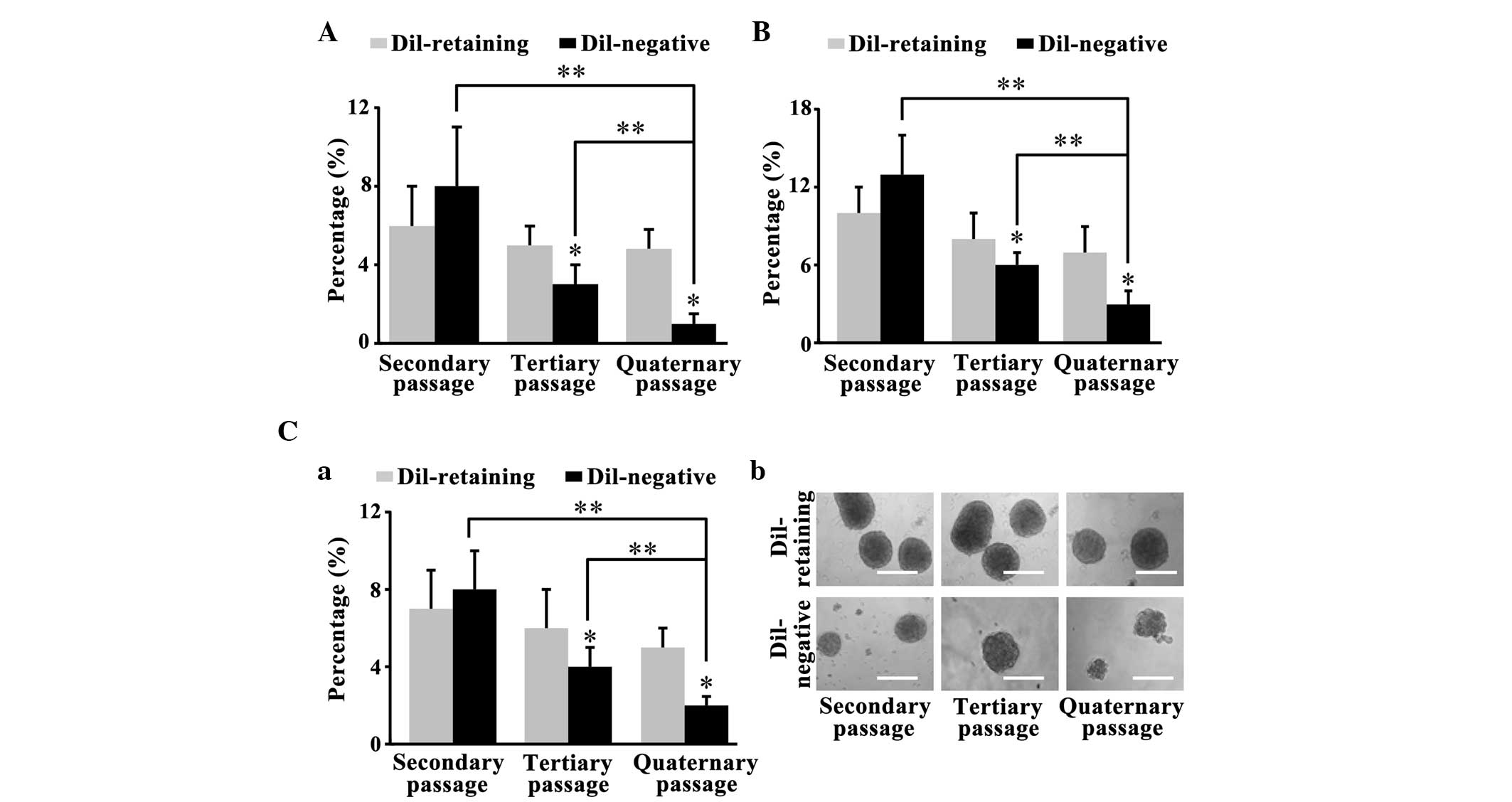
DiI-retaining cells possess increased
self-renewal capacity in vitro compared with DiI-negative
cells
The primary DiI-retaining and DiI-negative cells
separated by FACS analysis formed glioma spheres after 2–4 weeks.
The clonogenicity assay performed on the primary spheres revealed
that 2nd passage DiI-retaining cells and DiI-negative cells were
able to reform clones. The sphere-forming frequencies of
DiI-retaining cells from the NCH441, NCH644 and NCH421k cell lines
were 7±2, 10±2 and 6±2%, respectively; these frequencies were lower
than the sphere-forming frequencies in DiI-negative cells (8±2,
13±3 and 8±3% for the NCH441, NCH644 and NCH421k cell lines,
respectively; P=0.089, P=0.455 and P=0.653, respectively; Fig. 2A, B and C-a). The spheres were
harvested again and cultured in serum-free medium for 2 weeks. The
secondary clonogenicity assay performed on the secondary spheres
revealed that the sphere-forming frequency of the tertiary
DiI-retaining cells was significantly increased compared with that
of the tertiary DiI-negative cells (P=0.002, P=0.034 and P=0.016,
respectively; Fig. 2A, B and C-a).
Similarly, the tertiary clonogenicity assay performed on the
tertiary spheres revealed that the sphere-forming frequency in the
quaternary DiI-retaining cells was significantly increased compared
with that of the quaternary DiI-negative cells (P=0.003, P=0.027
and P=0.007, respectively; Fig. 2A, B and
C-a). Furthermore, the clonogenicity of the DiI-negative cells
decreased significantly with serial passaging (secondary vs.
quaternary passages, P=0.013 in NCH441 cells, P=0.003 in NCH644
cells and P=0.028 in NCH421k cells; tertiary vs. quaternary
passages, P=0.001 in NCH441 cells, P=0.017 in NCH644 cells and
P=0.042 in NCH421k cells), whereas the clonogenicity of the
DiI-retaining cells remained stable (Fig.
2A-C). Therefore, DiI-retaining cells possessed significantly
increased self-renewal capacity in vitro compared with
DiI-negative cells.
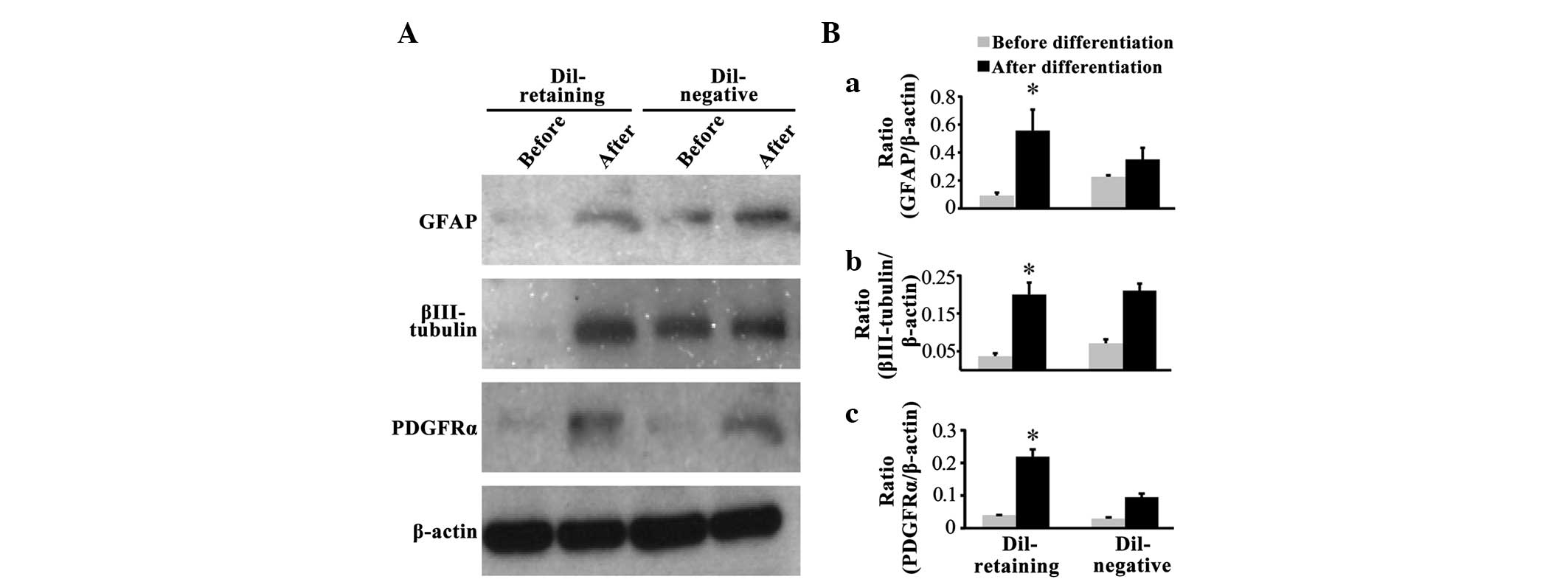
DiI-retaining cells possess
multipotency in vitro
Western blot assays revealed that the glial cell
marker GFAP, neural marker βIII-tubulin and oligodendrocyte marker
PDGFRα were not expressed in the DiI-retaining group prior to
differentiation, as no bands for these three markers were observed
(Fig. 3A); following differentiation,
GFAP, PDGFRα and βIII-tubulin expression was induced (Fig. 3A). In the DiI-negative group,
increased expression of these three lineage markers was
additionally observed following differentiation compared with that
prior to differentiation (Fig. 3A).
Notably, two weaker bands for GFAP and βIII-tubulin were observed
prior to differentiation in the DiI-negative group (Fig. 3A). Quantification of the band density
on the blots revealed that GFAP, βIII-tubulin and PDGFRα expression
increased significantly following differentiation in the
DiI-retaining group (P=0.027, P=0.006 and P=0.004, respectively;
Fig. 3B-a, B-b and B-c), whereas the
increased expression of these three markers was not statistically
significant in the DiI-negative group (P=0.789, P=0.658 and
P=0.089, respectively; Fig. 3B-a, B-b and
B-c).
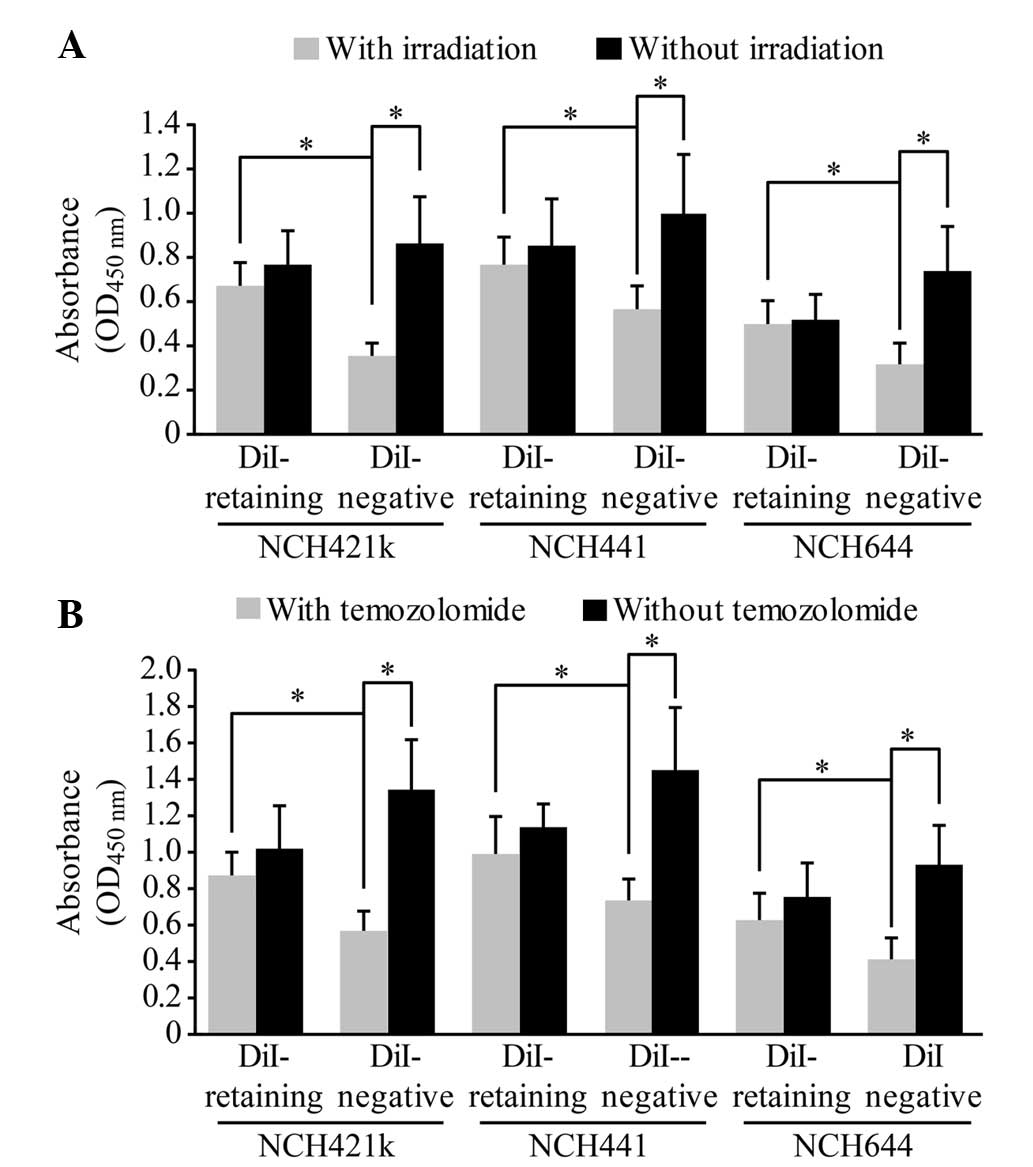
DiI-retaining cells exhibit
significant resistance to irradiation and temozolomide
treatment
The dissociated DiI-retaining and DiI-negative cells
were irradiated with a single dose of 10 Gy radiation or were
treated with 500 µmol/l temozolomide, a representative first-line
drug against malignant glioma. A total of 72 h later, the cell
proliferation assay revealed significantly increased proliferation
of DiI-retaining cells compared with that of DiI-negative cells
(following irradiation, P=0.012 in NCH421k cells, P=0.024 in NCH441
cells and P=0.036 in NCH644 cells; following temozolomide, P=0.003
in NCH421k cells, P=0.005 in NCH441 cells and P=0.029 in NCH644
cells; Fig. 4A and B), indicating
that DiI-negative cells were more sensitive to radiation or to
temozolomide treatment than DiI-retaining cells. In addition, no
significant differences in the proliferation of DiI-retaining cells
were observed prior to and following radiation or temozolomide
treatment, whereas the proliferation of DiI-negative cells
decreased significantly following radiation or temozolomide
treatment compared to that prior to radiation or temozolomide
treatment (following irradiation, P=0.0002 in NCH421k cells,
P=0.005 in NCH441 cells and P=0.003 in NCH644 cells; following
temozolomide, P=0.019 in NCH421k cells, P=0.027 in NCH441 cells and
P=0.004 in NCH644 cells; Fig. 4A and
B). Therefore, DiI-retaining cells exhibited significant
resistance to radiotherapy and temozolomide treatment.
The difference in CD133 expression
between DiI-retaining cells and DiI-negative cells is not
significant
Flow cytometric analysis revealed that
CD133-positive cells accounted for ~74±12% of the DiI-retaining
cells in the NCH421k cell line, 34±10% of those in the NCH441 cell
line and 60±15% of those in the NCH644 cell line. These percentages
were slightly higher than the percentages of CD133-positive cells
among the DiI-negative populations (64±11% of NCH421k cells, 21±7%
of NCH441 cells and 52±13% of NCH644 cells). These differences were
not statistically significant (P=0.553, P=0.624 and P=0.867,
respectively).
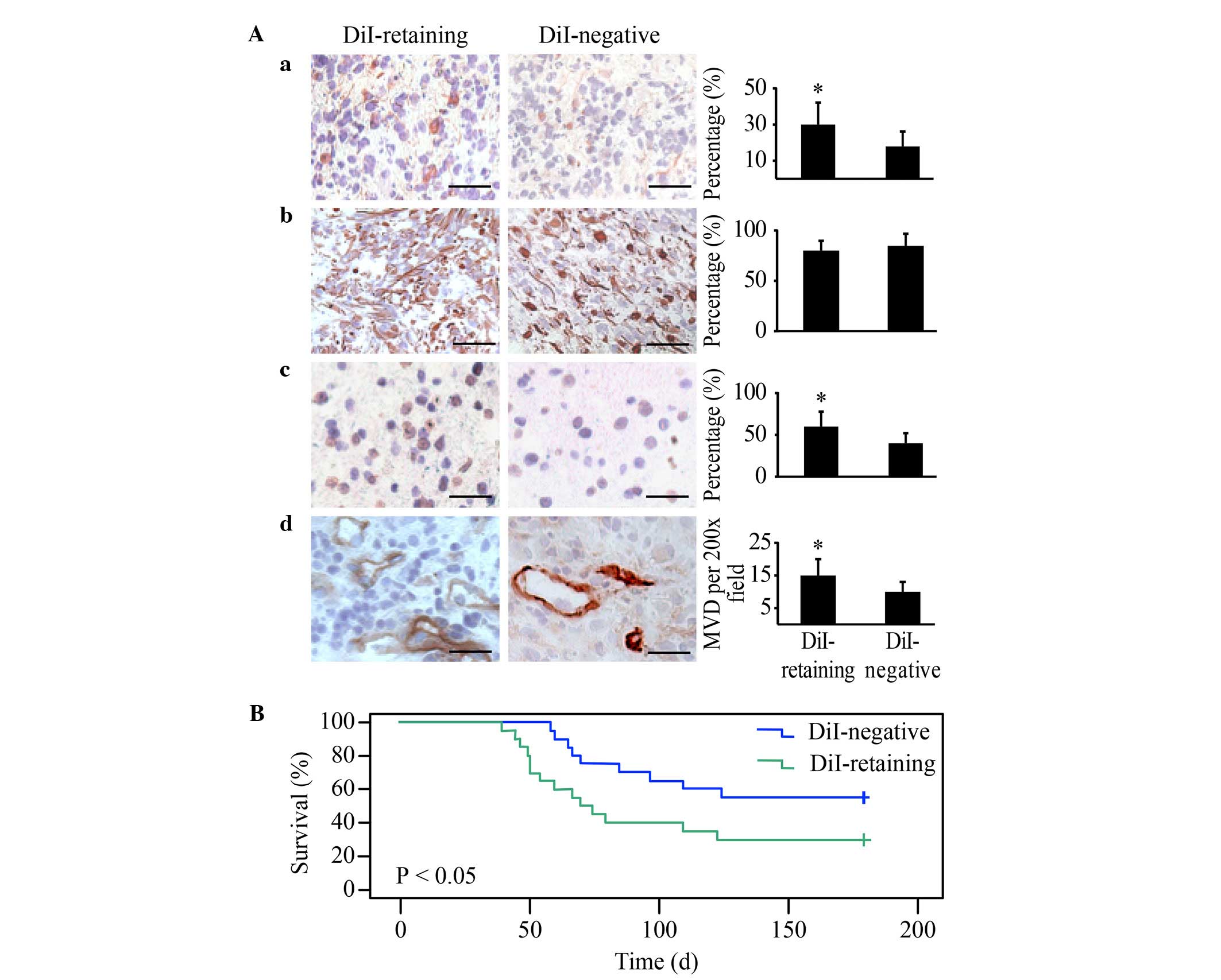
DiI-retaining cells demonstrate
significantly increased tumorigenicity in vivo compared with
DiI-negative cells
DiI-retaining cells and DiI-negative cells were
implanted into the brains of NOD/SCID mice at a serial gradient of
102, 103, 104 and 105.
DiI-retaining cells and DiI-negative cells (105
cells/implantation) were able to generate tumors in all treated
mice. As the number of implanted cells decreased, higher tumor
incidence was observed in the DiI-retaining group compared with the
DiI-negative group (Table I). With as
few as 102 implanted cells, only DiI-retaining cells
were able to effectively generate tumors; at this cell number, no
tumors were observed in the DiI-negative group 6 months subsequent
to implantation (Table I).
 | Table I.Tumor incidence and survival time in
non-obese diabetic/severe combined immunodeficiency mice (n=5 in
each group) implanted with DiI-retaining cells or DiI-negative
cells from the NCH421k cell line at a serial implantation
gradient. |
Table I.
Tumor incidence and survival time in
non-obese diabetic/severe combined immunodeficiency mice (n=5 in
each group) implanted with DiI-retaining cells or DiI-negative
cells from the NCH421k cell line at a serial implantation
gradient.
|
| Density of
implanted tumor cells |
|---|
|
|
|
|---|
| Parameter | 102 | 103 | 104 | 105 |
|---|
| DiI-retaining |
|
|
|
|
| Tumor
incidence, n | 2 | 3 | 4 | 5 |
|
Survival time,
daysa | 117±9 | 72±7 | 58±13 | 49±6 |
| DiI-negative |
|
|
|
|
| Tumor
incidence, n | 0 | 1 | 3 | 5 |
|
Survival time,
daysa | >180 | 125 | 97±13 | 64±5 |
Immunohistochemical analysis revealed strong
positive GFAP staining in the two groups and significantly
increased Nestin, Ki67 and CD31 in tumors generated from
DiI-retaining cells compared with those generated from DiI-negative
cells, with the same number of implanted cells (Fig. 5A). These data indicated that the
tumors generated from DiI-retaining cells were more enriched in
stem cells and exhibited a higher proliferation index and vessel
density, thereby more closely resembling the features of human
glioblastoma.
The Kaplan-Meier survival analysis revealed that the
overall survival of NOD/SCID mice harboring tumors from
DiI-retaining cells was significantly reduced compared with that of
mice harboring tumors from DiI-negative cells (P=0.016; Fig. 5B).
Discussion
Once cells are initially labeled with fluorescence
or with the DNA synthesis substrate bromodeoxyuridine (BrdU), the
proliferative status of these cells may be monitored based on
labeling intensity (14). In actively
proliferating cells, the labeling intensity decreases by half with
each division, whereas in dormant or slow-cycling cells, the
labeling intensity remains constant or decays marginally (14). Thus, label-retaining cells correspond
to dormant or slow-cycling cells (14).
In normal tissues, the label-retaining assay has
already been effectively used to isolate stem cells. In the
hematopoietic system, intestinal epithelium, hair follicles,
myocardium, prostate epithelium and central nervous system, BrdU
label-retaining cells manifest stem cell features (19,20). In a
number of human tumors, the label-retaining assay is effective at
isolating tumor stem cells (21–24).
Through initial carboxyfluorescein succinimidyl ester (CFSE)
fluorescence cell labeling and the subsequent tracking of
fluorescence intensity, Moore et al successfully confirmed
that the CFSE-retaining cells in the HCT116 colorectal cancer cell
line and in the MDA-MB-231 breast cancer cell line were tumor stem
cells (21). Using the fluorescence
dye PKH26, the long-term PKH26-retaining cells in melanoma and
ovarian cancer were demonstrated to be rich in cancer stem cells
(22,23). Dembinski and Krauss (24) reported that the population of
long-term DiI-retaining pancreatic cancer cells exhibited stem cell
features.
Similar to the present results, Deleyrolle et
al (25) used CFSE fluorescence
dye and Richichi et al (26)
used PKH26 dye to identify label-retaining stem cells in glioma
spheres successfully. These cells constitute only a small
population within the glioma spheres. Subsequent to tracking CFSE
fluorescence for 1 week, Deleyrolle et al (25) reported that the label-retaining cells
with marked characteristics of stem cells accounted for ~5% of the
population. Richichi et al (26) reported that the PKH26-retaining cell
population ranged between 1 and 3% following 2 weeks of
observation. The results of the present study suggested that the
DiI-retaining cell population in each of the three glioblastoma
sphere cell lines was <10% following 2 weeks of observation.
Notably, among the three studies, several fluorescent dyes were
employed. PKH26 and DiI are lipophilic carbocyanine dyes that
intercalate into the lipid bilayer of the cell membrane, whereas
CFSE binds to cytoplasmic components (18). Compared with PKH26, DiI fluorescence
is more uniformly distributed in the cell membrane; therefore, the
fluorescence decay of DiI more accurately reflects cell division
(18). Compared with CFSE, the cell
membrane distribution of DiI is less toxic to cells (18). Thus, DiI staining appears to be
superior to the other two dyes and more conducive to long-term cell
tracking.
Through serial clonogenic assays, the present study
demonstrated that the clonogenicity of the label-retaining cells
remained relatively constant despite serial passaging, whereas the
clonogenicity of the label-negative cells decreased significantly,
indicating that label-negative cells possess a limited self-renewal
capacity. Approximately 100 label-retaining cells were necessary to
generate tumors effectively in immunocompromised mice over 6
months, whereas no tumors were generated with the same number of
label-negative cells over the same time interval, highlighting the
different degrees of tumorigenicity between the two cell
populations. Therefore, the label-retaining cells were greatly
enriched in glioma stem cells compared with the label-negative
cells.
The present study did not identify any significant
differences between label-retaining cells and label-negative cells
regarding CD133 expression. Controversies regarding CD133 as a
cancer stem cell marker have existed for a long time. A number of
studies have demonstrated the tumorigenicity of CD133-negative
cells (5,6,27).
CD133-positive and CD133-negative stem cells may originate from
different cells (6,28). These stem cells are able to coexist in
an individual tumor tissue or be differentially distributed in
different individuals, reflecting the heterogeneity of glioblastoma
(27,29). The label-retaining assay for screening
cancer stem cells avoids the diversity and uncertainty of cell
surface markers (5,6,27–29). However, certain problems regarding
sensitivity and specificity may be encountered in label-retaining
assays. Certain dormant cells may enter the proliferative stage and
subsequently return to the dormant stage (12), thereby significantly decreasing the
labeling intensity and leading to false sorting into the
label-negative group. This false sorting may be responsible for the
self-renewal capacity of label-negative cells that has been
observed in serial clonogenic assays in the present study, which
remained significantly weaker compared with that of label-retaining
cells. Certain terminally differentiated cells may no longer divide
but may retain strong labeling intensity following the initial
staining, thus leading to false sorting into the label-retaining
group (30). However, as these cells
undergo apoptosis and death, their labeling intensities will
diminish and finally disappear, whereas the labeling intensity in
the dormant stem cells always remains constant during tracking
(21–26). Furthermore, the stem cell permissive
serum-free condition makes the survival of differentiated cells
difficult.
The present study demonstrated that label-retaining
cells exhibited significant resistance to radiotherapy and
chemotherapy. The dormant cells in numerous human tumors, including
ovarian, breast and colorectal cancer, are resistant to
conventional radiotherapies or chemotherapies, leading to tumor
recurrence or metastasis, sometimes decades later (21,31–33). In
glioma, Hussein et al (15)
reported that suspended glioma sphere cells were resistant to
etoposide treatment, which specifically kills cells at the
proliferative stages (S and G2) of the cell cycle. Additional cell
cycle analysis revealed that certain cells in glioma spheres reside
at the dormant stage of the cell cycle (G0/G1), thus providing a
reason for the etoposide resistance (15). Chen et al (34) constructed a Nestin-Δthymidine kinase
(ΔTK)-green fluorescence protein (GFP) transgenic mouse model that
developed glioma. In this model, the
Nestin+/GFP+ progenitor tumor cells were
almost Ki67−, indicating that the majority of the
progenitor cells were dormant (34).
Treating these mice with temozolomide only kills the proliferating
Ki67+ tumor cells and therefore only arrests tumor
growth (34). As a result, following
the conclusion of temozolomide treatment, marked tumor regrowth
originating from the dormant Nestin+/GFP+
subpopulation is observed (34).
Taken together, the results of these two previous studies and the
present study suggest that dormant stem cells may be resistant to
radiotherapy and chemotherapy.
The clarification of the regulation of stem cell
proliferation and dormancy enables an improved understanding of the
mechanism of tumor cell resistance (35–37). The
regulation of the proliferation and dormancy of mouse hematopoietic
stem cells has been well characterized (35,36).
Proliferative and dormant stem cells reside in distinct niches,
including a hypoxic osteoblastic niche that maintains the dormant
state and a normoxic vascular endothelial niche that maintains the
proliferative state of stem cells (35). Upon bone marrow injury or growth
factor stimulus with granulocyte-colony stimulating factor, the
dormant cells from the osteoblastic niche migrate into the vascular
endothelial niche and enter the proliferative state in response to
complex extra- and intracellular signal transduction (35). Numerous molecules participate in this
transition, including extracellular factors, for example,
thrombopoietin, angiopoietin, transforming growth factor β and
Notch1 and intracellular factors, for example, forkhead box O
proteins, mammalian target of rapamycin complex 1, F-box and WD
repeat domain-containing 7, early growth response protein 1,
pre-B-cell leukemia homeobox 1, retinoblastoma protein, c-Cbl, Myc
and Bmi1 (36). In glioma, the niche
responsible for maintaining the dormant state of glioma stem cells
and the dormancy-associated signaling pathways remains to be
elucidated. If the dormant stem cells could be induced into the
proliferative state, which is sensitive to radiotherapy and
chemotherapy, or if the dormant state could be permanently
maintained regardless of environmental stimuli (37), then significant progress would be
achieved in glioma therapy.
In conclusion, the present study successfully
identified a group of dormant or slow-cycling label-retaining cells
in glioblastoma spheres cultured in serum-free medium. Compared
with label-negative cells, label-retaining cells possessed
significant stem cell properties including self-renewal capacity,
multipotency in vitro and tumorigenicity in vivo. In
addition, these cells demonstrated significant resistance to
radiotherapy and chemotherapy. Label-retaining assays may be
considered as an effective way to enrich glioma stem cells for
additional study.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (Beijing, China; grant no.
81201681).
References
|
1
|
Rock K, McArdle O, Forde P, Dunne M,
Fitzpatrick D, O'Neill B and Faul C: A clinical review of treatment
outcomes in glioblastoma multiforme - the validation in a non-trial
population of the results of a randomised Phase III clinical trial:
Has a more radical approach improved survival? Br J Radiol.
85:e729–e733. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jackson M, Hassiotou F and Nowak A:
Glioblastoma stem-like cells: At the root of tumor recurrence and a
therapeutic target. Carcinogenesis. 36:177–185. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stopschinski BE, Beier CP and Beier D:
Glioblastoma cancer stem cells - from concept to clinical
application. Cancer Lett. 338:32–40. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
5
|
Wang J, Sakariassen PØ, Tsinkalovsky O,
Immervoll H, Bøe SO, Svendsen A, Prestegarden L, Røsland G, Thorsen
F, Stuhr L, et al: CD133 negative glioma cells form tumors in nude
rats and give rise to CD133 positive cells. Int J Cancer.
122:761–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Beier D, Hau P, Proescholdt M, Lohmeier A,
Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U and Beier
CP: CD133(+) and CD133(−) glioblastoma-derived cancer stem cells
show differential growth characteristics and molecular profiles.
Cancer Res. 67:4010–4015. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ogden AT, Wziri AE, Lochhead RA, Fusco D,
Lopez K, Ellis JA, Kang J, Assanah M, McKhann GM, Sisti MB, et al:
Identification of A2B5+CD133-tumor-initiating cells in adult human
gliomas. Neurosurgery. 62:505–514. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mao XG, Zhang X, Xue XY, Guo G, Wang P,
Zhang W, Fei Z, Zhen HN, You SW and Yang H: Brain tumor stem-like
cells identified by neural stem cell marker CD15. Transl Oncol.
2:247–257. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma I and Allan AL: The role of human
aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell
Rev. 7:292–306. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meyer M, Reimand J, Lan X, Head R, Zhu X,
Kushida M, Bayani J, Pressey JC, Lionel AC, Clarke ID, et al:
Single cell-derived clonal analysis of human glioblastoma links
functional and genomic heterogeneity. Proc Natl Acad Sci USA.
112:851–856. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Clément V, Marino D, Cudalbu C, Hamou MF,
Mlynarik V, de Tribolet N, Dietrich PY, Gruetter R, Hegi ME and
Radovanovic I: Marker-independent identification of
glioma-initiating cells. Nat Methods. 7:224–228. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wilson A, Laurenti E, Oser G, van der Wath
RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L,
Bockamp E, et al: Hematopoietic stem cells reversibly switch from
dormancy to self-renewal during homeostasis and repair. Cell.
135:1118–1129. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Radford IR and Lobachevsky PN: An
enteroendocrine cell-based model for a quiescent intestinal stem
cell niche. Cell Prolif. 39:403–414. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Glauche I, Moore K, Thielecke L, Horn K,
Loeffler M and Roeder I: Stem cell proliferation and quiescence -
two sides of the same coin. PLoS Comput Biol. 5:e10004472009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hussein D, Punjaruk W, Storer LC, Shaw L,
Othman R, Peet A, Miller S, Bandopadhyay G, Heath R, Kumari R, et
al: Pediatric brain tumor cancer stem cells: Cell cycle dynamics,
DNA repair, and etoposide extrusion. Neuro Oncol. 13:70–83. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Weidner N, Semple JP, Welch WR and Folkman
J: Tumor angiogenesis and metastasis - correlation in invasive
breast carcinoma. N Engl J Med. 324:1–8. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Progatzky F, Dallman MJ and Lo Celso C:
From seeing to believing: Labelling strategies for in vivo
cell-tracking experiments. Interface Focus. 3:201300012013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fernandez-Gonzalez R, Illa-Bochaca I,
Shelton DN, Welm BE, Barcellos-Hoff MH and Ortiz-de-Solorzano C:
In situ analysis of cell populations: Long-term
label-retaining cells. Methods Mol Biol. 621:1–28. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
van der Wath RC, Wilson A, Laurenti E,
Trumpp A and Liò P: Estimating dormant and active hematopoietic
stem cell kinetics through extensive modeling of bromodeoxyuridine
label-retaining cell dynamics. PLoS One. 4:e69722009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moore N, Houghton J and Lyle S:
Slow-cycling therapy-resistant cancer cells. Stem Cells Dev.
21:1822–1830. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Roesch A, Fukunaga-Kalabis M, Schmidt EC,
Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T
and Herlyn M: A temporarily distinct subpopulation of slow-cycling
melanoma cells is required for continuous tumor growth. Cell.
141:583–594. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao MQ, Choi YP, Kang S, Youn JH and Cho
NH: CD24+ cells from hierarchically organized ovarian cancer are
enriched in cancer stem cells. Oncogene. 29:2672–2680. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dembinski JL and Krauss S:
Characterization and functional analysis of a slow cycling stem
cell-like subpopulation in pancreas adenocarcinoma. Clin Exp
Metastasis. 26:611–623. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deleyrolle LP, Harding A, Cato K,
Siebzehnrubl FA, Rahman M, Azari H, Olson S, Gabrielli B, Osborne
G, Vescovi A and Reynolds BA: Evidence for label-retaining
tumour-initiating cells in human glioblastoma. Brain.
134:1331–1343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Richichi C, Brescia P, Alberizzi V,
Fornasari L and Pelicci G: Marker-independent method for isolating
slow-dividing cancer stem cells in human glioblastoma. Neoplasia.
15:840–847. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen R, Nishimura MC, Bumbaca SM,
Kharbanda S, Forrest WF, Kasman IM, Greve JM, Soriano RH, Gilmour
LL, Rivers CS, et al: A hierarchy of self-renewing tumor-initiating
cell types in glioblastoma. Cancer Cell. 17:362–375. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lottaz C, Beier D, Meyer K, Kumar P,
Hermann A, Schwarz J, Junker M, Oefner PJ, Bogdahn U, Wischhusen J,
et al: Transcriptional profiles of CD133+ and CD133-
glioblastoma-derived cancer stem cell lines suggest different cells
of origin. Cancer Res. 70:2030–2040. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Soeda A, Hara A, Kunisada T, Yoshimura S,
Iwama T and Park DM: The evidence of glioblastoma heterogeneity.
Sci Rep. 5:79792015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Q, Nguyen DH, Dong Q, Shitaku P, Chung
K, Liu OY, Tso JL, Liu JY, Konkankit V, Cloughesy TF, et al:
Molecular properties of CD133+ glioblastoma stem cells derived from
treatment-refractory recurrent brain tumors. J Neurooncol. 94:1–19.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fillmore CM and Kuperwasser C: Human
breast cancer cell lines contain stem-like cells that self-renew,
give rise to phenotypically diverse progeny and survive
chemotherapy. Breast Cancer Res. 10:R252008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Allan AL, Vantyghem SA, Tuck AB and
Chambers AF: Tumor dormancy and cancer stem cells: Implications for
the biology and treatment of breast cancer metastasis. Breast Dis.
26:87–98. 2007. View Article : Google Scholar
|
|
33
|
Udagawa T: Tumor dormancy of primary and
secondary cancers. APMIS. 116:615–628. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen J, Li Y, Yu TS, McKay RM, Burns DK,
Kernie SG and Parada LF: A restricted cell population propagates
glioblastoma growth after chemotherapy. Nature. 488:522–526. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Trumpp A, Essers M and Wilson A: Awakening
dormant haematopoietic stem cells. Nat Rev Immunol. 10:201–209.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wilson A, Laurenti E and Trumpp A:
Balancing dormant and self-renewing hematopoietic stem cells. Curr
Opin Genet Dev. 19:461–468. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schillert A, Trumpp A and Sprick MR: Label
retaining cells in cancer - the dormant root of evil? Cancer Lett.
341:73–79. 2013. View Article : Google Scholar : PubMed/NCBI
|