Introduction
Breast cancer is one of the most common types of
cancer and the leading cause of cancer-associated mortality among
women, worldwide (1). At present, the
incidence of breast cancer in China is increasing: Breast cancer
has become the second most frequently diagnosed cancer, accounting
for 248,620 novel cases in 2011, with an annual incidence rate of
37.86 cases per 100,000 individuals. In 2011, breast cancer
accounted for 60,473 mortalities with a mortality rate of 9.21
mortalities per 100,000 individuals in China (2). An increasing number of studies have
focused on the identification of novel therapeutics for the
treatment of breast cancer, including novel anti-metabolic therapy
(3). Aberrant metabolism is a
hallmark of cancer; metabolic reprogramming of cancer cells is
observed in various cancers, including breast cancer (4). Mammalian cell proliferation uses
glutamine as an important anaplerotic source to replenish
metabolites in the tricarboxylic acid cycle (TCA) for biosynthesis
(5). Glutamine metabolism is one of
the central metabolic pathways in cancer, and thus targeting
glutamine metabolism may present a novel therapeutic approach for
the treatment of breast cancer patients with different molecular
subtypes of the disease (5–7). Glutamine is the main source for TCA
anaplerosis in proliferating cells: Glutamine is firstly converted
to glutamate by glutaminase (GLS) followed by conversion to
α-ketoglutarate by either glutamate dehydrogenase (GDH) or
transamination-coupled reactions. Sirtuin 4 (SIRT4) directly
downregulates GDH activity via adenosine diphosphate
(ADP)-ribosylation, which regulates glutamine catabolism. SIRT4 is
an important gene that mediates the blockade of glutamine
catabolism and therefore, it is considered a ‘glutamine gatekeeper’
(7,8).
The human SIRT4 gene is located on chromosome 12.
SIRT4 is a member of the SIRT family (SIRT1-7) of protein
deacetylases and ADP-ribosylases that are involved in multiple
cellular processes, including the maintenance of genomic stability
and regulation of metabolism (9).
SIRT1, −6 and −7 are localized to the nucleus, SIRT2 resides in the
cytosol and SIRT3, −4, and −5 are usually located in the
mitochondria. SIRT4 predominantly acts as an
ADP-ribosyltransferase, exhibiting demalonylase, desuccinylase and
deacetylase activities in specific tissues (9–11). The
SIRT4 gene encodes a member of the SIRT family of proteins, which
are grouped into four types that are characterized by a SIRT core
domain (12). The functions of SIRT4
outside pancreatic β-cells are unknown. At present, the function of
SIRT4 enzymatic activity in the mitochondrion and in nuclear gene
transcription remains unclear. A recent study (10) indicated that reduced SIRT4 expression
causes both increased glutamine-dependent proliferation and
stress-induced genomic instability, resulting in tumorigenic
phenotypes. Furthermore, SIRT4-knockout mice spontaneously develop
lung tumors (10). Another study
(5) revealed that SIRT4 mRNA
expression is lower in bladder, breast, colon, stomach, ovarian and
thyroid carcinomas compared with normal tissues. In addition, loss
of SIRT4 mRNA expression is associated with shorter time to
metastasis in breast cancer patients (5), suggesting that SIRT4 expression
correlates with the development and progression of breast cancer
and that targeting SIRT4 may present a therapeutic target for the
treatment of the disease. However, the protein levels of SIRT4 in
breast cancer and its significance remain unclear. In the present
study, immunohistochemical (IHC) analysis of SIRT4 protein
expression in breast cancer tissues was performed to investigate
the association between SIRT4 protein expression and
clinicopathological features and survival of advanced breast cancer
patients.
Materials and methods
Patient samples
A total of 409 histologically confirmed breast
cancer and 241 adjacent non-cancerous breast tissues obtained
during surgery between January 2003 and December 2007 and assessed
in the Department of Pathology of The Affiliated Tumor Hospital of
Harbin Medical University (Harbin, China) were retrospectively
reviewed. Patients selected in this study were aged from 28 to 76
years (median, 49.66 years) Adjacent non-cancerous breast tissues
located 2 cm from the tumor tissue were resected. The cancer and
non-cancerous breast tissues were formalin-fixed, paraffin-embedded
and cut into 4-µm sections for IHC analysis. The inclusion criteria
were as follows: Presence of primary, unilateral and operable
invasive breast cancer and available data regarding initial
diagnosis and clinical follow-up. The exclusion criteria were as
follows: Locally advanced disease with recurrent tumors, metastatic
disease and neoadjuvant therapy. The estrogen receptor (ER),
progesterone receptor (PR), human epidermal growth factor receptor
2 (HER2), tumor protein p53 (p53) and nuclear-associated antigen
Ki-67 (Ki-67) expression status was analyzed in all patients;
tissue sections were incubated with antibodies to the proteins ER
(catalog no. TA506414; dilution, 1:150; incubation time, 20 min;
temperature, 25°C), PR (catalog no. TA802606; dilution, 1:150;
incubation time, 10 min; temperature, 25°C), HER2 (catalog no.
TA503443; dilution, 1:50; incubation time, 30 min; temperature,
25°C), p53 (catalog no. TA502780; dilution, 1:100; incubation time,
15 min; temperature, 25°C) and Ki-67 (catalog no. TA500265;
dilution, 1:50; incubation time, 20 min; temperature, 25°C)
antibodies (Origene Technologies Inc., Rockville, MD, USA).
Individual samples with ≥14% Ki-67+ tumor cells were
considered to exhibit high levels of proliferation (13). The St. Gallen International Expert
Consensus system was used to grade the tissues (14). The present study was approved by the
Ethical Committee of Harbin Medical University and written informed
consent for the analysis of tissue specimens was obtained from all
patients.
Patient follow-up
All patients were followed up every 3 months
post-surgery for the first 5 years and every 12 months thereafter.
The follow-up was conducted at The Affiliated Hospital of Harbin
Medical University and patient clinical records were periodically
reviewed. In addition, the overall survival (OS) was assessed:
Overall survival time was defined as the time from the initial
diagnosis to the date of death or the study endpoint (December 31,
2012).
Tissue microarray (TMA) and IHC
analysis
Tissue blocks containing 409 invasive breast
carcinoma samples and 241 paired adjacent normal breast tissue
samples were arrayed. The breast cancer TMA for each tissue sample
was prepared from the paraffin blocks using a thin-walled needle
(inner diameter, 2 mm). All tissue blocks were cut with a microtome
to 4-µm and affixed to slides. IHC analysis of SIRT4 was performed
using a standard two-step method (15). Tissue specimens underwent antigen
retrieval in citrate buffer (pH 6.0) followed by incubation
overnight at 4°C with polyclonal rabbit SIRT4 antibody (catalog no.
ab105039; dilution, 1:150; Abcam, Cambridge, MA, USA). Secondary
antibodies were of the Anti-Rabbit IgG Detection system (catalog
no. SP-9001; ZSGB-Bio, Beijing, China). Slides incubated with
phosphate-buffered saline and rabbit serum served as the blank and
negative control groups, respectively. Positively immunostained
slides were used as positive controls.
SIRT4 protein expression was only analyzed in the
cytoplasm (as an indirect measure of the mitochondrial protein
expression). The percentage and intensity of positive SIRT4
staining was evaluated in 10 randomly selected high-power fields
(magnification, ×400; Olympus BX53; Olympus, Tokyo, Japan) by two
investigators blinded to the clinical characteristics and outcomes
of the patients. The proportion of positively-stained tumor cells
in a field was scored as follows: 0, none; 1, <10%; 2, 10–50%;
and 3, >50%. The staining intensity in a field was scored as
follows: 0, no staining; 1, weak staining appearing as a light
yellow color; 2, moderate staining appearing as a yellowish-brown
color; and 3, strong staining appearing as a brown color. The
staining index (SI) was calculated as follows: SI = average
staining intensity score × proportion score. Cut-off values for
protein expression were determined by measuring heterogeneity.
Scores of <4 and ≥4 indicated low and high SIRT4 expression,
respectively (16).
Statistical analysis
All data analysis was performed using MedCalc
statistical software (version 11.2.0.0; MedCalc Software, Oostende,
Belgium). The χ2 test was used to analyze the
associations between SIRT4 expression and clinicopathological
features, including age, lymph node metastasis (LNM), pathological
stage, ER, PR, HER2 and Ki-67 status and molecular subtype. The OS
times of the different patient groups were evaluated using the
log-rank test. The Cox proportional hazards model was used for
univariate and multivariate regression analyses. P<0.05 was
considered to indicate a statistically significant difference.
Results
SIRT4 protein expression is
significantly higher in breast cancer tissues than adjacent
non-cancerous tissues
SIRT4 protein was examined in 409 breast cancer
tissues and 241 paired adjacent non-tumor tissues. The cytoplasmic
expression of SIRT4 was significantly higher in cancer tissues
compared with adjacent non-cancerous tissues (P<0.0001). Among
the 409 breast cancer specimens, 94 (22.98%) tissues exhibited
positive SIRT4 expression and 315 (77.02%) tissues exhibited
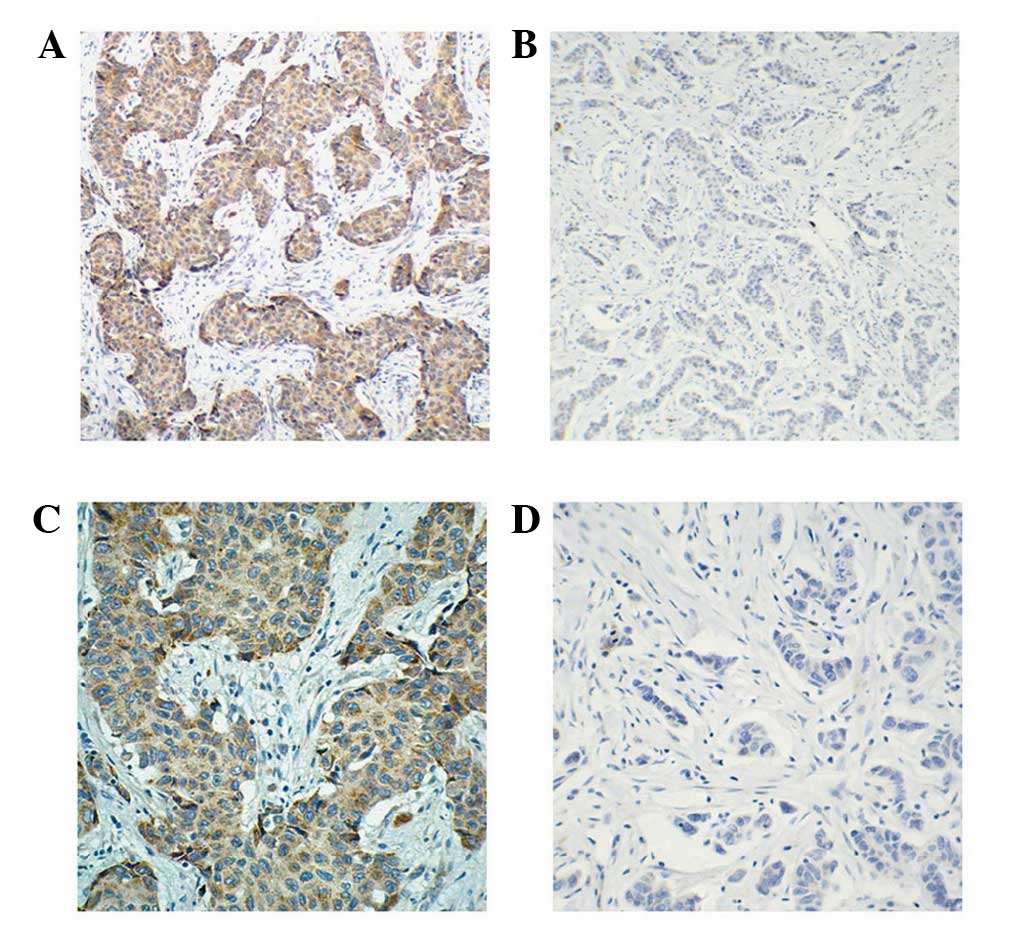
negative SIRT4 expression. Representative images of IHC SIRT4
staining are shown in Fig. 1. Of the
241 paired cancer and non-cancerous tissues, 48 (19.92%) breast
cancer tissue samples were SIRT4 positive whereas only 29 (12.03%)
adjacent non-cancerous tissue samples were SIRT4 positive
(P<0.001).
SIRT4 expression is associated with
ER, PR and p53 status and molecular subtypes of breast cancer
To investigate the association between SIRT4
expression and clinicopathological parameters, the 409 breast
cancer tissue samples were divided into two subgroups based on
their SIRT4 expression (negative and positive SIRT4 expression).
The differences in clinical parameters, including age, LNM, TNM
pathological stage and grade, ER, PR, HER2, Ki-67 and p53 status
and molecular subtypes were compared between the two groups
(Table I). Significant differences
between the SIRT4 positive and negative expression groups were
identified for the following parameters: ER status (P=0.0068), PR
status (P=0.0073), p53 status (P=0.0040) and molecular subtypes
(P=0.0404). SIRT4 protein expression rates were significantly
different in the different subtypes of breast cancer (P=0.0404).
The overall positive expression rate in each subtype was extremely
low. SIRT4 protein expression rates were higher in Luminal B
(27.97%) and Luminal A (20.00%) subtypes than the HER2 (13.73%) and
triple negative breast cancer (TNBC) (16.09%) subtypes. Notably,
the negative SIRT4 expression rates were significantly higher in
TNBC (83.91%) and HER2-positive subtypes (86.27%) than that of the
Luminal A (72.03%) and Luminal B (80%) subtypes (P=0.0404). The
positive SIRT4 protein expression rates were significantly higher
in ER or PR positive patients than that in ER or PR negative
patients (P=0.0111). The expression of SIRT4 was significantly
higher in the p53-positive cancer tissues compared with the
p53-negative tissues (P=0.0040). However, no significant
correlation was identified between SIRT4 expression and patient
age, LNM, tumor size, TNM stage, pathological grade, HER2 status or
Ki-67 status (P>0.05; Table
I).
 | Table I.Comparison of clinicopathological
features between SIRT4 positive and negative breast cancer patients
(n=409). |
Table I.
Comparison of clinicopathological
features between SIRT4 positive and negative breast cancer patients
(n=409).
|
|
| Cytoplasmic SIRT4
expression |
|
|---|
|
|
|
|
|
|---|
| Parameter | Patients, n | Negative, n (%) | Positive, n (%) | P-value |
|---|
| Age, years |
|
|
| 0.3759 |
|
<50 | 223 | 176 (78.92) | 47 (21.08) |
|
| ≥50 | 186 | 139 (74.73) | 47 (25.27) |
|
| Tumor size, cm |
|
|
| 0.2251 |
|
<2 | 141 | 114 (80.85) | 27 (19.15) |
|
| ≥2 | 268 | 201 (75.00) | 67 (25.00) |
|
| LNM |
|
|
| 0.9020 |
|
Negative | 187 | 143 (76.47) | 44 (23.53) |
|
|
Positive | 222 | 172 (77.48) | 50 (22.52) |
|
| TNM stage |
|
|
| 0.1713 |
| I–II | 78 | 55 (70.51) | 23 (29.49) |
|
| III | 331 | 260 (78.55%) | 71 (21.45) |
|
| Histological
grade |
|
|
| 0.1649 |
| G1-2 | 143 | 104 (72.73) | 39 (27.27) |
|
| G3 | 266 | 211 (79.32) | 55 (20.68) |
|
| ER status |
|
|
| 0.0068 |
|
Negative | 226 | 186 (82.30) | 40 (17.70) |
|
|
Positive | 183 | 129 (70.49%) | 54 (29.51) |
|
| PR status |
|
|
| 0.0073 |
|
Negative | 164 | 138 (84.15) | 26 (15.85) |
|
|
Positive | 245 | 177 (72.24) | 68 (27.76) |
|
| HER2 status |
|
|
| 0.1447 |
|
Negative | 324 | 244 (75.31) | 80 (24.69) |
|
|
Positive | 85 | 71 (83.53) | 14 (16.47) |
|
| Ki-67 status,
% |
|
|
| 0.4060 |
|
<14 | 222 | 175 (78.83) | 47 (21.17) |
|
|
≥14 | 187 | 140 (74.87) | 47 (25.13) |
|
| p53 status |
|
|
| 0.0040 |
|
Negative | 79 | 71 (89.87) | 8 (10.13) |
|
|
Positive | 330 | 244 (73.94) | 86 (26.06) |
|
| Molecular
subtype |
|
|
| 0.0404 |
| Luminal
A | 236 | 170 (72.03) | 66 (27.97) |
|
| Luminal
B | 35 | 28 (80.00) | 7 (20.00) |
|
|
HER2 | 51 | 44 (86.27) | 7 (13.73) |
|
| Triple
negative | 87 | 73 (83.91) | 14 (16.09) |
|
| ER/PR status |
|
|
| 0.0111 |
| Double
negative | 94 | 73 (77.66) | 21 (22.34) |
|
|
Positive | 315 | 198 (62.86) | 117 (37.14) |
|
High SIRT4 expression is a favorable
prognostic factor for breast cancer
Kaplan-Meier survival analysis was performed to
investigate the association between SIRT4 expression and the
survival time of patients with certain subtypes of invasive breast
cancer. As shown in Table II,
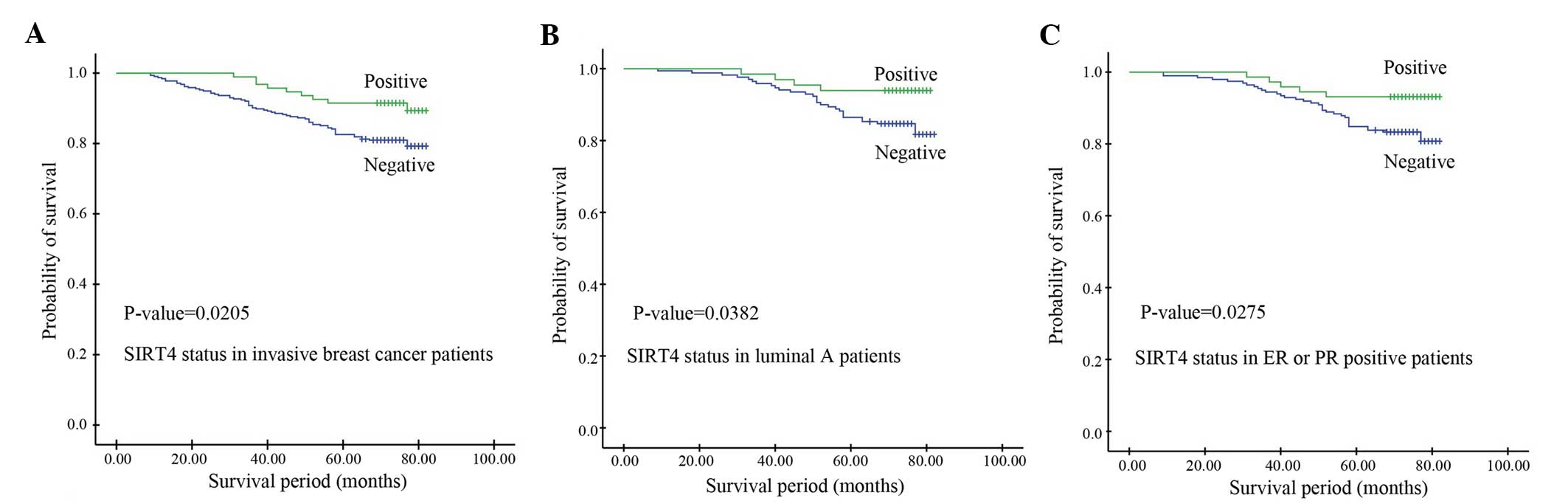
univariate analyses identified SIRT4 expression (P=0.0205),
pathological stage (P=0.0019), PR status (P=0.0127), Ki-67 status
(P=0.0003) and LNM (P<0.0001) as significant prognostic
predictors. The median OS time of the low SIRT4 expression group
was 67.8 months, which was significantly shorter than that of the
high SIRT4 expression group (73.2 months) (P=0.0205; Fig. 2A). Multivariate analysis identified
that SIRT4 expression (P=0.02985), Ki-67 status (P=0.01767) and LNM
(P=0.001393) were independent prognostic factors (Table II). The results of this study
indicate that high SIRT4 expression is a favorable prognostic
factor for patients with invasive breast cancer.
 | Table II.Univariate and multivariate analysis
of prognostic factors in invasive breast cancer patients. |
Table II.
Univariate and multivariate analysis
of prognostic factors in invasive breast cancer patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameter | Risk ratio | 95% CI | P-value | Risk ratio | 95% CI | P-value |
|---|
| Age, years |
|
| 0.5538 |
|
|
|
| ≥50 vs.
<50 | 1.1505 | 0.7213–1.8350 |
|
|
|
|
| Pathological
stage |
|
| 0.0019 |
|
| 0.3856 |
| I–II
vs. III | 4.3055 | 2.4185–7.6647 |
| 1.6719 | 0.5267–5.3077 |
|
| Tumor size, cm |
|
| 0.9110 |
|
|
|
| ≥2 vs.
<2 | 1.0284 | 0.6303–1.6779 |
|
|
|
|
| LNM |
|
| 0.0001 |
|
| 0.0014 |
|
Positive vs. negative | 3.8147 | 2.3950–6.0759 |
| 3.0141 | 1.5375–5.9088 |
|
| Histological
grade |
|
| 0.1086 |
|
| 0.3856 |
| G1-2
vs. G3 | 1.5313 | 0.9429–2.4870 |
| 1.6719 | 0.5267–5.3077 |
|
| ER status |
|
| 0.3940 |
|
|
|
|
Positive vs. negative | 0.8146 | 0.5108–1.2992 |
|
|
|
|
| PR status |
|
| 0.0127 |
|
| 0.2652 |
|
Positive vs. negative | 0.5585 | 0.3457–0.9023 |
| 0.7465 | 0.4476–1.2451 |
|
| HER2 status |
|
| 0.0046 |
|
| 0.2387 |
|
Positive vs. negative | 2.0190 | 1.1183–3.6452 |
| 1.3858 | 0.8076–2.3781 |
|
| Ki-67 status,
% |
|
| 0.0003 |
|
| 0.0177 |
| ≥14 vs.
<14 | 2.3919 | 1.4961–3.8240 |
| 1.8769 | 1.1186–3.1492 |
|
| p53 status |
|
| 0.1445 |
|
|
|
|
Positive vs. negative | 0.6739 | 0.3708–1.2249 |
|
|
|
|
| SIRT4 status |
|
| 0.0205 |
|
| 0.0299 |
|
Positive vs. negative | 0.4480 | 0.2608–0.7696 |
| 0.4586 | 0.2277–0.9234 |
|
Positive SIRT4 expression is
associated with longer OS times in patients with Luminal A breast
cancer and ER/PR positive patients
Kaplan-Meier OS curves were established to determine
the effects of SIRT4 expression on survival in patients with
different molecular subtypes of breast cancer. No significance
differences in survival were identified between the Luminal B, HER2
and TNBC subtypes (P>0.05). In the Luminal A group, the OS time
of patients with negative SIRT4 expression (70.35 months) was
significantly shorter than that of patients with positive SIRT4
expression (73.64 months) (P=0.0382; Fig.
2B). Similarly, in ER or PR positive patients, the OS time of
patients with negative SIRT4 expression was significantly shorter
than that of patients with positive SIRT4 expression, indicating
that patients with positive SIRT4 expression exhibit a longer
survival time than those with negative SIRT4 expression (P=0.0275;
Fig. 2C).
Discussion
SIRT4 exhibits a crucial function in carcinogenesis
as a tumor suppressor (5). The
present study revealed that SIRT4 expression in breast cancer
tissues was significantly higher than that in adjacent
non-cancerous tissues. Univariate and multivariate analyses
demonstrated that high SIRT4 expression is a protective factor in
breast cancer patients. Notably, the positive expression rates of
SIRT4 were low in both control and breast cancer tissues. Jeong
et al (10) performed a
meta-analysis to compare SIRT4 expression in human cancers and
normal tissues, which demonstrated that SIRT4 mRNA expression
levels were significantly lower in cancers of the bladder, breast,
colon, stomach, ovarian and thyroid than normal tissue. In
addition, low expression of SIRT4 mRNA was significantly correlated
with early metastasis in breast cancer patients (10). In the present study, high levels of
SIRT4 protein expression were not identified in the adjacent
non-tumor tissues. We hypothesize that post-transcription
regulation may affect SIRT4 expression and translation. Thus,
further study is required to confirm this and to elucidate the
possible underlying mechanisms.
The majority of previous studies regarding SIRT4
gene function have been performed in normal tissues (17–19). SIRTs
are nicotinamide adenine dinucleotide+-dependent deacylases and
ADP-ribosyltransferases that are involved in numerous cellular
processes (17–19). SIRT4 inhibition increases fat
oxidative capacity and mitochondrial function in liver and muscle
cells (20). A limited number of
genes directly interact with SIRT4, including cyclic adenosine
monophosphate response element-binding protein 2 (CREB2), malonyl
CoA decarboxylase and GDH (5,18). In the present study, SIRT4 expression
was significantly correlated with ER, PR and p53 status. Previously
it has been demonstrated that SIRT4 regulates ADP-ribosylates and
inhibits GDH to regulate glutamine metabolism (8). A gas chromatography-time-of-flight mass
spectrometry-based metabolomics study revealed that the glutamate
metabolite changes are increased in the ER-negative subtype of
human breast cancer (21). The
detected changes included the metabolism of glutamine with a
2.2-fold decrease in the concentration of glutamine and an increase
in the concentration of glutamate. The highest glutamine metabolic
activity is observed in HER2+ breast cancer and TNBC
(22). In the present study, positive
SIRT4 expression was identified in 54/183 (29.51%) ER-positive
patients compared with 40/226 (17.70%) ER-negative patients.
Furthermore, the OS time of patients with negative SIRT4 expression
was significantly shorter than patients with positive SIRT4
expression in ER or PR-positive and double-negative patients.
ER-negative breast cancers exhibit a particular type of
glutamine-dependent anaplerosis that is characterized by elevated
levels of the gene encoding phosphoglycerate dehydrogenase (PHGDH)
(23). Notably, inhibition of PHGDH
in breast cancer cell lines induces a metabolic collapse in TCA
cycle intermediates, which is also observed during DNA damage
response-induced SIRT4 upregulation and GDH inhibition (10,23). Thus,
SIRT4 may affect the functional correlation between GDH and ER and
PR.
Glutamine exhibits a critical function in cancer
cell protein synthesis by supplying cellular adenosine triphosphate
and serving as a metabolic intermediate for nucleotide synthesis
(24). Glutamine dependence leads to
growth restriction and cell death in glutamine limiting conditions,
indicating that the glutamate metabolic pathway may present a
potential therapeutic target (24–26). In
the present study, low SIRT4 expression was associated with Luminal
A breast cancer, which is a subtype of ER and PR positive breast
cancer that is treated using anti-hormone receptor therapy. Among
Luminal A breast cancer patients, the levels of SIRT4 expression
were significantly associated with OS: Low SIRT4 expression was
correlated with worse OS. In the present study, all patients with
the luminal subtype of breast cancer were treated with anti-hormone
receptor therapy. The drug sensitivity of anti-hormone receptors
may be associated with SIRT4 expression levels. Therefore, the use
of anti-hormone receptor therapy in combination with drugs that
inhibit GDH or elevate SIRT4 protein levels may be beneficial in
Luminal A patients. Although, no significant differences between
SIRT4 expression and the OS of luminal B patients were identified,
anti-HER2 therapy may affect the OS of these patients. Future
studies are required to elucidate the potential association between
anti-hormone drugs and SIRT4 expression.
In the present study, the positive expression rates
of SIRT4 were significantly different between p53-positive and
p53-negative patients. The p53 gene affects glutamine metabolism by
targeting phosphate-activated mitochondrial GLS2 in tumor cells.
GLS2 is a key enzyme involved in the conversion of glutamine to
glutamate. It controls the apoptotic response and the cellular
levels of reactive oxygen species (ROS). The tumor suppressor
function of p53 may involve the interaction between glutamine
metabolism, energy and ROS homeostasis (27). SIRT4 directly downregulates GDH
activity via ADP-ribosylation to regulate the conversion of
glutamate to α-ketoglutaric acid (8),
indicating that SIRT4 and p53 interact indirectly. However, in
another study, DNA-damaging agents increased SIRT4 levels in
p53-inactive HEK293T cells and p53-null human prostate cancer PC3
cells, indicating that SIRT4 is induced in a p53-independent
manner. After cell DNA damage, glutamine metabolism is repressed by
SIRT4 expression in the mitochondria, which leads to inhibition of
cell proliferation (10).
SIRT4 exhibits a critical function in cellular
metabolism by regulating mitochondrial glutamine metabolism. SIRT4
represses tumor formation in vivo. A reduction in SIRT4
expression results in larger tumors compared with controls
(5). In the phosphatidylinositide
3-kinase/protein kinase B/mammalian target of rapamycin (mTOR)
pathway, the mTOR complex 1 (mTORC1) gene negatively regulates the
transcription of SIRT4. mTORC1 increases the binding of CREB2 to
β-transducin repeat-containing protein leading to CREB2
ubiquitination (5). The transcription
factor CREB2 regulates the transcription of SIRT4. The mTORC1 gene
indirectly regulates SIRT4 transcription to inhibit GDH, affecting
glutamine metabolism. SIRT3, −4 and −5 are all expressed in the
mitochondria. SIRT3 also regulates genomic instability to repress
tumorigenesis (28,29). SIRT4 and −3 appear to coordinately
regulate tumor cell anabolism (30).
The present study demonstrated that negative SIRT4
protein expression was an independent predictor of a worse
prognosis in invasive breast carcinoma. High SIRT4 expression
levels were associated with a better OS in breast cancer patients.
Based on the correlation between SIRT4 protein levels and the
survival status in ER and PR-positive breast cancer patients, we
postulate that low SIRT4 protein expression is Luminal A patients
is a more effective anti-hormone therapy when used in combination
with the glutamine metabolic inhibitor. Future study is required to
determine the molecular mechanism by which SIRT4 regulates the
progression of breast cancer.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81172498/H1622).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zeng H, Zhang S and He J:
Annual report on status of cancer in China, 2011. Chin J Cancer
Res. 27:2–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bange J, Zwick E and Ullrich A: Molecular
targets for breast cancer therapy and prevention. Nat Med.
7:548–552. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kung HN, Marks JR and Chi JT: Glutamine
synthetase is a genetic determinant of cell type-specific glutamine
independence in breast epithelia. PLoS Genet. 7:e10022292011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Csibi A, Fendt SM, Li C, Poulogiannis G,
Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T,
et al: The mTORC1 pathway stimulates glutamine metabolism and cell
proliferation by repressing SIRT4. Cell. 153:840–854. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fernandez-Marcos PJ and Serrano M: Sirt4:
The glutamine gatekeeper. Cancer Cell. 23:427–428. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
DeBerardinis RJ, Lum JJ, Hatzivassiliou G
and Thompson CB: The biology of cancer: Metabolic reprogramming
fuels cell growth and proliferation. Cell Metab. 7:11–20. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Haigis MC, Mostoslavsky R, Haigis KM,
Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos
GD, Karow M, Blander G, et al: SIRT4 inhibits glutamate
dehydrogenase and opposes the effects of calorie restriction in
pancreatic beta cells. Cell. 126:941–954. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sebastián C, Satterstrom FK, Haigis MC and
Mostoslavsky R: From sirtuin biology to human diseases: An update.
J Biol Chem. 287:42444–42452. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jeong SM, Xiao C, Finley LW, Lahusen T,
Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, et al:
SIRT4 has tumor-suppressive activity and regulates the cellular
metabolic response to DNA damage by inhibiting mitochondrial
glutamine metabolism. Cancer Cell. 23:450–463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He W, Newman JC, Wang MZ, Ho L and Verdin
E: Mitochondrial sirtuins: Regulators of protein acylation and
metabolism. Trends Endocrinol Metab. 23:467–476. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vassilopoulos A, Fritz K S, Petersen DR
and Gius D: The human sirtuin family: Evolutionary divergences and
functions. Hum Genomics. 5:485–496. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cheang MC, Chia SK, Voduc D, Gao D, Leung
S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, et al: Ki67
index, HER2 status, and prognosis of patients with luminal B breast
cancer. J Natl Cancer Inst. 101:736–750. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Goldhirsch A, Wood WC, Coates AS, Gelber
RD, Thurlimann B and Senn HJ: Panel members: Strategies for
subtypes - dealing with the diversity of breast cancer: Highlights
of the St. Gallen International Expert Consensus on the Primary
Therapy of Early Breast Cancer 2011. Ann Oncol. 22:1736–1747. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shaoqiang C, Yue Z, Yang L, Hong Z, Lina
Z, Da P and Qingyuan Z: Expression of HOXD3 correlates with shorter
survival in patients with invasive breast cancer. Clin Exp
Metastasis. 30:155–163. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu T, Zhang X, Shang M, Zhang Y, Xia B,
Niu M, Liu Y and Pang D: Dysregulated expression of Slug, vimentin,
and E-cadherin correlates with poor clinical outcome in patients
with basal-like breast cancer. J Surg Oncol. 107:188–194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lombard DB, Tishkoff DX and Bao J:
Mitochondrial sirtuins in the regulation of mitochondrial activity
and metabolic adaptation. Handb Exp Pharmacol. 206:163–188. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Laurent G, German NJ, Saha AK, de Boer VC,
Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran
B, et al: SIRT4 coordinates the balance between lipid synthesis and
catabolism by repressing malonyl CoA decarboxylase. Mol Cell.
50:686–698. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Houtkooper RH, Pirinen E and Auwerx J:
Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol
Cell Biol. 13:225–238. 2012.PubMed/NCBI
|
|
20
|
Nasrin N, Wu X, Fortier E, Feng Y, Bare'
OC, Chen S, Ren X, Wu Z, Streeper RS and Bordone L: SIRT4 regulates
fatty acid oxidation and mitochondrial gene expression in liver and
muscle cells. J Biol Chem. 285:31995–32002. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Budczies J, Brockmöller SF, Müller BM,
Barupal DK, Richter-Ehrenstein C, Kleine-Tebbe A, Griffin JL,
Orešič M, Dietel M, Denkert C and Fiehn O: Comparative metabolomics
of estrogen receptor positive and estrogen receptor negative breast
cancer: Alterations in glutamine and beta-alanine metabolism. J
Proteomics. 94:279–288. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McGuirk S, Gravel SP, Deblois G,
Papadopoli DJ, Faubert B, Wegner A, Hiller K, Avizonis D, Akavia
UD, Jones RG, et al: PGC-1α supports glutamine metabolism in breast
cancer. Cancer Metab. 1:222013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Possemato R, Marks KM, Shaul YD, Pacold
ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et
al: Functional genomics reveal that the serine synthesis pathway is
essential in breast cancer. Nature. 476:346–350. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wise DR and Thompson CB: Glutamine
addiction: A new therapeutic target in cancer. Trends Biochem Sci.
35:427–433. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tennant DA, Durán RV and Gottlieb E:
Targeting metabolic transformation for cancer therapy. Nat Rev
Cancer. 10:267–277. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
DeBerardinis RJ and Cheng T: Q's next: The
diverse functions of glutamine in metabolism, cell biology and
cancer. Oncogene. 29:313–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Suzuki S, Tanaka T, Poyurovsky MV, Nagano
H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y,
et al: Phosphate-activated glutaminase (GLS2), a p53-inducible
regulator of glutamine metabolism and reactive oxygen species. Proc
Natl Acad Sci USA. 107:7461–7466. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim HS, Patel K, Muldoon-Jacobs K, Bisht
KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage
J, Owens KM, et al: SIRT3 is a mitochondria-localized tumor
suppressor required for maintenance of mitochondrial integrity and
metabolism during stress. Cancer Cell. 17:41–52. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Finley LW, Carracedo A, Lee J, Souza A,
Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish
CB, et al: SIRT3 opposes reprogramming of cancer cell metabolism
through HIF1α destabilization. Cancer Cell. 19:416–428. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kumar S and Lombard DB: Mitochondrial
sirtuins and their relationships with metabolic disease and cancer.
Antioxid Redox Signal. 22:1060–1077. 2015. View Article : Google Scholar : PubMed/NCBI
|