Introduction
Alcohol and tobacco use are associated with ≥75% of
all head and neck squamous cell carcinomas (HNSCCs) (1). In addition, alcohol has been reported to
independently increase the risk of cancer (2). Despite compelling evidence suggests that
alcohol plays a key role in the pathogenesis of HNSCC, its
molecular mechanism remains poorly understood (2). Alcohol is speculated to increase the
risk of cancer by impairing DNA-repair genes or folate metabolism
(2). However, previous studies have
suggested that it is not alcohol, but instead its metabolite
acetaldehyde, the major contributor to HNSCC progression (2,3).
Acetaldehyde is produced by alcohol dehydrogenases in the
intestine, kidney, liver and oral cavity, and has been proposed to
exert mutagenic effects, including DNA cross-linking, chromatid
exchange, aneuploidy and other chromosomal abnormalities (3). The present study specifically focuses on
the effect of alcohol exposure on HNSCC, and proposes that ethanol
or its derivative acetaldehyde alters the expression of key long
non-coding RNAs (lncRNAs) that may be critical in the pathogenesis
of HNSCC.
The human genome sequence is composed primarily of
non-coding RNAs (ncRNAs), with only 2% of RNAs coding for proteins
(4,5).
Previously, ncRNAs were considered to be transcriptional noise
(4). However, it has been recently
reported that ncRNAs are important in transcriptional and
post-transcriptional processes (4).
Recent studies have revealed that ncRNAs influence messenger RNA
(mRNA) translation and chromatin modifications (5). Since previous studies have demonstrated
that alcohol is able to regulate ncRNAs (6,7), it is
possible that the role of ncRNAs as epigenetic regulators could
account for the effects of alcohol in the pathogenesis of
HNSCC.
The present study focused on lncRNAs, which are
ncRNAs of >200 nucleotides in length. lncRNAs have been reported
to play a critical role in cancer progression through the
modification of transcription factors associated with regulation of
oncogenes, tumor suppressor proteins, self-renewal and
differentiation (8,9). lncRNAs regulate transcription factors by
acting as chromatin modifiers and direct transcriptional regulators
(8,10,11). This
regulation has been demonstrated to occur either in cis (in
close proximity to the transcribed lncRNA) or in trans (far
from the transcription site) (12).
Therefore, alcohol-dysregulated lncRNAs could play critical roles
in the inhibition of tumor suppressors or the activation of
oncogenes that are required for the malignant transformation of
normal oral epithelial cells.
Using RNA sequencing (RNA-seq) technology, the
present study identified a panel of lncRNAs that were
differentially expressed between alcohol drinkers and non-alcohol
drinkers among HNSCC patients. This panel was partially validated
in vitro in normal oral keratinocytes treated with
clinically relevant levels of ethanol and acetaldehyde.
Materials and methods
RNA-seq analysis
An in silico differential expression analysis
was conducted utilizing publicly available RNA-seq libraries
obtained from The Cancer Genome Atlas (TCGA; https://tcga-data.nci.nih.gov/tcga), which comprised
34 HNSCC patients, 17 of which were alcohol drinkers and 17
non-alcohol drinkers. Within each category of alcohol drinkers and
non-alcohol drinkers, 12 patients were tobacco smokers and 5 were
non-tobacco smokers (Table I). The
libraries were generated by TCGA utilizing a Genome Analyzer IIx
(Illumina Inc., San Diego, CA, USA), which resulted in paired-end
RNA-seq libraries with insert sizes of 200 bp-5 kb. These RNA-seq
files were next position-sorted, indexed and aligned to a human
reference genome (hg19). The files were then annotated with a
browser extensible data file containing 32,108 human lncRNA
transcripts, which was downloaded from LNCipedia (http://www.lncipedia.org/) (13). The bedtools (http://bedtools.readthedocs.io/en/latest/) (14) utility coverageBed was then used to
generate lncRNA read counts (integer values of expression levels)
by calculating the number of alignments from each RNA-seq file that
overlapped with each individual lncRNA provided by the annotation
file from LNCipedia. A total of 13,338 lncRNAs displayed reads
generated in the RNA-seq libraries.
 | Table I.Demographic characteristics of 34
patients with head and neck squamous cell carcinoma included in the
present in silico analysis, with categorical breakdowns of
drinking status, smoking status, vital state, gender, tumor site,
stage and grade. |
Table I.
Demographic characteristics of 34
patients with head and neck squamous cell carcinoma included in the
present in silico analysis, with categorical breakdowns of
drinking status, smoking status, vital state, gender, tumor site,
stage and grade.
| Variables | Total patients (%)
n=34 | Alcohol drinkers
and tobacco smokers (%) n=12 | Alcohol drinkers
but non-tobacco smokers (%) n=5 | Non-alcohol
drinkers but tobacco smokers (%) n=12 | Non-alcohol
drinkers or tobacco smokers (%) n=5 |
|---|
| Gender |
|
|
Male | 25 (74) | 10 (83) | 3 (60) | 8 (67) | 4 (80) |
|
Female | 9 (26) | 2 (17) | 2 (40) | 4 (33) | 1 (20) |
| Drinks per day |
|
|
None | 17 (50) | 0 (0) | 0 (0) | 12 (100) | 5 (100) |
|
0–2 | 14 (41) | 11 (92) | 3 (60) | 0 (0) | 0 (0) |
|
>2 | 3 (9) | 1 (8) | 2 (40) | 0 (0) | 0 (0) |
| Vital state |
|
|
Deceased | 11 (32) | 4 (33) | 0 (0) | 6 (50) | 1 (20) |
|
Alive | 23 (68) | 8 (67) | 5 (100) | 6 (50) | 4 (80) |
| Tumor site |
|
|
Oral | 24 (70) | 8 (67) | 3 (60) | 9 (75) | 4 (80) |
|
Pharyngeal | 4 (12) | 1 (8) | 2 (40) | 0 (0) | 1 (20) |
|
Laryngeal | 6 (18) | 3 (25) | 0 (0) | 3 (25) | 0 (0) |
| Stage |
|
| Low (I,
II) | 5 (15) | 2 (17) | 2 (40) | 1 (8) | 0 (0) |
| High
(III, IV) | 29 (85) | 10 (83) | 3 (60) | 11 (92) | 5 (100) |
| Grade |
|
| GX | 1 (3) | 0 (0) | 1 (20) | 0 (0) | 0 (0) |
|
G1-G2 | 26 (76) | 7 (58) | 4 (80) | 10 (83) | 5 (100) |
|
G3-G4 | 7 (21) | 5 (42) | 0 (0) | 2 (17) | 0 (0) |
These read counts were then utilized for lncRNA
differential expression analysis, which compared alcohol drinkers
vs. non-alcohol drinkers using the R/Bioconductor software package
edgeR (version 3.4.2; http://www.bioconductor.org/packages). The read counts
were normalized within edgeR based on the relative library sizes of
each cohort. The differential expression analysis implemented in
edgeR utilized an empirical Bayes estimation and exact tests based
on the negative binomial distribution of the reads (15). From this comparison, a list of
differentially expressed lncRNAs with false discovery rates <5%
was compiled in HNSCC patients who were alcohol drinkers vs. those
who were non-alcohol drinkers (Table
II).
 | Table II.Differentially expressed lncRNAs with
FDR<0.05, including expression log fold change between drinkers
and nondrinkers, gene log counts per million, and forward and
reverse primers used for in vitro verification.
lnc-NETO1-1 and lnc-SLC39A11-2 are represented by
different splice variants and with one common forward and reverse
primer. |
Table II.
Differentially expressed lncRNAs with
FDR<0.05, including expression log fold change between drinkers
and nondrinkers, gene log counts per million, and forward and
reverse primers used for in vitro verification.
lnc-NETO1-1 and lnc-SLC39A11-2 are represented by
different splice variants and with one common forward and reverse
primer.
| lncRNAs | log2
FC | log2
CPM | P-value | FDR | Forward primer | Reverse primer |
|---|
|
lnc-SLC39A11-2:7 | 4.194151 | 3.878865 | 1.94E-07 | 0.001085 |
5′-CGATGTGTCTCTTTTTCCCGT-3′ |
5′-AGAAGGCTGAACCAGACGAC-3′ |
|
lnc-SLC39A11-2:5 | 4.140367 | 3.885084 | 2.44E-07 | 0.001085 |
5′-CGATGTGTCTCTTTTTCCCGT-3′ |
5′-AGAAGGCTGAACCAGACGAC-3′ |
|
lnc-SLC39A11-2:6 | 4.142305 | 3.884648 | 2.46E-07 | 0.001085 |
5′-CGATGTGTCTCTTTTTCCCGT-3′ |
5′-AGAAGGCTGAACCAGACGAC-3′ |
|
lnc-LAMB3-1:1 | −2.581367 | 6.606811 | 2.54E-06 | 0.008414 |
5′-TAAGGCTGTGTGTCCTGGTT-3′ |
5′-TCCTCTGTTCCATACCATCACT-3′ |
|
lnc-CCL18-1:1 | −2.190396 | 5.522936 | 5.69E-06 | 0.015064 |
5′-AGAAGTCATACCCCAACCCAA-3′ |
5′-CGAATCAGTTAGCAAGAGGCA-3′ |
|
lnc-NETO1-1:9 | 3.351077 | 3.585717 | 1.56E-05 | 0.022272 |
5′-TCTGCCCCCACATCATTTCT-3′ |
5′-TCCAGTGATTAGGGCTTGAACT-3′ |
|
lnc-NETO1-1:3 | 3.364047 | 3.615927 | 1.63E-05 | 0.022272 |
5′-TCTGCCCCCACATCATTTCT-3′ |
5′-TCCAGTGATTAGGGCTTGAACT-3′ |
|
lnc-NETO1-1:2 | 3.362404 | 3.616193 | 1.64E-05 | 0.022272 |
5′-TCTGCCCCCACATCATTTCT-3′ |
5′-TCCAGTGATTAGGGCTTGAACT-3′ |
|
lnc-NETO1-1:4 | 3.347929 | 3.625721 | 1.68E-05 | 0.022272 |
5′-TCTGCCCCCACATCATTTCT-3′ |
5′-TCCAGTGATTAGGGCTTGAACT-3′ |
|
lnc-NETO1-1:6 | 3.34735 | 3.624247 | 1.68E-05 | 0.022272 |
5′-TCTGCCCCCACATCATTTCT-3′ |
5′-TCCAGTGATTAGGGCTTGAACT-3′ |
|
lnc-PSD4-1:14 | 1.669653 | 1.020086 | 2.74E-05 | 0.032967 |
5′-GCTGATGGCAAGGGATAGCA-3 |
5′-CTGGCTTCCTTCACCCAAAA-3′ |
|
lnc-SPANXA2-2:1 | 2.367287 | 2.927082 | 3.36E-05 | 0.037051 |
5′-ACCAACTCTCCTGATTTCCTCA-3′ |
5′-CTGGGGCTGTCCTGTTTTTA-3′ |
|
lnc-AC002472.13.1-1:1 | −1.862271 | 1.547773 | 4.26E-05 | 0.041177 |
5′-CAGGATGGAGTGGAGCCTTC-3′ |
5′-TCTGGTAGAAAAAGGGATGGGT-3′ |
|
lnc-ERC1-1:2 | −3.066857 | 4.256589 | 4.98E-05 | 0.041177 |
5′-TAGCAAGAGAGCGAAGTCCC-3′ |
5′-GTGTTTGGAGGAGGAAGGGT-3′ |
|
lnc-FBXL14-1:1 | −3.066857 | 4.256589 | 4.98E-05 | 0.041177 |
5′-GTGTTTGGAGGAGGAAGGGT-3′ |
5′-TAGCAAGAGAGCGAAGTCCC-3′ |
|
lnc-ERC1-1:3 | −3.066857 | 4.256589 | 4.98E-05 | 0.041177 |
5′-TAGCAAGAGAGCGAAGTCCC-3′ |
5′-GTGTTTGGAGGAGGAAGGGT-3′ |
|
lnc-KTN1-AS1-1:6 | −2.463648 | 1.922281 | 5.81E-05 | 0.042703 |
5′-GCTCCAGGCTAAGGTAATGAGA-3′ |
5′-CTGTGGCTCTATTCCCCATCT-3′ |
Cell culture
In vitro experiments were performed on OKF4
and OKF6, two noncancerous cell lines obtained from the laboratory
of Dr James Rheinwald at Harvard Medical School (Harvard
University, Boston, MA, USA). Normal oral keratinocytes from the
floor of the mouth were used, since the hypothesis proposed by the
present authors concerns the initial steps in the pathogenesis of
alcohol-induced oropharyngeal cancer.
The oral keratinocytes were cultured in 1X
Keratinocyte-serum-free medium (SFM) with L-glutamine (catalogue
no. 17005-042), supplemented with 0.2 ng/ml human recombinant
epidermal growth factor (EGF) type B (amino acids 1–53), 25 µg/ml
bovine pituitary extract (BPE), 0.3 mM calcium chloride, 100 U/ml
penicillin and 100 µg/ml streptomycin (all from Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), at 37°C and 5%
CO2. Upon reaching 30% confluency, OKF4 and OKF6 cells
were cultured with equal parts of supplemented Keratinocyte-SFM and
DFK medium, which was prepared with equal parts of Dulbecco's
modified Eagle's medium (catalogue no. 21068-028; Gibco; Thermo
Fisher Scientific, Inc.) and Ham's F-12 nutrient mixture (catalogue
no. 11765-054; Thermo Fisher Scientific, Inc.), and supplemented
with 0.2 ng/ml EGF type B (amino acids 1–53), 25 µg/ml BPE, 2 mM
L-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin.
Ethanol/acetaldehyde treatments
Two independent experiments regarding cell
treatments were conducted in the present study, one for ethanol and
one for acetaldehyde. For both, two biological replicates were
performed. For the alcohol experiments, the two cell lines were
treated with increasing dosages of 200 proof ethanol (0, 20, 50 and
170 µM). The concentrations of ethanol were selected based on their
toxicity, and an
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay was performed to determine the levels of cell proliferation
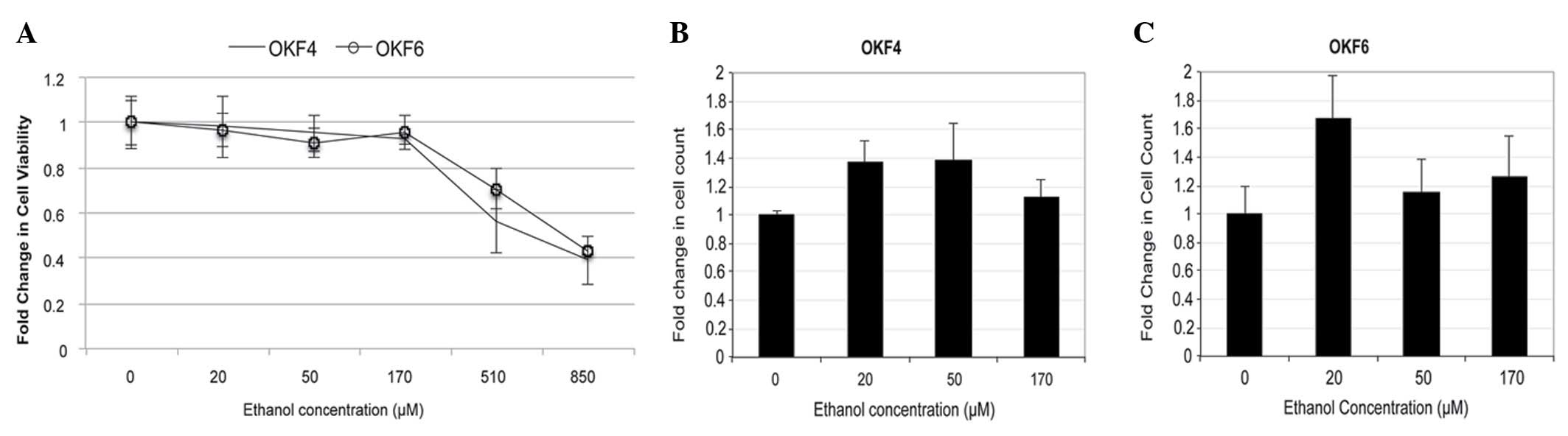
at varying ethanol concentrations (Fig.
1). To represent long-term alcohol use, cells were treated
every 24 h with ethanol diluted in medium (equal parts
Keratinocyte-SFM and DFK) for a period of 28 days. The ethanol and
the medium were replaced daily. The ethanol-treated culture plates
were covered with plastic paraffin film while incubating at 37°C to
minimize ethanol evaporation.
The above treatment was repeated with 17.82 M
acetaldehyde (catalogue no. SHBD3908V; Sigma-Aldrich, St. Louis,
MO, USA), since this is the first metabolite of ingested alcohol in
the human body (3). OKF4 and OKF6
cells were treated with acetaldehyde at concentrations of 0, 75,
150, 300 and 1,000 µM for 48 h, with acetaldehyde added every 4 h
and the medium (equal parts Keratinocyte-SFM and DFK) replaced
every 8 h. The appropriate concentrations of acetaldehyde were
determined from previous studies (16). Due to the volatility, toxicity and
short half-life of acetaldehyde, cells could not be treated for 28
days (as they had been for ethanol). The plates were also covered
with plastic paraffin film to minimize evaporation of acetaldehyde
while incubated at 37°C. Cell lines were passaged at 30–80%
confluency prior to harvesting.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Upon completion of alcohol and acetaldehyde
treatments, cells were harvested, and total cell lysates were
collected. RNA was extracted using SurePrep RNA Isolation kit
(Thermo Fisher Scientific, Inc.). Complementary DNA was synthesized
according to the manufacturer's protocol, using LncProfiler qPCR
Array kit (catalogue no. RA900A-1; System Biosciences, Mountain
View, CA, USA). qPCR was performed using SYBR Green reagent
(Applied Biosystems; Thermo Fisher Scientific, Inc.) in a
StepOnePlus Real-time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) for 2 min at 50°C, 95°C for 10 min, and 40 cycles
of 95°C for 15 sec and 60°C for 1 min. Gene expression levels with
gene-specific primers (Eurofins MWG Operon, Louisville, KY, USA;
Table II) and error bars were
calculated utilizing the 2−ΔΔCq method (17), with non-treated cells acting as a
control for the ethanol and acetaldehyde treatments, and
glyceraldehyde 3-phosphate dehydrogenase serving as a
control for endogenous gene expression.
Survival data analysis
Utilizing the original 34 patient cohort and
clinical information provided by the TCGA, the expression levels of
key dysregulated lncRNAs were correlated with the patients'
long-term survival. The expression levels of the lncRNAs were
classified as either increased or decreased depending on whether
they fell above or below the median value. A Cox proportional
hazards regression model was then applied to determine both
univariate and multivariate survival hazard ratios (HRs) for the
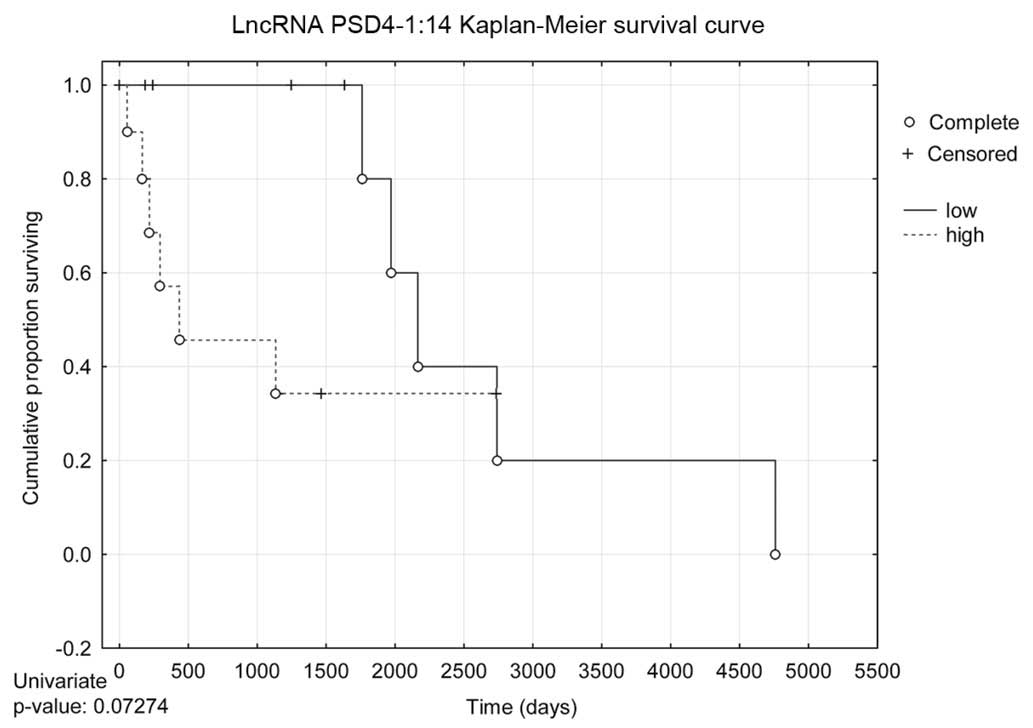
lncRNAs, based on a decreased expression (Table III). A Kaplan-Meier survival curve
was used to illustrate the correlation between lncRNA expression
and survival.
 | Table III.Cox proportional hazards regression
model for the lncRNA PSD4-1:14. Survival information,
including HR and P-value, for lnc-PSD4-1:14 in both
univariate and multivariate models demonstrates a strong
correlation between low expression of PSD4-1:14 and improved
overall survival. |
Table III.
Cox proportional hazards regression
model for the lncRNA PSD4-1:14. Survival information,
including HR and P-value, for lnc-PSD4-1:14 in both
univariate and multivariate models demonstrates a strong
correlation between low expression of PSD4-1:14 and improved
overall survival.
| Low expression | Univariate HR (95%
CI) | P-value | Multivariate HR
(95% CI) | P-value |
|---|
|
lnc-PSD4-1:14 | 0.267150 | 0.047926 | 0.236208 | 0.034013 |
|
|
(0.072234–0.988021) |
|
(0.062212–0.896836) |
|
Results and Discussion
The purpose of the present study was to identify key
lncRNAs implicated in the pathogenesis of HNSCC, since lncRNAs are
known to regulate transcription factors associated with regulation
of oncogenes, tumor suppressor proteins, self-renewal and
differentiation (8,9). Recent studies have demonstrated that
dysregulated lncRNA expression could mark the progression of a
disease (18). lncRNAs may also serve
as an indicator of patient survival independent of other variables
(19). In addition, lncRNAs have
previously been implicated in HNSCC and other types of epithelial
cancer (20). Therefore, lncRNAs may
aid the understanding of the molecular basis of HNSCC, thus
enabling advances in early detection and identification of novel
therapeutic targets aimed to improve patient prognosis.
Determination of ethanol concentrations
MTT assays were performed in the present study to
determine the ethanol concentrations required for cell treatments
and to evaluate cell proliferation prior and subsequent to
treatment. Concentrations of 0, 20, 50 and 170 µM ethanol were
selected for the experimental assays (Fig. 1). However in cell culture, 170 µM
ethanol exhibited high toxicity against normal oral keratinocytes,
and therefore was not used in subsequent experiments.
Identification of alcohol-dysregulated
lncRNAs from HNSCC patient samples
The expression patterns of 32,108 lncRNAs were
examined in the present study, 13,338 of which were detected within
the present cohort of 34 HNSCC patients. Of those 13,338 lncRNAs,
11 were differentially expressed between alcohol drinkers and
non-alcohol drinkers, with several lncRNAs represented by multiple
isoforms (Table II). Of the
identified lncRNAs, 4 were upregulated and 7 were downregulated,
with fold-changes ranging from ≥3.5 to 18.5 and false discovery
rates <5%. The small panel of lncRNAs differentially expressed
between alcohol drinkers and non-alcohol drinkers suggests that the
mechanism by which alcohol contributes to the pathogenesis of HNSCC
involves certain lncRNAs.
In vitro validation of lncRNAs
differentially expressed in clinical samples
The lncRNAs identified by RNA-seq analysis in the
present study were evaluated in vitro by measuring their
relative expression levels in two normal keratinocyte cell lines,
which were exposed to ethanol concentrations of 0, 20, 50 and 170
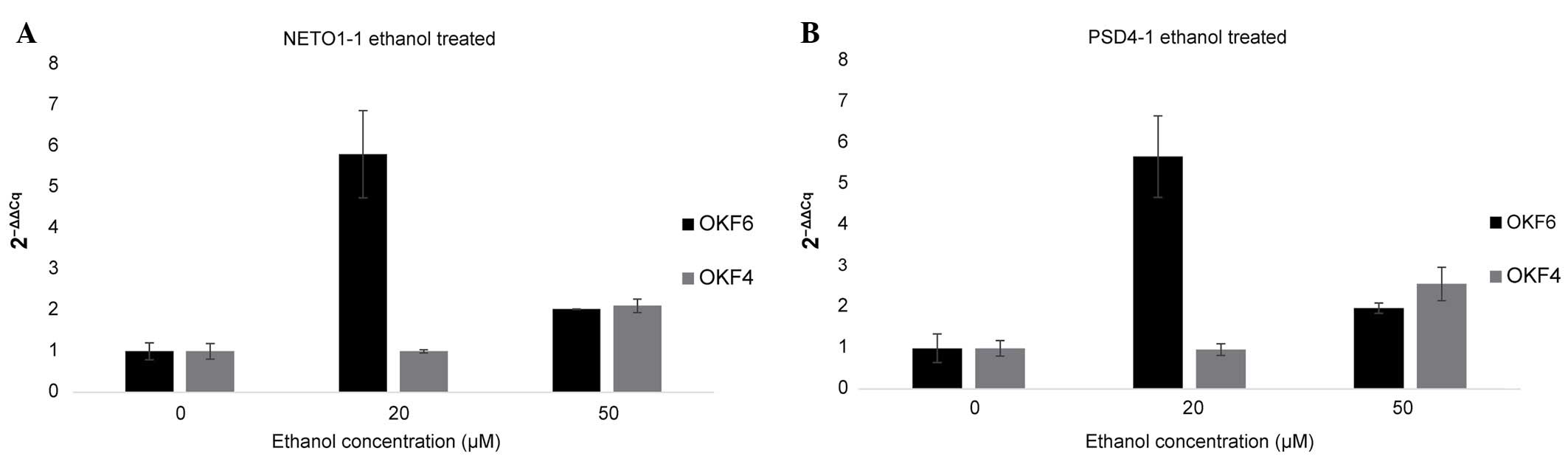
µM (Fig. 3) and acetaldehyde
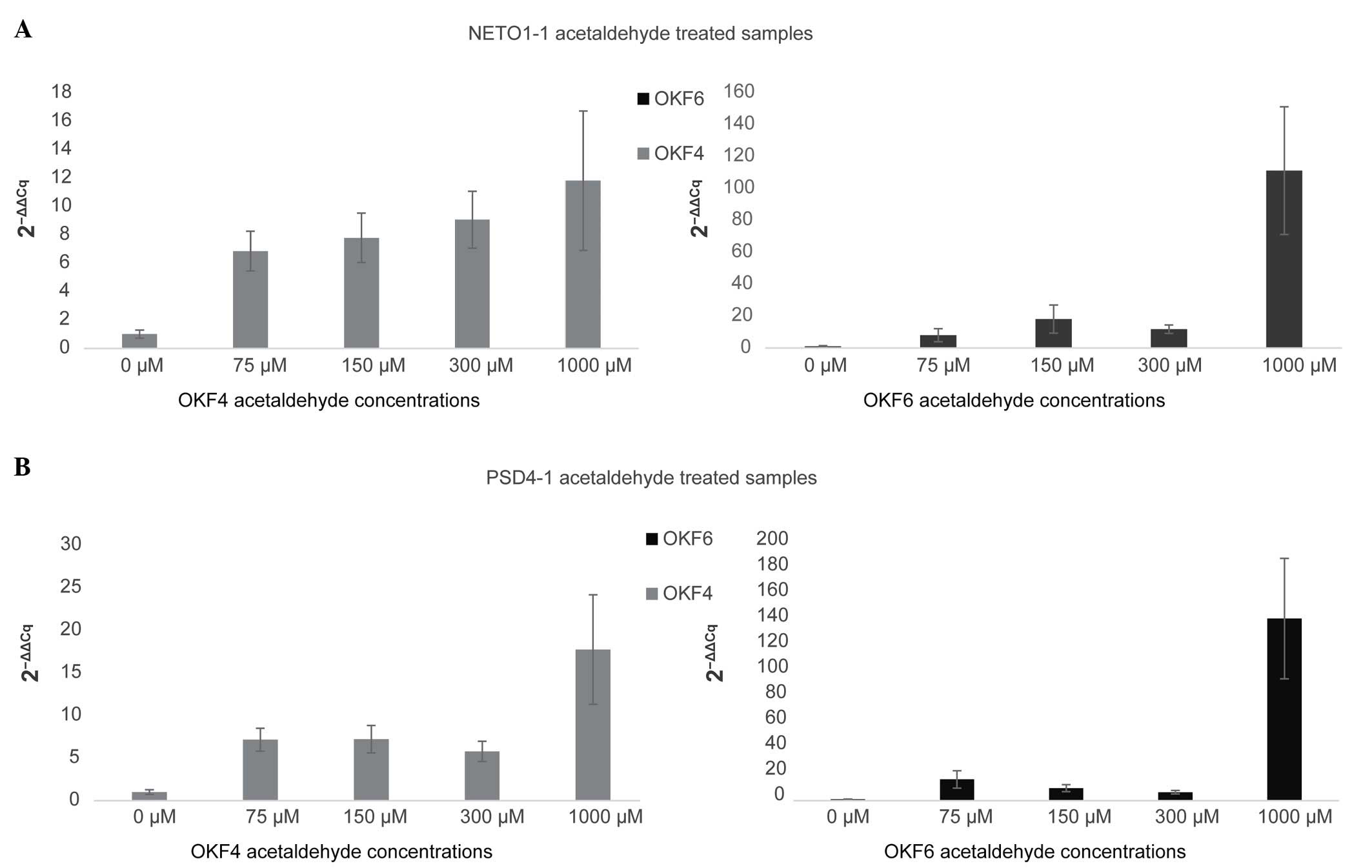
concentrations of 0, 75, 150, 300 and 1,000 µM (Fig. 4), via RT-qPCR. Of the 11 lncRNAs
identified in the RNA-seq analysis, two were verified in
vitro: lnc-PSD4-1 (including the isoform,
lnc-PSD4-1:14) and lnc-NETO1-1, whose expression
levels were increased in the treated samples, compared with the
non-treated controls. These results suggest that the above lncRNAs
may be important in the pathogenesis and progression of
alcohol-associated HNSCC.
For the ethanol-treated OKF4 cell line, both
PSD4-1 and NETO1-1 exhibited negligible changes in
their expression levels when 20 µM ethanol was used, while a 2-fold
increase in their expression levels was observed in 50 µM
ethanol-treated cells. For OKF6 cells, 20 µM ethanol exposure
resulted in a 5-fold increase in the expression levels of
PSD4-1 and NETO1-1, while 50 µM ethanol exposure
resulted in a 2-fold increase in their expression levels, compared
with the control. The higher expression levels observed in the 20
µM ethanol-treated cells vs. the 50 µM ethanol-treated cells may be
due to the 50 µM ethanol concentration being too toxic for OKF6
cells, in contrast to the OKF4 cell line, where 50 µM ethanol
demonstrated the largest effect.
While the ethanol treatment used in the present
study mimics the physiological levels of alcohol in the body,
ethanol is less carcinogenic than its metabolic derivative
acetaldehyde (3). Therefore,
acetaldehyde treatment may be a more accurate in vitro model
of the role of alcohol in HNSCC than ethanol treatment. In general,
in acetaldehyde-treated samples, both PSD4-1 and
NETO1-1 displayed higher expression levels, correlating with
higher concentrations of acetaldehyde. In OKF4 cells treated with
1,000 µM acetaldehyde, NETO1-1 and PSD4-1
demonstrated a 12-fold and 16-fold increase in expression,
respectively, compared with the control. In OKF6 cells treated with
the same concentration of acetaldehyde, both PSD4-1 and
NETO1-1 demonstrated a >100-fold increase in expression.
In the present study, 1,000 µM was selected as the upper range of
acetaldehyde concentration, since the treatment of the OKF4 and
OKF6 cell lines was limited to only 48 h due to the acetaldehyde's
volatility. This concentration is considered to be more
representative of the actual exposure to alcohol experienced by
patients with a long history of alcohol use.
Survival data
In addition to the aforementioned in vitro
tests, long-term survival analysis correlating NETO1-1 and
PSD4-1:14 expression levels and patient outcomes was
conducted. NETO1-1 did not exhibit any significant
correlation with patient survival. By contrast, low expression
levels of PSD4-1:14 were highly correlated with overall
better patient survival in both univariate (HR, 0.267150;
P=0.047926) and multivariate (HR, 0.236208; P=0.034013) Cox
proportional hazards regression models (Table III). The univariate Kaplan-Meier
survival curve in Fig. 5 demonstrates
that low expression levels of PSD4-1:14 correlate with
better patient survival. Although the Kaplan-Meier survival curve
is not statistically significant, it approached ~P=0.05, and would
likely be statistically significant in a larger sample size.
In summary, the present findings have demonstrated
an association between alcohol-associated HNSCC and increased
expression of the lncRNAs NETO1-1 and PSD4-1:14. The
increased expression of NETO1-1 and PSD4-1:14 in both
HNSCC patient clinical samples and in vitro models of
alcohol usage suggest that these lncRNAs may act as activators of
oncogenes. Previous studies have demonstrated that lncRNAs in an
antisense orientation are able to control the transcription of
mRNAs and oncogenes (21–23). PSD4-1:14 overlaps in an
antisense orientation with paired box 8 (PAX8), which
belongs to the PAX gene family, which plays a critical role
in the formation of tissues and organs during embryonic development
(24). PAX8 specifically is
considered to activate genes involved in the formation of the
thyroid gland and kidney (25,26).
PAX8 has been previously characterized as a potential
oncogene whose expression has been positively correlated with
various types of epithelial and ovarian cancer (27–29). In
addition, its overexpression has been associated with high levels
of p53 (30). In those previous
studies, PAX8 was identified as a biomarker that could be
used to differentiate between different types of epithelial tumors
(31). It is possible that
PSD4-1:14 acts as a cis-regulator of PAX8, resulting
in increased transcription of PAX8, although this may not be
the case, and the position of PAX8 in the genome may not be
associated with its interacting genes. To the best of our
knowledge, PAX8 has not been extensively studied in HNSCC;
however, its role in other types of epithelial cancer suggests that
it is candidate oncogene involved in the pathogenesis and
progression of HNSCC. The exact molecular nature of the association
between PSD4-1:14 and PAX8 has not been addressed in
the present study, and further characterization of this association
is a potential avenue of future research.
The cause of alcohol-associated HNSCC has not been
previously characterized. Based on the results of the present
study, it could be proposed that NETO1-1 and PSD4-1
may be partially responsible for the pathogenesis of
alcohol-associated HNSCC, which highlights the importance of
lncRNAs in the molecular mechanisms underlying the pathogenesis of
HNSCC. While further studies are required to understand the exact
mechanisms by which these lncRNAs function, PSD4-1 and
NETO1-1 may be considered promising potential biomarkers and
therapeutic targets of HSNCC. Further studies on these lncRNAs
could potentially lead to innovations in the prevention and
treatment of alcohol-induced HNSCC.
Acknowledgements
The clinical results described in the present study
are in whole or partly based on data generated by the TCGA Research
Network (http://cancergenome.nih.gov).
References
|
1
|
Hashibe M, Brennan P, Benhamou S,
Castellsague X, Chen C, Curado MP, Dal Maso L, Daudt AW, Fabianova
E, Fernandez L, et al: Alcohol drinking in never users of tobacco,
cigarette smoking in never drinkers and the risk of head and neck
cancer: Pooled analysis in the International Head And Neck Cancer
Epidemiology Consortium. J Natl Cancer Inst. 99:777–789. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boffetta P and Hashibe M: Alcohol and
cancer. Lancet Oncol. 7:149–156. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Homann N, Jousimies-Somer H, Jokelainen K,
Heine R and Salaspuro M: High acetaldehyde levels in saliva after
ethanol consumption: Methodological aspects and pathogenetic
implications. Carcinogenesis. 18:1739–1743. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rahimy E, Kuo SZ and Ongkeko WM:
Evaluation of non-coding RNAs as potential targets in head and neck
squamous cell carcinoma cancer stem cells. Curr Drug Targets.
15:1247–1260. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Deng G and Sui G: Noncoding RNA in
oncogenesis: A new era of identifying key players. Int J Mol Sci.
14:18319–18349. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reilly M: Role of non-coding RNAs in the
neuroadaptation to alcoholism and fetal alcohol exposure. Front
Genet. 3:702012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Laufer BI, Mantha K, Kleiber ML, Diehl EJ,
Addison SM and Singh SM: Long-lasting alterations to DNA
methylation and ncRNAs could underlie the effects of fetal alcohol
exposure in mice. Dis Model Mech. 6:977–992. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dinger ME, Amaral PP, Mercer TR, Pang KC,
Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C,
et al: Long noncoding RNAs in mouse embryonic stem cell
pluripotency and differentiation. Genome Res. 18:1433–1445. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Saxena A and Carninci P: Long non-coding
RNA modifies chromatin: Epigenetic silencing by long non-coding
RNAs. BioEssays. 33:830–839. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Volders PJ, Helsens K, Wang X, Menten B,
Martens L, Gevaert K, Vandesompele J and Mestdagh P: LNCipedia: A
database for annotated human lncRNA transcript sequences and
structures. Nucleic Acids Res. 41:(Database issue). D246–D251.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Quinlan AR and Hall IM: BEDTools: A
flexible suite of utilities for comparing genomic features.
Bioinformatics. 26:841–842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Homann N, Tillonen J, Meurman JH,
Rintamäki H, Lindqvist C, Rautio M, Jousimies-Somer H and Salaspuro
M: Increased salivary acetaldehyde levels in heavy drinkers and
smokers: A microbiological approach to oral cavity cancer.
Carcinogenesis. 21:663–668. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-δδCT method. Methods 25.4. 402–408. 2001.
|
|
18
|
Prensner JR, Iyer MK, Balbin OA,
Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso
CS, Kominsky HD, et al: Transcriptome sequencing across a prostate
cancer cohort identifies PCAT-1, an unannotated lincRNA implicated
in disease progression. Nat Biotechnol. 29:742–749. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang QQ and Deng YF: Long non-coding RNAs
as novel biomarkers and therapeutic targets in head and neck
cancers. Int J Clin Exp Pathol. 7:1286–1292. 2014.PubMed/NCBI
|
|
21
|
Morris KV: Long antisense non-coding RNAs
function to direct epigenetic complexes that regulate transcription
in human cells. Epigenetics. 4:296–301. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Carrieri C, Cimatti L, Biagioli M, Beugnet
A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C,
et al: Long non-coding antisense RNA controls Uchl1 translation
through an embedded SINEB2 repeat. Nature. 491:454–457. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
di Magliano M Pasca, Di Lauro R and
Zannini M: Pax8 has a key role in thyroid cell differentiation.
Proc Natl Acad Sci USA. 97:13144–13149. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Narlis M, Grote D, Gaitan Y, Boualia SK
and Bouchard M: Pax2 and pax8 regulate branching morphogenesis and
nephron differentiation in the developing kidney. J Am Soc Nephrol.
18:1121–1129. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Muratovska A, Zhou C, He S, Goodyer P and
Eccles MR: Paired-Box genes are frequently expressed in cancer and
often required for cancer cell survival. Oncogene. 22:7989–7997.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Laury AR, Perets R, Piao H, Krane JF,
Barletta JA, French C, Chirieac LR, Lis R, Loda M, Hornick JL, et
al: A comprehensive analysis of PAX8 expression in human epithelial
tumors. Am J Surg Pathol. 35:816–826. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liliac L, Carcangiu ML, Canevari S,
Căruntu ID, Apostol DG Ciobanu, Danciu M, Onofriescu M and Amălinei
C: The value of PAX8 and WT1 molecules in ovarian cancer diagnosis.
Rom J Morphol Embryol. 54:17–27. 2013.PubMed/NCBI
|
|
30
|
Brunner AH, Riss P, Heinze G, Meltzow E
and Brustmann H: Immunoexpression of PAX 8 in endometrial cancer:
Relation to high-grade carcinoma and p53. Int J Gynecol Pathol.
30:569–575. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xiang L and Kong B: PAX8 is a novel marker
for differentiating between various types of tumor, particularly
ovarian epithelial carcinomas. Oncol Lett. 5:735–738.
2013.PubMed/NCBI
|