Introduction
The alteration of important molecules involved in
signal transduction is a common characteristic of all malignant
cells (1). The mitogen activated
protein kinase (MAPK) pathway, which is crucially involved in
balancing cell death and differentiation on one side and
proliferation and migration on the other, is one of the best
described signalling routes that is deregulated in cancer (2). Due to the vital role of the MAPK
pathway, numerous feedback loops are present to allow a fine-tuning
of signal intensity and duration (3).
Certain molecules acting in such a regulatory loop are the members
of the Sprouty (Spry) protein family. First discovered in
Drosophila (4,5), the Spry proteins counteract processes
induced by the presence of various growth factors. In humans, 4
Spry family members are known; all of which share a conserved Spry
domain at the C-terminus and 2 homolog N-terminal boxes (6). As indicated by knockout studies in mice,
the functions of Sprouty proteins overlap but are not redundant
(7–10). Although Spry1, Spry2 and Spry4 can be
detected in all organs, their expression pattern in the various
cell types is clearly distinguishable (11). Like in Drosophila, mammalian
Spry proteins interfere with signal transduction, particularly in
the MAPK pathway, in response to various mitogens, such as the
members of the fibroblast growth factors (6). In contrast to Drosophila, in
mammalian systems, signalling activated by epidermal growth factor
is increased with augmented Spry expression. Therefore, the Spry
proteins in humans are generally considered as modulators of signal
transduction (12).
Accordingly, dependent on the tumour type, Spry
proteins are found to fulfil multiple roles in cancer. For example,
Spry2 has been indicated to function as an oncogene in colon cancer
(13,14) and Spry1 has a tumour promoting role in
rhabdomyosarcoma (15). However, Spry
proteins have been primarily shown to suppress the tumorigenic
process (12). Spry2 is a
tumour-suppressor in lung (16),
prostate (17,18), breast (19), liver (20) and ovarian cancer (21). In addition, Spry1 has also been
identified as negative regulator of cancerogenesis in numerous
types of tumour, including breast (19), prostate (22) and ovarian cancer (23). Although Spry4 has been less
intensively studied in comparison to Spry2 and Spry1, the
literature documents that the likelihood of Spry4 to act as a
tumour suppressor may be dependent on the cancer origin. Spry4 has
previously failed to significantly influence malignant phenotypes
in cancer originating from the bones (24), ovary (21) or pancreas (25), while it fulfils all features of a
tumour suppressor when the cancer has developed from lung (26) or breast tissue (27) cells. This includes an overall decrease
in Spry4 expression in cancerous tissues compared with the normal
tissue counterparts (26,28). However, the mechanisms responsible for
this decreased expression are required to be elucidated.
The gene encoding Spry4 is localized on chromosome
5q and is organized in 3 exons (29).
Due to alternative splicing, two messenger RNA (mRNA) variants can
be generated. Spry4.1 is missing the second exon, while the longer
variant Spry4.2 consists of all 3 exons, and the two variants are
shown to be expressed in most organs (30). Since alternative splicing is often
involved in modulating gene expression and function during the
cancerogenic process, the present study aimed to analyse the
expression pattern of the two Spry4 variants in breast and lung
cancer-derived cells.
Materials and methods
Cell culture
All lung adenocarcinoma-derived (Calu3, Calu6, A549,
A427 and SKLU-1) and breast cancer-derived (MDA-MB231, BT20,
MDA-MB468, ZR-75, MCF7 and SKBR3) cell lines were purchased from
American Type Culture Collection (ATCC; Rockville, MD, USA). The
squamous cell carcinoma-derived cell lines (VL-7, VL-8 and VL-10)
were established at the Institute of Cancer Research, Comprehensive
Cancer Center in the Department of Medicine I, Medical University
of Vienna (Vienna, Austria) (31). Of
the lung cancer-derived cell lines, 3 (A549, Calu6 and A427) were
known to harbour K-Ras mutations (16). Additionally, the embryonic lung
fibroblasts (WI-38) and the fibrosarcoma-derived HT-1080 cell line
were obtained from ATCC. All the cells were maintained in
Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
calf serum (FCS; Thermo Fisher Scientific, Inc.), penicillin (100
units/ml) and streptomycin (100 µg/ml) at 37°C in 7.5%
CO2. Primary normal human mammary epithelial cells
(nHMECs) were purchased from Thermo Fisher Scientific, Inc. and
normal human bronchial epithelial cells (nHBEpCs) from Pelobiotech
GmbH (Planegg, Germany). These cells were maintained according to
the providers' recommendations.
Reverse transcription-polymerase chain
reaction (RT-PCR)
RNA was extracted as previously described (32). Total RNA (3 µg), random hexamers (100
ng) and moloney-murine leukemia virus reverse transcriptase were
used to generate of complementary DNA, according to manufacturer's
protocol (Promega, Madison, USA). Semi-quantitative PCR was
performed as previously described (31). For Spry4.1, the primer
5′-CCCGGCTTCAGGATTTAC-3′ (forward) was used, and for Spry4.2, the
primer 5′-AGCCTGTATTGAGCGGTTTG-3′ (forward) was used. The primer
5′-GCTGGACCATGACTGAGTTG-3′ (reverse) was used for the two variants.
As a control for standardisation, β-microglobulin levels were
measured using the following primer pair:
5′-ACCCCAACTGAAAAAGATGA-3′ (forward) and 5′-ATCTTCAAACCTCCATGATG-3′
(reverse). The thermocycling conditions were as follows: Initial
denaturation step at 95°C for 2 min, followed by different numbers
of cycles (30 and 33 cycles for Spry4.1, 35 and 38 cycles for
Spry4.2 and 30 cycles for β-microglobulin) consisting of 95°C for
30 sec, annealing at 58°C for 30 sec and extension at 72°C for 1
min. The PCR reaction was terminated with a final step at 72°C for
5 min. For analysis, the PCR products were separated by
polyacrylamide gel electrophoresis, stained with ethidium bromide
and scanned using the Gel Doc™ XR Imaging System (Bio-Rad
Laboratories, Hercules, CA, USA). Quantification of the obtained
signals was performed using Image Quant 5.0 software (GE Healthcare
Life Sciences, Chalfont, UK). β-microglobulin was used to
standardise the RNA input. For normalisation, RNA of an arbitrarily
chosen, external cell line (HT-1080) was included in each
experiment and set as 1 (31).
Immunoblot
Protein levels were determined and extracted as
previously described (33).
Immunoblotting was carried out using affinity-purified antibodies
against Spry4 (1:100) that were generated as previously described
(30). Antibodies against
glyceraldehyde 3-phosphate dehydrogenase (GAPDH; clone, G-9; mouse
monoclonal; catalog no., sc-365062; 1:30,000; Santa Cruz
Biotechnology Inc., Dallas, TX, USA) and horseradish
peroxidase-coupled sheep anti-mouse (catalog no., NA931; 1:5,000)
and donkey anti-rabbit (catalog no., NA934; 1:5,000) IgG secondary
antibodies (GE Healthcare Life Sciences) were also used.
Statistical analysis
All data were presented as the mean value ± standard
error of the mean. GraphPad Prism 5 software (GraphPad Software
Inc., La Jolla, CA, USA) was used to perform all statistical
analyses. Expression differences were calculated using an unpaired
Student's t-test. Associations were analysed assuming a Gaussian
population. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression of Spry4.1 and Spry4.2 in
breast-derived cells
In order to analyse the expression pattern of the
two reported Spry4 splice variants, RNA from 6 breast
cancer-derived cell lines and nHMECs was isolated. Using RT-PCR,
the levels of the two Spry mRNA variants were determined. As
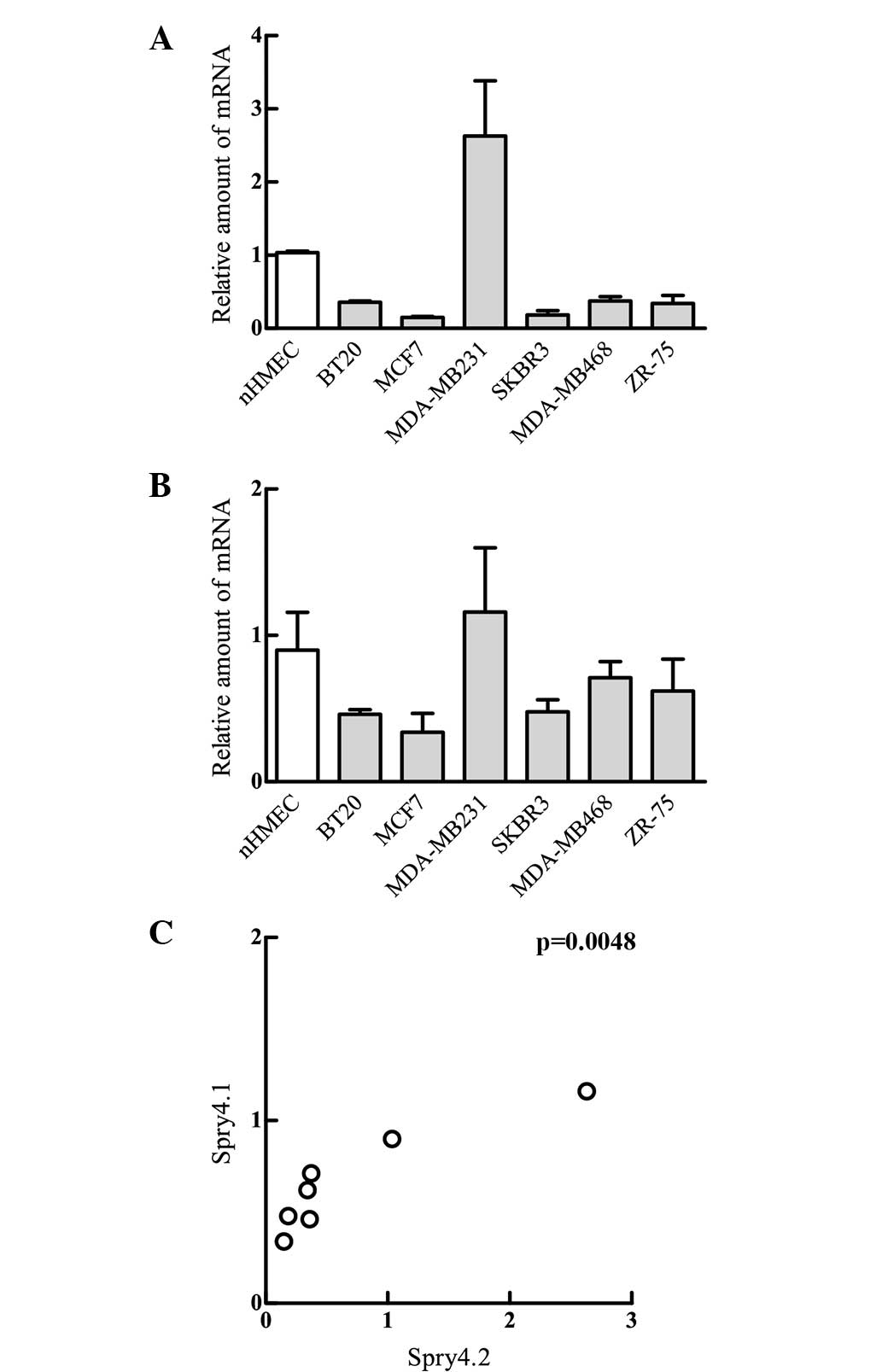
depicted in Fig. 1, both Spry4
variants are expressed in all investigated cell lines. Compared
with the levels detected in the non-malignant nHMEC cells, Spry4.1
expression in MDA-MB231 was elevated by approximately 2–3 fold. In
all other breast tumour cell lines, the levels of the Spry4.1
variant were reduced by a third (BT20, MDA-MB468, ZR-75) or even a
fifth (MCF7, SKBR3) (Fig. 1A). The
longer Spry4 variant (Spry4.2), which includes exon 2, demonstrated
similar results to the Spry4.1 variant, as the expression levels
were decreased in all tumour-derived cell lines with the exception
of MDA-MB231, although the differences were less pronounced
(Fig. 1B). Consequently, the
statistical analysis revealed that expression patterns of Spry4.1
and Spry4.2 are associated.
These data indicate that the mechanisms involved in
repression of Spry4 mRNA during malignant transformation of breast
cells may not discriminate between the two Spry4 variants.
Expression of Spry4.1 and Spry4.2 in
lung-derived cells
In addition to its involvement in breast cancer
(27), Spry4 has also been shown to
function as a tumour suppressor in lung cancer (26). Therefore, the present study
subsequently analysed the expression of each Spry4 variant in lung
cancer-derived cell lines and compared the determined levels with
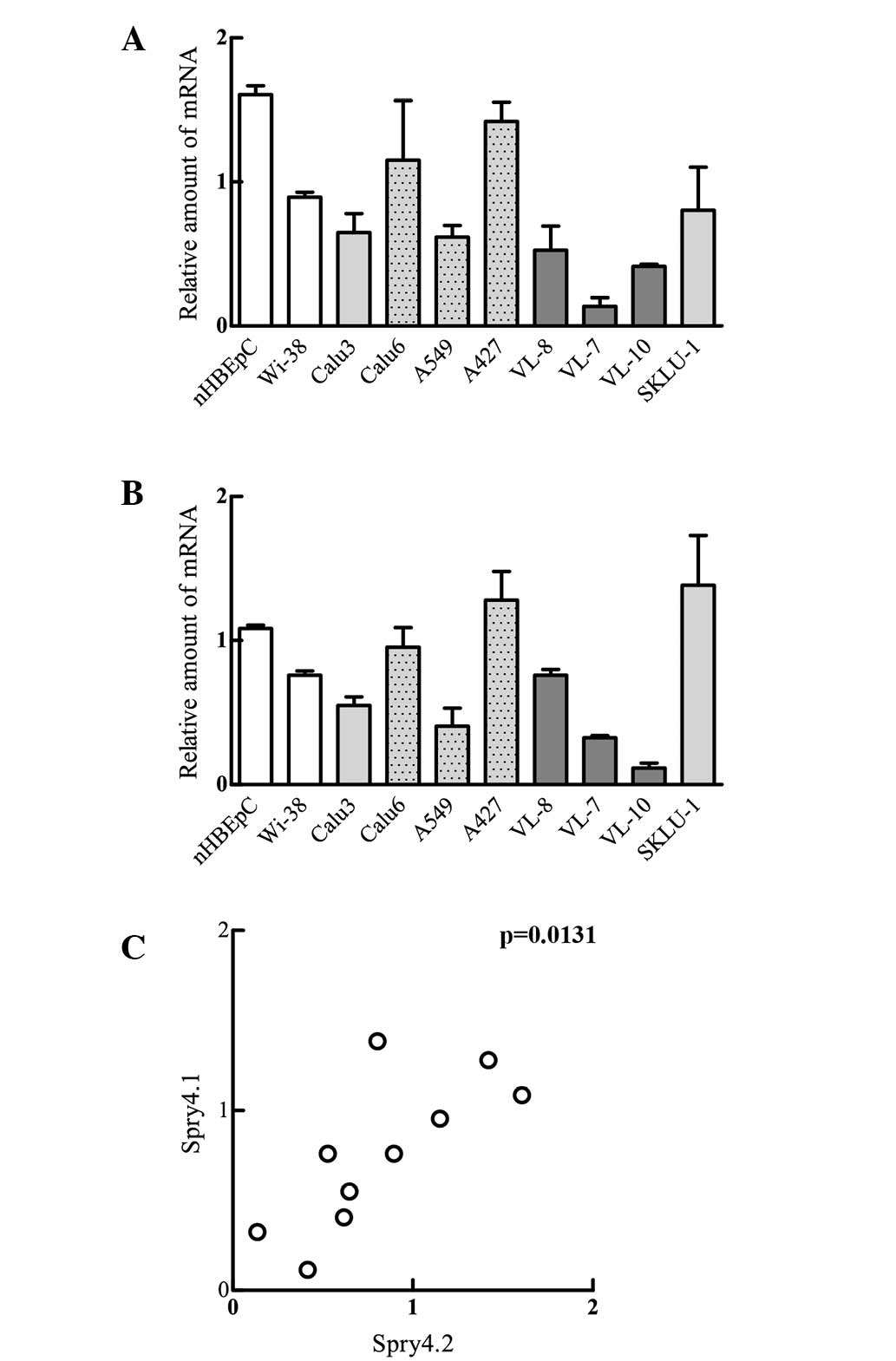
those evaluated for nHBEpCs. The nHBEpCs were observed to express
significantly more of the two Spry4 variants (Fig. 2A and B). Analysis of the Spry4.1
expression revealed that all tumour-derived cell lines irrespective
of their histological subtype and the occurrence of K-Ras mutations
show reduced Spry4 levels (Fig. 2A).
Additionally, Spry4.1 levels determined for the lung
adenocarcinoma-derived cell lines (Calu3, Calu6, A549, A427 and
SKLU-1) were significantly increased compared with the levels
measured in squamous cell carcinoma cell lines (VL-8, VL-7 and
VL-10). Analogue to the Spry4.1 variant, Spry4.2 mRNA levels in
nHBEpCs were increased compared with the levels measured in the
embryonic lung fibroblasts (WI-38) and in the majority of the
tumour-derived cell lines. Only A427 and SKLU-1 had levels
comparable to the normal counterpart (Fig. 2B). Hence the expression levels of the
Spry4 splicing variants are also significantly associated in
lung-derived cells (Fig. 2C).
Therefore, the two Spry4 variants are hypothesized to be
co-regulated.
Comparison of Spry4 mRNAs with protein
levels in breast-derived cells
The second step of gene expression is the conversion
of the mRNA to the protein. Due to numerous regulative processes,
the patterns measured at the RNA level are not always reflected at
the protein stage. In the case of Spry4 expression in
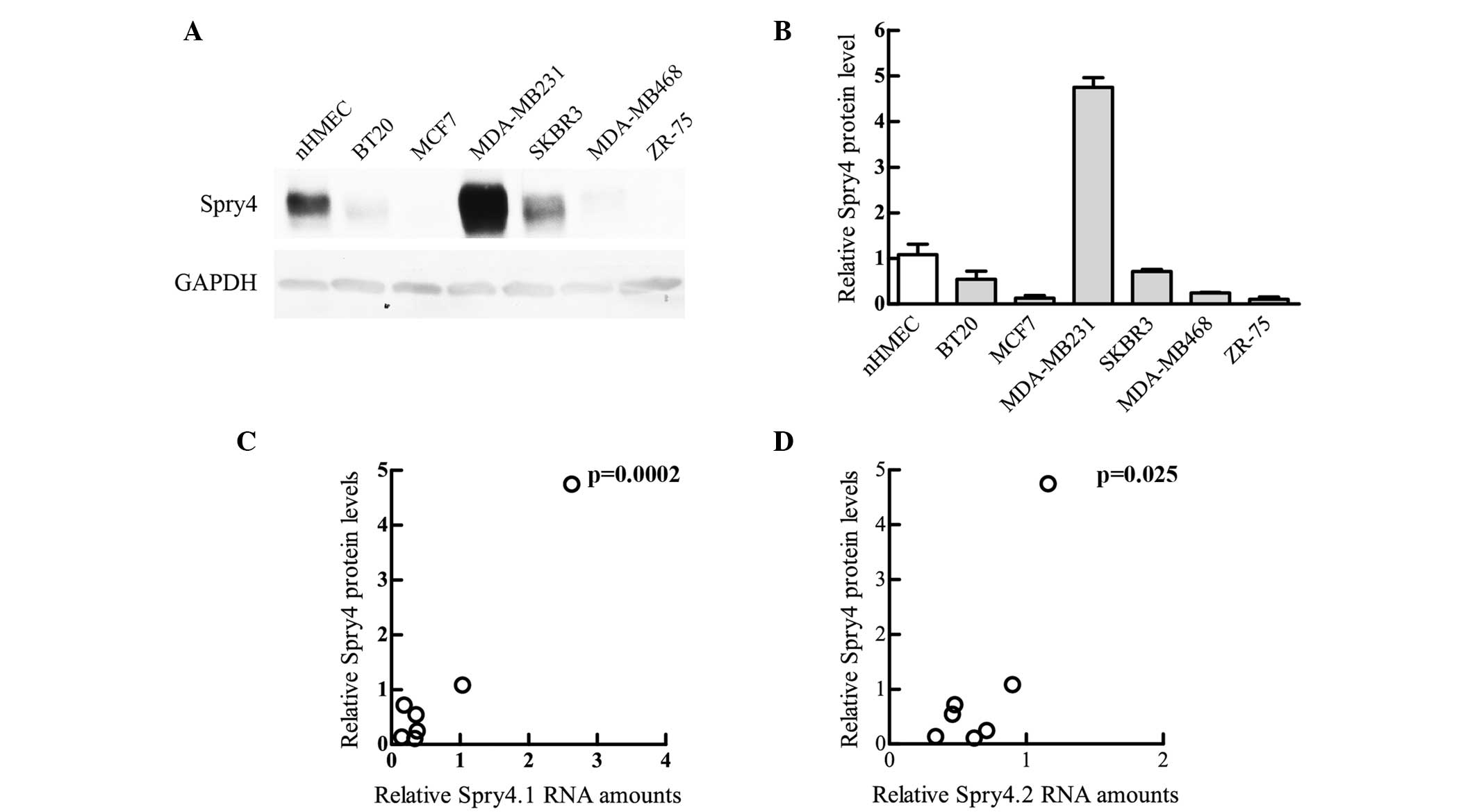
breast-derived cells, the protein levels determined by
immunoblotting were similar to those determined in RT-PCR. In
nHMECs, Spry4 was found to be more abundant compared with all other
breast cancer-derived cell lines, with the exception of MDA-MB231
(Fig. 3A and B). Although the protein
levels were associated with the expression of the two Spry4
variants, the differences were more similar to the amounts of
Spry4.1 measured (Fig. 3C and D).
This comparison suggests that the mechanisms regulating Spry4 gene
expression subsequent to mRNA production are not involved in the
malignant transformation process of breast cells.
Comparison of Spry4 mRNAs with protein
levels in lung-derived cells
Determination of Spry4 protein levels in
lung-derived cells revealed that only the squamous cell
carcinoma-derived cell lines have clearly decreased Spry4 protein
levels compared with nHBEpCs (Fig. 4A and
B). Two of the cell lines (Calu6 and A427) expressed
definitively higher levels of Spry4 compared with nHBEpCs (Fig. 4A). The protein expression levels of
the other adenocarcinoma cells were comparable to those measured in
nHBEpC cells (Fig. 4A and B). This is
in clear contrast to the observations made at mRNA level (Fig. 2). Accordingly, the expression pattern
of Spry4 protein, Spry4.1 (Fig. 4C)
and Spry4.2 (Fig. 4D) are not
associated. Notably, the two cell lines (Calu6 and A427) showing
clear elevated Spry4 protein expression are known to harbour an
activating K-Ras mutation. Although the Spry4 protein levels in the
third cell line with a mutated K-Ras, A549, are only comparable to
the levels measured in normal cells, the protein expression is
relatively high considering that the mRNA levels are decreased by
half compared with that of the normal cells (compare Fig. 2A and B with Fig. 4B). If only the cells with an unaltered
K-Ras are analysed, the protein levels are least significantly
associated with the Spry4.2 variant. These results indicate that a
constitutive signal transduction via K-Ras may affect the
mechanisms involved in modulating Spry4 protein levels.
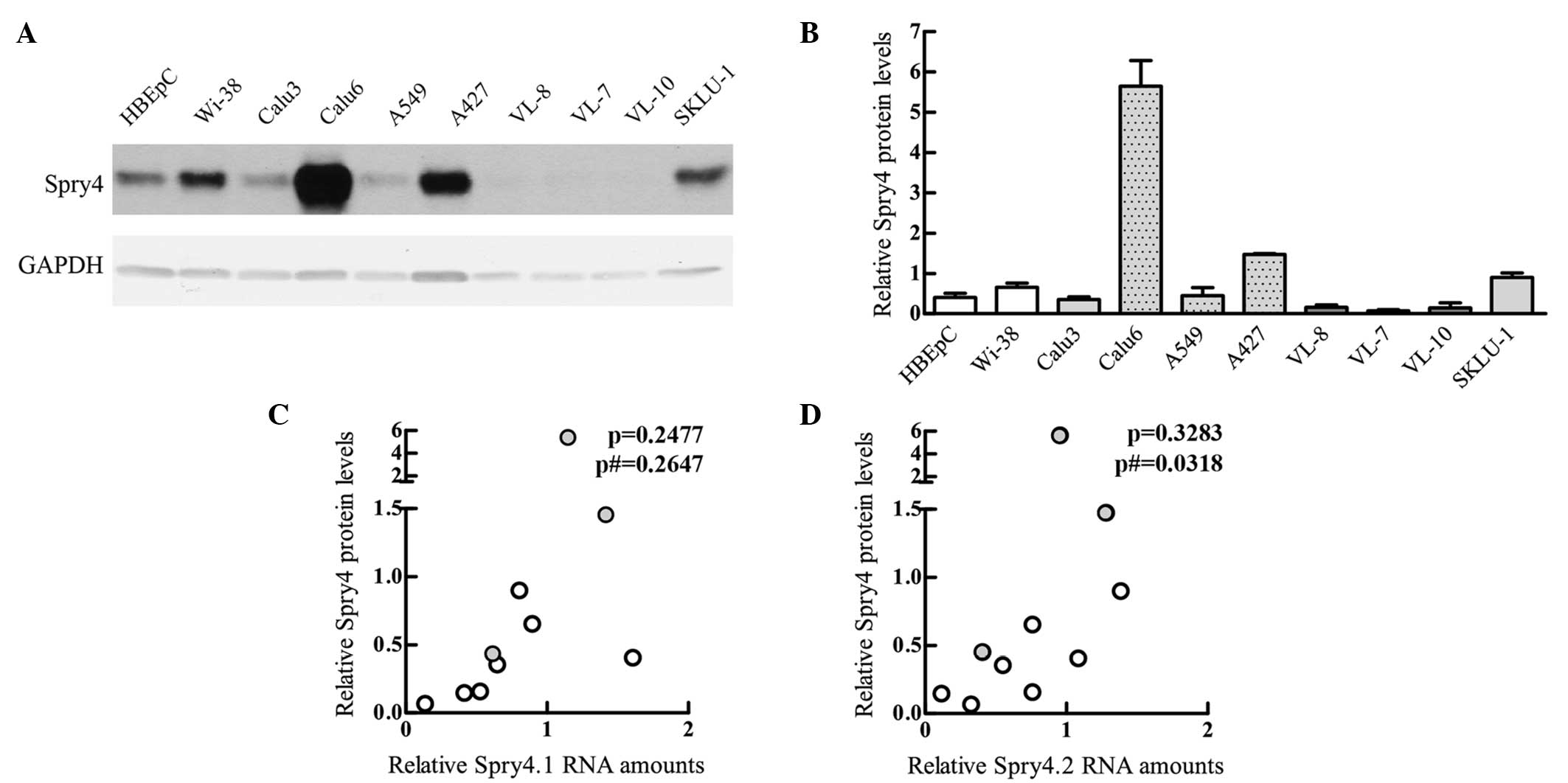 | Figure 4.Levels of Spry4 protein in lung cells
were compared with the respective Spry4.1 and Spry4.2 mRNA
variants. (A) A representative immunoblot measuring Spry4 protein
expression in various lung and lung cancer-derived cells in the
logarithmic stage of growth is depicted. GAPDH was used as loading
control. (B) Using Image Quant 5.0 software, Spry4 and GAPDH
protein signals were quantified. The relative Spry4 protein levels
were calculated using the GAPDH signals as a standardised protein
loading and HT-1080 as an external standard set as 1. Mean values ±
standard error of the mean are depicted. nHBEpCs (white bars) were
compared with cell lines established from adenocarcinoma with
wild-type K-Ras (light grey), mutated K-Ras (light grey with a
dotted pattern) and from squamous cell carcinoma (dark grey). (C)
Mean protein levels were compared with Spry4.1 mRNA levels or (D)
Spry4.2 variant expression. The cell lines harbouring mutated K-Ras
are indicated in grey. Using GraphPad Prism 5 software, the
association was calculated. In addition to the P-value summarizing
all lung cells, the P-value excluding the K-Ras mutated cell lines
is indicated. Spry, sprouty; mRNA, messenger RNA; nHBEpC, normal
human bronchial epithelial cell; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase; p, P-value for all lung cells; p#, P-value excluding
K-Ras mutated cell lines.. |
Discussion
During cancer development, signalling transduction
cascades are often targeted by various alterations. Spry proteins
modulate the intensity and duration of the transmitted signals
within such cascades, and therefore are important for the
tumorigenesis of various tissues. Spry4 has been previously shown
to fulfil a tumour suppressive function in lung (26) and breast (27) cancer.
The present study investigated the expression levels
of the two known Spry4 mRNA splice variants in cells that were
cultured from the normal and malignant tissues of the lungs and
breast. All of the cell lines used in the study expressed both
forms. In accordance with a previous study (30), which investigated the Spry4 mRNA forms
in various tissues, the shorter Spry4.1 mRNA was the more abundant
variant in the present study. With regards to normal cells,
expression of the two Spry4 forms was higher in lung-derived cells
compared with the breast-derived cells. Although in a previous
study the breast tissue was not included, Spry4 expression in the
lung tissue was indicated to be increased compared with the
majority of other organ tissues (30).
Furthermore, the present study compared the
expression of Spry4 variants in nHMECs with the expression in
breast cancer-derived cell lines and found that, with one
exception, the expression of both variants was decreased in the
malignant cells. In agreement with an earlier study (27), the Spry4 levels were found to be
differentially expressed depending on the subtype. The luminal
subtypes of breast cancer demonstrated a decreased expression of
the two Spry4 variants compared with the basal subtype.
Additionally, the present observations revealed that Spry4 mRNA
levels are repressed in malignant lung cells in comparison to their
normal counterparts, which reinforces the data from Tennis et
al (26). Supplementary to the
observations regarding breast and lung malignancies, repressed
Spry4 mRNA levels have already been found in the liver (34) and in approximately half of the
prostate cancers investigated (35).
In a previous study that investigated the Spry4 mRNA
variants, qRT-PCR of 20 types of normal tissues showed that the
levels of the two Spry4 mRNA variants were associated (29). Accordingly, in the lung and breast
cells used in the present study, a statistically significant
association between the short Spry4.1 and the slightly longer
Spry4.2 version could be observed. These data indicate that the
repression of Spry4 levels in malignant cells is not mainly
achieved by alterations to the components that regulate
splicing.
The present study also compared the mRNA and protein
levels in the investigated cell lines. In breast-derived cells,
mRNA and protein expression were associated. Although, to the best
of our knowledge, the present study is the first to report that
Spry4 mRNA and protein levels are associated across various cell
lines that originate from one tissue, a concomitant increase of
Spry4 mRNA production and protein expression in response to serum
induction of malignant (36) and
normal cells (37) has already been
reported. Additionally, elevated Spry4 expression following the
application of amphiregulin (38) and
hypoxia (32) has been observed at
the mRNA and protein levels.
The immunoblot analysis confirms that in breast
tissues, the Spry4 protein is repressed in the cancer cells
compared with their normal cell counterparts. In addition, previous
results generated by immunohistochemistry have shown that, in the
endometrium, Spry4 is decreased in the malignant tissue compared
with normal tissues (39).
In contrast to the significant association of RNA
and protein expression in breast cells, the mRNA levels in
lung-derived cells do not determine the amount of Spry4 protein,
indicating that posttranscriptional mechanisms have an important
affect on Spry4 expression in lung tissue. Earlier observations
support that Spry4 expression is also controlled at a protein
level, since the amplitude of Spry4 protein increase in response to
serum was more intense compared with at mRNA level, and epidermal
growth factor-mediated signalling promoted Spry4 protein expression
without affecting its mRNA levels (36). Particularly in cell lines harbouring a
mutated version of the K-Ras gene, the Spry4 protein levels were
notably higher than expected from the detected mRNA levels. This
potentially suggests that one of the components involved in the
regulation of the Spry4 at the protein level is controlled by
K-Ras-connected pathways.
In conclusion, the presented study shows that, in
breast and lung tissues, the expression of Spry4 splice variants is
co-regulated, and is repressed in malignant cells. At protein
level, the expression of Spry4 may be additionally upregulated by a
K-Ras dependent mechanism.
Acknowledgements
The present study was supported by the
Herzfelder'sche Family Foundation, Vienna, Austria (grant no.
2015).
Glossary
Abbreviations
Abbreviations:
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
|
Spry
|
sprouty
|
|
nHMECs
|
normal human mammary epithelial
cells
|
|
nHBEpCs
|
normal human bronchial epithelial
cells
|
References
|
1
|
Hahn WC and Weinberg RA: Rules for making
human tumor cells. N Engl J Med. 347:1593–1603. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hommes DW, Peppelenbosch MP and van
Deventer SJ: Mitogen activated protein (MAP) kinase signal
transduction pathways and novel anti-inflammatory targets. Gut.
52:144–151. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Freeman M: Feedback control of
intercellular signalling in development. Nature. 408:313–319. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Casci T, Vinós J and Freeman M: Sprouty,
an intracellular inhibitor of Ras signaling. Cell. 96:655–665.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hacohen N, Kramer S, Sutherland D, Hiromi
Y and Krasnow MA: sprouty encodes a novel antagonist of FGF
signaling that patterns apical branching of the Drosophila airways.
Cell. 92:253–263. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guy GR, Jackson RA, Yusoff P and Chow SY:
Sprouty proteins: Modified modulators, matchmakers or missing
links? J Endocrinol. 203:191–202. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Basson MA, Akbulut S, Watson-Johnson J,
Simon R, Carroll TJ, Shakya R, Gross I, Martin GR, Lufkin T,
McMahon AP, et al: Sprouty1 is a critical regulator of
GDNF/RET-mediated kidney induction. Dev Cell. 8:229–239. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shim K, Minowada G, Coling DE and Martin
GR: Sprouty2, a mouse deafness gene, regulates cell fate decisions
in the auditory sensory epithelium by antagonizing FGF signaling.
Dev Cell. 8:553–564. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taketomi T, Yoshiga D, Taniguchi K,
Kobayashi T, Nonami A, Kato R, Sasaki M, Sasaki A, Ishibashi H,
Moriyama M, et al: Loss of mammalian Sprouty2 leads to enteric
neuronal hyperplasia and esophageal achalasia. Nat Neurosci.
8:855–857. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Taniguchi K, Ayada T, Ichiyama K, Kohno R,
Yonemitsu Y, Minami Y, Kikuchi A, Maehara Y and Yoshimura A:
Sprouty2 and Sprouty4 are essential for embryonic morphogenesis and
regulation of FGF signaling. Biochem Biophys Res Commun.
352:896–902. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang S, Lin Y, Itäranta P, Yagi A and
Vainio S: Expression of Sprouty genes 1, 2 and 4 during mouse
organogenesis. Mech Dev. 109:367–370. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Masoumi-Moghaddam S, Amini A and Morris
DL: The developing story of Sprouty and cancer. Cancer Metastasis
Rev. 33:695–720. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barbáchano A, Ordóñez-Morán P, Garcia JM,
Sánchez A, Pereira F, Larriba MJ, Martínez N, Hernández J, Landolfi
S, Bonilla F, et al: SPROUTY-2 and E-cadherin regulate reciprocally
and dictate colon cancer cell tumourigenicity. Oncogene.
29:4800–4813. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Holgren C, Dougherty U, Edwin F, Cerasi D,
Taylor I, Fichera A, Joseph L, Bissonnette M and Khare S: Sprouty-2
controls c-Met expression and metastatic potential of colon cancer
cells: Sprouty/c-Met upregulation in human colonic adenocarcinomas.
Oncogene. 29:5241–5253. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schaaf G, Hamdi M, Zwijnenburg D, Lakeman
A, Geerts D, Versteeg R and Kool M: Silencing of SPRY1 triggers
complete regression of rhabdomyosarcoma tumors carrying a mutated
RAS gene. Cancer Res. 70:762–771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sutterluty H, Mayer CE, Setinek U, Attems
J, Ovtcharov S, Mikula M, Mikulits W, Micksche M and Berger W:
Down-regulation of Sprouty2 in non-small cell lung cancer
contributes to tumor malignancy via extracellular signal-regulated
kinase pathway-dependent and -independent mechanisms. Mol Cancer
Res. 5:509–520. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McKie AB, Douglas DA, Olijslagers S,
Graham J, Omar MM, Heer R, Gnanapragasam VJ, Robson CN and Leung
HY: Epigenetic inactivation of the human sprouty2 (hSPRY2)
homologue in prostate cancer. Oncogene. 24:2166–2174. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fritzsche S, Kenzelmann M, Hoffmann MJ,
Müller M, Engers R, Gröne HJ and Schulz WA: Concomitant
down-regulation of SPRY1 and SPRY2 in prostate carcinoma. Endocr
Relat Cancer. 13:839–849. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lo TL, Yusoff P, Fong CW, Guo K, McCaw BJ,
Phillips WA, Yang H, Wong ES, Leong HF, Zeng Q, et al: The
ras/mitogen-activated protein kinase pathway inhibitor and likely
tumor suppressor proteins, sprouty 1 and sprouty 2 are deregulated
in breast cancer. Cancer Res. 64:6127–6136. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fong CW, Chua MS, McKie AB, Ling SH, Mason
V, Li R, Yusoff P, Lo TL, Leung HY, So SK and Guy GR: Sprouty 2, an
inhibitor of mitogen-activated protein kinase signaling, is
down-regulated in hepatocellular carcinoma. Cancer Res.
66:2048–2058. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Masoumi-Moghaddam S, Amini A, Wei AQ,
Robertson G and Morris DL: Sprouty 2 protein, but not Sprouty 4, is
an independent prognostic biomarker for human epithelial ovarian
cancer. Int J Cancer. 137:560–570. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kwabi-Addo B, Wang J, Erdem H, Vaid A,
Castro P, Ayala G and Ittmann M: The expression of Sprouty1, an
inhibitor of fibroblast growth factor signal transduction, is
decreased in human prostate cancer. Cancer Res. 64:4728–4735. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Masoumi-Moghaddam S, Amini A, Wei AQ,
Robertson G and Morris DL: Sprouty 1 predicts prognosis in human
epithelial ovarian cancer. Am J Cancer Res. 5:1531–1541.
2015.PubMed/NCBI
|
|
24
|
Rathmanner N, Haigl B, Vanas V, Doriguzzi
A, Gsur A and Sutterlüty-Fall H: Sprouty2 but not Sprouty4 is a
potent inhibitor of cell proliferation and migration of
osteosarcoma cells. FEBS Lett. 587:2597–2605. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jäggi F, Cabrita MA, Perl AK and
Christofori G: Modulation of endocrine pancreas development but not
beta-cell carcinogenesis by Sprouty4. Mol Cancer Res. 6:468–482.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tennis MA, Van Scoyk MM, Freeman SV,
Vandervest KM, Nemenoff RA and Winn RA: Sprouty-4 inhibits
transformed cell growth, migration and invasion, and
epithelial-mesenchymal transition, and is regulated by Wnt7A
through PPARgamma in non-small cell lung cancer. Mol Cancer Res.
8:833–843. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vanas V, Mühlbacher E, Kral R and
Sutterlüty-Fall H: Sprouty4 interferes with cell proliferation and
migration of breast cancer-derived cell lines. Tumour Biol.
35:4447–4456. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Faratian D, Sims AH, Mullen P, Kay C, Um
I, Langdon SP and Harrison DJ: Sprouty 2 is an independent
prognostic factor in breast cancer and may be useful in stratifying
patients for trastuzumab therapy. PLoS One. 6:e237722011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ding W, Bellusci S, Shi W and Warburton D:
Genomic structure and promoter characterization of the human
Sprouty4 gene, a novel regulator of lung morphogenesis. Am J
Physiol Lung Cell Mol Physiol. 287:52–59. 2004. View Article : Google Scholar
|
|
30
|
Khaitan D, Dinger ME, Mazar J, Crawford J,
Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long
noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer
Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Berger W, Elbling L, Hauptmann E and
Micksche M: Expression of the multidrug resistance-associated
protein (MRP) and chemoresistance of human non-small-cell lung
cancer cells. Int J Cancer. 73:84–93. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Haigl B, Mayer CE, Siegwart G and
Sutterlüty H: Sprouty4 levels are increased under hypoxic
conditions by enhanced mRNA stability and transcription. Biol Chem.
391:813–821. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kral RM, Mayer CE, Vanas V, Gsur A and
Sutterlüty-Fall H: In non-small cell lung cancer mitogenic
signaling leaves Sprouty1 protein levels unaffected. Cell Biochem
Funct. 32:96–100. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sirivatanauksorn Y, Sirivatanauksorn V,
Srisawat C, Khongmanee A and Tongkham C: Differential expression of
sprouty genes in hepatocellular carcinoma. J Surg Oncol.
105:273–276. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang J, Thompson B, Ren C, Ittmann M and
Kwabi-Addo B: Sprouty4, a suppressor of tumor cell motility, is
down regulated by DNA methylation in human prostate cancer.
Prostate. 66:613–624. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Doriguzzi A, Haigl B, Gsur A and
Sutterlüty-Fall H: The increased Sprouty4 expression in response to
serum is transcriptionally controlled by Specific protein 1. Int J
Biochem Cell Biol. 64:220–228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mayer CE, Haigl B, Jantscher F, Siegwart
G, Grusch M, Berger W and Sutterlüty H: Bimodal expression of
Sprouty2 during the cell cycle is mediated by phase-specific
Ras/MAPK and c-Cbl activities. Cell Mol Life Sci. 67:3299–3311.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
So WK, Cheng JC, Liu Y, Xu C, Zhao J,
Chang VT and Leung PC: Sprouty4 mediates amphiregulin-induced
down-regulation of E-cadherin and cell invasion in human ovarian
cancer cells. Tumour Biol. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang H, Guo Q, Wang X, Wang C, Zhao X and
Li M: Aberrant expression of hSef and Sprouty4 in endometrial
adenocarcinoma. Oncol Lett. 11:45–50. 2016.PubMed/NCBI
|