Introduction
The osteogenic differentiation process of
mesenchymal stem cells involves either systemic hormones or
specific local molecules, including transforming growth factor-β
1/2 (TGF-β), fibroblast growth factor-2 (FGF-2), bone morphogenic
proteins (BMPs), insulin-like growth factor (IGF), prostaglandins,
vascular endothelial growth factors (VEGFs) and the Wnt/β-catenin
pathway (1). As a result,
intracellular signalling promotes the expression of transcription
factors. Among these, runt-related transcription factor 2 (Runx2)
serves a pivotal role and it is considered a master gene for
osteogenic differentiation (1). Runx2
induces the expression of specific downstream genes, including
collagen type I, bone alkaline phosphatase, osteopontin and
osteocalcin (2), and it is essential
for terminal chondrocyte differentiation (3). Runx2 knock-out mice are affected by
cleidocranial dysplasia syndrome (3),
while Runx2 overexpression in mice impairs mineralization,
suggesting that this gene affects bone formation in different ways
(4). A previous study demonstrated
that the expression of Runx2 in circulating mesenchymal stem cells
was lower in patients with osteoporosis when compared with normal
donors (5). Runx2 expression is
modulated by several regulatory pathways. Important negative
regulators include histone deacetylases (HDACs), in particular
HDAC3, HDAC4, HDAC5, HDAC6 and HDAC7 (6). Twist proteins (7), activator protein 1, transcription factor
4 and osterix are additional regulators of Runx2 expression
(2). Furthermore, it has been
demonstrated that Runx2 function may be downmodulated by microRNA
(miR) action, in particular miR-3960 (8), and phosphorylation induced by the
extracellular signal-regulated kinase/mitogen-activated protein
kinase pathway results in Runx2 activation (9). The involvement of Runx2 in the oncogenic
process has been recently suggested to occur in human osteosarcoma
(10), in addition to other forms of
malignancy such as pancreatic and thyroid cancer, and increased
expression correlates with a poor prognosis (11,12).
Epithelial-mesenchymal transition (EMT) is involved
in carcinogenesis and promotes metastatic spreading (13–15).
Following its recognition as a regulator gene in transformed
epithelial cells in breast, lung and thyroid carcinoma (13–15), it
has been suggested that Runx2 may promote breast cancer metastasis
by EMT (13). The cancer caused by
EMT is a consequence of complicated reprogramming process involving
differentiation, epigenetics and metabolic balance disruption
(16). In this scenario, Runx2 has
been identified as a regulator gene of transformed epithelial cells
in breast, lung and thyroid carcinoma (13–15), and
it has been suggested that this gene promotes breast cancer
metastasis via EMT (13).
A number of researchers have focused on identifying
cancer markers that may provide clinical information a less
invasive way. A previous study reported that Runx2 expression was
elevated in the tissue, serum and circulating cells of patients
with thyroid cancer suggesting that Runx2 may serve as a useful
biomarker for thyroid malignancies (17).
On the basis of these findings, the present study
speculated that Runx2 may be expressed in cells derived from
malignancies other than bone tumours. Therefore, the expression of
this gene was analysed in pancreatic, melanoma, breast and prostate
cancer cell lines. In addition, in order to evaluate potential
applications in oncological malignancies, Runx2 cell-free RNA was
examined in sera obtained from patients affected by various forms
of cancer.
Materials and methods
Cell culture
A total of 4 pancreatic, 2 breast, 3 prostate and 2
bone human cancer cell lines, purchased by American Type Culture
Collection (Rockville, MD, USA), were used in the present study
(Table I). Table I specified the previous applications
of these cell lines studies (18–28). The
pancreatic cancer cell lines were cultured in RPMI 1640
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) with 10%
foetal bovine serum (FBS) (Sigma-Aldrich; Merck Millipore), whereas
the breast, prostate and bone cell lines were cultured as
previously described (29–33). For cell synchronization, cell cycles
were arrested at G1 phase by adding 400 mM mimosine
(Sigma-Aldrich; Merck Millipore) for 24 h as previously described
(34). Cells subsequently underwent
three washes with PBS (Sigma-Aldrich; Merck Millipore) and were
cultured in serum-free RPMI 1640 medium for 3 days. Finally, cells
were cultured in fresh RPMI 1640 medium with 10% FBS (plus 2 mM
L-glutamine and penicillin/streptomycin) until they reached 70%
confluence. Adherent cells and supernatants for each cell line were
harvested to perform expression analyses. For each cell line, three
different cultures were tested.
 | Table I.Cancer cell lines. |
Table I.
Cancer cell lines.
| Author, year | Cell line | Source | Tumour | Refs. |
|---|
| Morgan et
al, 1980 | Colo357 | Metastatic | Pancreatic | (18) |
| Kim et al,
1989 | HPAF | Metastatic | Pancreatic | (19) |
| Lieber et
al, 1975 | Panc1 | Primary | Pancreatic | (20) |
| Parekh et
al, 1994 | BON | Metastatic | Pancreatic | (21) |
| Soule et al,
1973 | T47D | Metastatic | Breast | (22) |
| Keydar et
al, 1979 | MCF7 | Metastatic | Breast | (23) |
| Stone et al,
1978 | DU145 | Metastatic | Prostatic | (24) |
| Tai et al,
2011 | PC3 | Primary | Prostatic | (25) |
| Horoszewicz et
al, 1983 | LNCaP | Metastatic | Prostatic | (26) |
| Niforou et
al, 2008 | U2OS | Primary | Osteosarcoma | (27) |
| Billiau et
al, 1977 | MG63 | Primary | Osteosarcoma | (28) |
Patients
Characteristics of the population analysed are
presented in Table II. A total of 41
patients with cancer were positively diagnosed from 2010 to 2013 by
pathologists (Pancreas Institute; Integrated University Hospital of
Verona, Verona, Italy) prior to providing blood samples, and 41
age-matched donors, who were hospitalized in Clinic of Internal
Medicine, Integrated University Hospital of Verona for
cardiovascular or metabolic diseases, were recruited as controls.
Bone metastases were present in 17 patients. All subjects had
provided written informed consent and the study was approved by the
local Institutional Ethics Committee of the Integrated University
Hospital of Verona.
 | Table II.Characteristics of the study
population. |
Table II.
Characteristics of the study
population.
| N | Gender | Age, years | Diagnosis | TNM |
|---|
| 1 | M | 65 | Neuroendocrine
adenocarcinoma | TxN1M1 |
| 2 | M | 80 | Intestinal
adenocarcinoma | T1-2N0M0 |
| 3 | M | 71 |
Hepatocarcinoma | T3N1M0 |
| 4 | M | 55 | Prostatic
adenocarcinoma |
T2N0M0 |
| 5 | M | 83 | Prostatic
adenocarcinoma | T3N0M1 |
| 6 | M | 87 | Prostatic
adenocarcinoma | T4N1M0 |
| 7 | M | 66 | Prostatic
adenocarcinoma | T2N0M0 |
| 8 | M | 67 | Kidney
adenocarcinoma | T4N0M0 |
| 9 | M | 73 | Intestinal
adenocarcinoma | TxN0M0 |
| 10 | M | 94 | Gastric
adenocarcinoma | T3N0M0 |
| 11 | M | 81 | Gastric
adenocarcinoma | T3N2M0 |
| 12 | M | 70 | Lung carcinoma | T1N1M1 |
| 13 | M | 70 | Mesenchymal
cancer | T4NxM1 |
| 14 | M | 81 | Prostatic
adenocarcinoma | T1N1M1 |
| 15 | M | 92 | Breast
carcinoma | T2N1M1 |
| 16 | M | 60 | Intestinal
adenocarcinoma | T1-2N0M0 |
| 17 | M | 75 | Pancreatic
adenocarcinoma | T3N1M1 |
| 18 | M | 60 | Pancreatic
adenocarcinoma | T3N1M1 |
| 19 | M | 64 | Pancreatic
adenocarcinoma | T3N0M0 |
| 20 | M | 67 | Pancreatic
adenocarcinoma | T1N0M0 |
| 21 | M | 78 | Bladder
carcinoma | T1N0M0 |
| 22 | F | 87 |
Hepatocarcinoma | T3N0M0 |
| 23 | F | 72 | Intestinal
adenocarcinoma | T1N0M0 |
| 24 | F | 27 | Adrenal
carcinoma | T3-4N1M1 |
| 25 | F | 82 | Intestinal
adenocarcinoma | TxN0M0 |
| 26 | F | 82 | Lung carcinoma | T2N0M0 |
| 27 | F | 52 | Esophageal
carcinoma | T4N1M1 |
| 28 | F | 68 | Ovarian
carcinoma | T3N2M1 |
| 29 | F | 80 | Breast
carcinoma | T2N1M1 |
| 30 | F | 86 | Breast
carcinoma | T0N1M1 |
| 31 | F | 75 | Lung carcinoma | T1NxM0 |
| 32 | F | 81 | Pancreatic
adenocarcinoma | T3N0M0 |
| 33 | F | 71 | Bladder
carcinoma | T3aN1M0 |
| 34 | F | 62 | Pancreatic
adenocarcinoma | T3N1M1 |
| 35 | F | 71 | Pancreatic
adenocarcinoma | T4N0M0 |
| 36 | F | 75 | Pancreatic
adenocarcinoma | T3N0M0 |
| 37 | F | 70 | Pancreatic
adenocarcinoma | T1N0M0 |
| 38 | F | 49 | Pancreatic
adenocarcinoma | T1N0M0 |
| 39 | M | 78 | Prostatic
adenocarcinoma | T2N0M1 |
| 40 | M | 80 | Prostatic
adenocarcinoma | T2N0M1 |
| 41 | M | 75 | Prostatic
adenocarcinoma | T3N1M1 |
Serum preparation
Serum samples were obtained following three rounds
of centrifugation (800 × g, 1,000 × g and 1,500 ×
g at 4°C) of collected blood to keep lymphocyte
contamination to a minimum as previously described (35).
RNA extraction and reverse
transcription
RNA from cancer cell lines was extracted using the
RNeasy® Mini kit (Qiagen, Hilden, Germany), and RNA
extraction from sera and culture supernatants was performed using
the QIAamp® UltraSens® Virus kit (Qiagen)
with DNAse I treatment according to the manufacturer's protocol.
First-strand cDNA was generated using the High-Capacity cDNA
Archive kit with random hexamers (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol. cDNA products were stored at −80°C until use.
Quantitative polymerase chain reaction
(qPCR)
PCR was performed in a total volume of 50 µl
containing 1X Taqman Universal PCR Master mix, No
AmpErase® UNG and 5 µl cDNA. The real time
amplifications included 10 min at 95°C, followed by 40 cycles at
95°C for 15 sec and at 60°C for 1 min. Predesigned, gene-specific
primers and a probe set for Runx2 were obtained from
Assay-on-Demand™ Gene Expression products (Applied Biosystems;
Thermo Fisher Scientific, Inc.). In order to normalize the results,
the following three housekeeping genes were used: β-actin
(structural gene), glyceraldehyde 3-phosphate dehydrogenase (GAPDH;
metabolism-related gene) and β-2 microglobulin (component of major
histocompatibility complex class I gene). The primer sequences were
pre-designed by the supplier (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The relative expression levels of the Runx2 gene
were calculated for each sample following normalization using the
2-∆∆Ct method for comparing differences in relative fold
expression (36). The data are
reported as mRNA fold expression.
Western blot analysis
Cells were lysed on ice for 45 min in a buffer
containing protease inhibitor cocktail [1% IGEPAL®, 1%
sodium dodecyl sulfate (SDS), 10% glycerol, 1 mM
ethylenediaminetetraacetic acid, 5% b-mercaptoethanol, 1.5% Triton
X-100 and 4% Protease Inhibitor Cocktail (Sigma-Aldrich; Merck
Millipore)]. Cell lysates were then centrifuged (10,000 × g)
for 15 min at 4°C to remove insoluble materials. Protein
concentration in the supernatants was measured using the Coomassie
Protein assay kit (Pierce Biotechnology, Inc., Rockford, IL, USA).
Proteins (70 µg) were separated by 10% SDS-polyacrylamide gel
electrophoresis and electrotransferred onto a polyvinylidene
fluoride membrane. The membrane was subsequently blocked for 30 min
with 3% bovine serum albumin (Sigma-Aldrich; Merck Millipore) in
0.05% Tween 20 with Tris-buffered saline (t-TBS) at room
temperature. For immunodetection, blots were incubated for 2 h at
room temperature on titer plate agitator with anti-Runx2 antibodies
(cat no. 05-1478; dilution 1:500; clone AS110; EMD Millipore,
Billerica, MA, USA). The membranes were washed three times in
t-TBS, incubated at room temperature with horseradish
peroxidase-conjugated anti-rabbit secondary antibodies (dilution,
1:2,500) in TBS for 1 h and washed in fresh t-TBS three times for a
total of 20 min. Bands were detected using Luminata™ Forte Western
HRP Substrate (Merck Millipore) and a G:BOX Chemi XX6 (Syngene,
Frederick, MD, USA).
Statistical analysis
Results are expressed as the mean ± standard error.
The Wilcoxon signed-ranked test was used for non-parametric data.
Analysis of variance followed by Bonferroni correction was
performed as a post-hoc analysis and the results are
expressed as the mean ± standard error of the mean. P<0.05 was
considered to indicate a statistically significant difference.
Analyses were applied to experiments carried out at least three
times, and statistical analyses were performed using SPSS v16.0
(SPSS, Inc., Chicago, IL, USA).
Results
Runx2 expression in cancer cell
lines
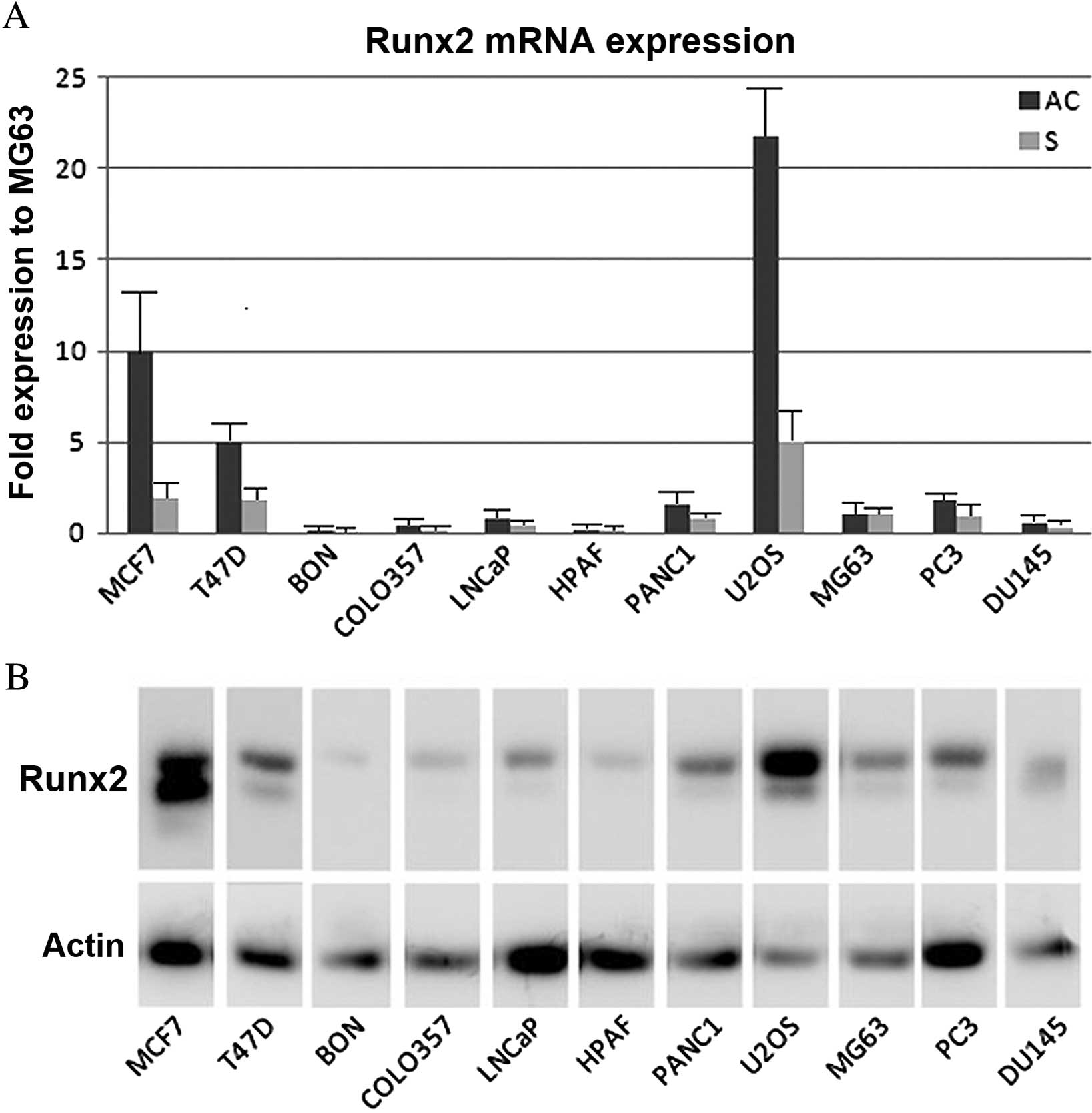
Runx2 gene expression was analysed in adherent cells
and in culture supernatants, and the MG63 cell line was used as a
calibrator (fold of expression). It was observed that Runx2 mRNA
was expressed in adherent cells and supernatants of the cancer cell
lines, although expression was largely varied across the different
cell types (Fig. 1A). In order to
analyse the expression of Runx2 protein in adherent cells,
immunoblotting using anti-Runx2 antibodies was performed. The
results demonstrated that the protein was also expressed in all
cell lines (Fig. 1B).
Runx2 gene expression in patients with
cancer
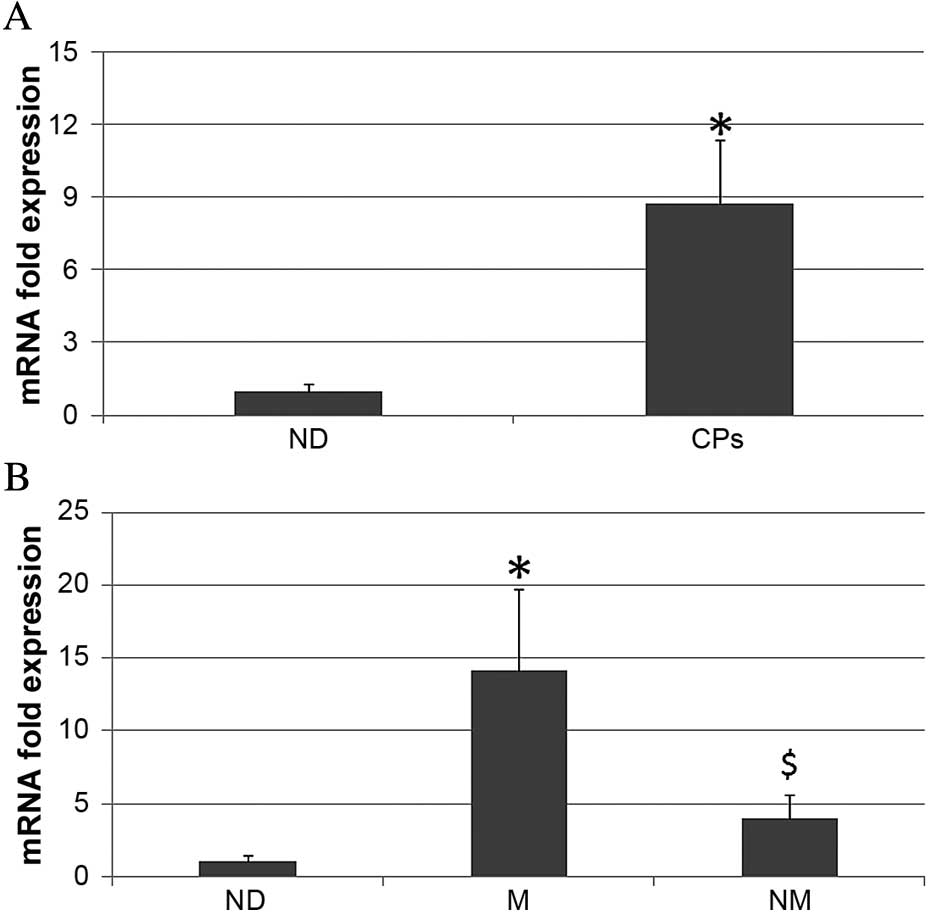
The expression data of patients with cancer was
reported as fold of expression in respect to a calibrator (40
normal donors). Patients with cancer and normal donors each
expressed Runx2 mRNA; however, their expression levels were
different. Notably, the expression of Runx2 in the patients with
cancer was 8.74 (±3.5)-fold higher than the normal donors
(P<0.01; Fig. 2A). In addition,
Runx2 mRNA expression in patients with bone metastases was higher
than in patients without metastases. Runx2 expression in patients
with metastases was 14.12 (±4.2)-fold higher than the normal donors
(P<0.01), whereas in patients without metastases Runx2
expression was 4.01 (±2.01)-fold higher than the normal donors
(P<0.05) (Fig. 2B).
Discussion
In order to establish less invasive methods for the
diagnosis and follow-up of patients with cancer, research has aimed
to identify cell-free RNA encoding for genes upregulated in cancer
malignancies (17,35,37).
Previous studies primarily focused on osteosarcoma and metastatic
breast and prostate cancer have linked Runx2 to neoplastic
transformation (38–41). The present study enrolled patients
affected by various types of tumours, including pancreatic,
prostatic, intestinal, lung, breast, gastric, liver,
neuroendocrine, kidney, mesenchymal, adrenal gland, oesophageal and
ovarian cancer. Notably, the results of the current study
demonstrated an increase in Runx2 circulating mRNA in multiple
forms of cancer, thus opening the possibility to investigate it as
a relatively comprehensive biomarker.
The Runx gene family is comprised of three related
transcription factors, which are involved in the differentiation of
multiple haematopoietic lineages (Runx1), cartilage and bone
(Runx2) and epithelial tissues (Runx3). However, all three genes
are implicated in cancer by promoting (Runx1 and Runx2) or
suppressing (Runx3) neoplastic transformation (42). Multiple mechanisms contribute to Runx2
functional modulation, including post-translational modification,
in addition to protein-protein interaction and direct stimulation
(11). Several hypotheses, such as
the involvement of integrin alpha5 (39), p53 (43)
or microRNA-205 (40) have been put
forward to describe the molecular process of Runx2 in
carcinogenesis. In osteosarcoma, loss of p53 upregulates Runx2
expression (43); this cause-effect
relationship may explain Runx2 ectopic expression in various forms
of cancer.
P53 and Runx2 have been demonstrated to be part of
the regulatory network controlling EMT (44). P53 controls miRNAs, major EMT-related
signalling pathways (TGF-β, Wnt, IGF, and signal transducer and
activator of transcription), and EMT-associated transcription
factors that promote a chemoresistant phenotype, invasion and loss
of cell polarity (44). The direct
involvement of Runx2 in cancer was demonstrated by downmodulation
experiments in thyroid carcinoma cells (15) and upregulation experiments in breast
cancer (45). EMT represents an early
event of tumour progression and is mediated by well-characterized
transcription factors (e.g. Snail and Twist family and
helix-loop-helix factors) (46). The
present study speculates that Runx2 participates in these events to
promote invasion and metastasis in a larger number of cancer forms
than previously anticipated. The data from the current study
demonstrated an increase in the concentration of circulating
cell-free Runx2 cell-free mRNA in patients with metastasis. In
agreement with these results, Runx2 has been repeatedly identified
as a regulator of bone metastases in breast and prostate cancer in
previous studies (47–49). Bone is particularly recurrent as a
target of metastasizing cells, thus a master skeletal transcription
factor like Runx2 may be extremely relevant in potentiating tumour
cell invasiveness of bone marrow, among others, contributing
directly to the osteolysis process (38). Further studies with a larger number of
patients should be performed in order to validate the predictive
value of minimally invasive tests based on Runx2 cell-free
mRNA.
In conclusion, the present study demonstrated that
Runx2 is expressed at high levels in osteosarcoma and expanded this
finding to non-osseous cells, thus supporting the possible use of
Runx2-related cell-free RNA as a cancer marker for screening
purposes. In addition, this useful, less invasive method may allow
clinicians to monitor the development of metastases in patients
with cancer.
References
|
1
|
Carbonare L Dalle, Innamorati G and
Valenti MT: Transcription factor Runx2 and its application to bone
tissue engineering. Stem Cell Rev. 8:891–897. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cohen MM Jr: Perspectives on RUNX genes:
An update. Am J Med Genet A 149A. 2629–2646. 2009. View Article : Google Scholar
|
|
3
|
Otto F, Thornell AP, Crompton T, Denzel A,
Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen
BR, et al: Cbfa1, a candidate gene for cleidocranial dysplasia
syndrome, is essential for osteoblast differentiation and bone
development. Cell. 89:765–771. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Otto F, Lübbert M and Stock M: Upstream
and downstream targets of RUNX proteins. J Cell Biochem. 89:9–18.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Valenti MT, Garbin U, Pasini A, Zanatta M,
Stranieri C, Manfro S, Zucal C and Carbonare L Dalle: Role of
ox-PAPCs in the differentiation of mesenchymal stem cells (MSCs)
and Runx2 and PPARγ2 expression in MSCs-like of osteoporotic
patients. PloS One. 6:e203632011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jensen ED, Schroeder TM, Bailey J,
Gopalakrishnan R and Westendorf JJ: Histone deacetylase 7
associates with Runx2 and represses its activity during osteoblast
maturation in a deacetylation-independent manner. J Bone Miner Res.
23:361–372. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yousfi M, Lasmoles F and Marie PJ: TWIST
inactivation reduces CBFA1/RUNX2 expression and DNA binding to the
osteocalcin promoter in osteoblasts. Biochem Biophys Res Commun.
297:641–644. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hu R, Liu W, Li H, Yang L, Chen C, Xia ZY,
Guo LJ, Xie H, Zhou HD, Wu XP and Luo XH: A Runx2/miR-3960/miR-2861
regulatory feedback loop during mouse osteoblast differentiation. J
Biol Chem. 286:12328–12339. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ge C, Xiao G, Jiang D, Yang Q, Hatch NE,
Roca H and Franceschi RT: Identification and functional
characterization of ERK/MAPK phosphorylation sites in the Runx2
transcription factor. J Biol Chem. 284:32533–32543. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lau CC, Harris CP, Lu XY, Perlaky L,
Gogineni S, Chintagumpala M, Hicks J, Johnson ME, Davino NA, Huvos
AG, et al: Frequent amplification and rearrangement of chromosomal
bands 6p12-p21 and 17p11.2 in osteosarcoma. Genes Chromosomes
Cancer. 39:11–21. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kayed H, Jiang X, Keleg S, Jesnowski R,
Giese T, Berger MR, Esposito I, Löhr M, Friess H and Kleeff J:
Regulation and functional role of the Runt-related transcription
factor-2 in pancreatic cancer. Br J Cancer. 97:1106–1115. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Endo T, Ohta K and Kobayashi T: Expression
and function of Cbfa-1/Runx2 in thyroid papillary carcinoma cells.
J Clin Endocrinol Metab. 93:2409–2412. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Owens TW, Rogers RL, Best SA, Ledger A,
Mooney AM, Ferguson A, Shore P, Swarbrick A, Ormandy CJ, Simpson
PT, et al: Runx2 is a novel regulator of mammary epithelial cell
fate in development and breast cancer. Cancer Res. 74:5277–5286.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hsu YL, Huang MS, Yang CJ, Hung JY, Wu LY
and Kuo PL: Lung tumor-associated osteoblast-derived bone
morphogenetic protein-2 increased epithelial-to-mesenchymal
transition of cancer by Runx2/Snail signaling pathway. J Biol Chem.
286:37335–37346. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Niu DF, Kondo T, Nakazawa T, Oishi N,
Kawasaki T, Mochizuki K, Yamane T and Katoh R: Transcription factor
Runx2 is a regulator of epithelial-mesenchymal transition and
invasion in thyroid carcinomas. Lab Invest. 92:1181–1190. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li L and Li W: Epithelial-mesenchymal
transition in human cancer: Comprehensive reprogramming of
metabolism, epigenetics, and differentiation. Pharmacol Ther.
150:33–46. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Carbonare L Dalle, Frigo A, Francia G,
Davì MV, Donatelli L, Stranieri C, Brazzarola P, Zatelli MC,
Menestrina F and Valenti MT: Runx2 mRNA expression in the tissue,
serum, and circulating non-hematopoietic cells of patients with
thyroid cancer. J Clin Endocrinol Metab. 97:E1249–E1256. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Morgan RT, Woods LK, Moore GE, Quinn LA,
McGavran L and Gordon SG: Human cell line (COLO 357) of metastatic
pancreatic adenocarcinoma. Int J Cancer. 25:591–598. 1980.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim YW, Kern HF, Mullins TD, Koriwchak MJ
and Metzgar RS: Characterization of clones of a human pancreatic
adenocarcinoma cell line representing different stages of
differentiation. Pancreas. 4:353–362. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lieber M, Mazzetta J, Nelson-Rees W,
Kaplan M and Todaro G: Establishment of a continuous tumor-cell
line (panc-1) from a human carcinoma of the exocrine pancreas. Int
J Cancer. 15:741–747. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Parekh D, Ishizuka J, Townsend CM Jr,
Haber B, Beauchamp RD, Karp G, Kim SW, Rajaraman S, Greeley G Jr
and Thompson JC: Characterization of a human pancreatic carcinoid
in vitro: Morphology, amine and peptide storage, and secretion.
Pancreas. 9:83–90. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Soule HD, Vazguez J, Long A, Albert S and
Brennan M: A human cell line from a pleural effusion derived from a
breast carcinoma. J Natl Cancer Inst. 51:1409–1416. 1973.PubMed/NCBI
|
|
23
|
Keydar I, Chen L, Karby S, Weiss FR,
Delarea J, Radu M, Chaitcik S and Brenner HJ: Establishment and
characterization of a cell line of human breast carcinoma origin.
Eur J Cancer. 15:659–670. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stone KR, Mickey DD, Wunderli H, Mickey GH
and Paulson DF: Isolation of a human prostate carcinoma cell line
(DU 145). Int J Cancer. 21:274–281. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tai S, Sun Y, Squires JM, Zhang H, Oh WK,
Liang CZ and Huang J: PC3 is a cell line characteristic of
prostatic small cell carcinoma. Prostate. 71:1668–1679. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Horoszewicz JS, Leong SS, Kawinski E, Karr
JP, Rosenthal H, Chu TM, Mirand EA and Murphy GP: LNCaP model of
human prostatic carcinoma. Cancer Res. 43:1809–1818.
1983.PubMed/NCBI
|
|
27
|
Niforou KM, Anagnostopoulos AK, Vougas K,
Kittas C, Gorgoulis VG and Tsangaris GT: The proteome profile of
the human osteosarcoma U2OS cell line. Cancer Genomics Proteomics.
5:63–78. 2008.PubMed/NCBI
|
|
28
|
Billiau A, Edy VG, Heremans H, Van Damme
J, Desmyter J, Georgiades JA and De Somer P: Human interferon: Mass
production in a newly established cell line, MG-63. Antimicrob
Agents Chemother. 12:11–15. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carbonare L Dalle, Valenti MT, Bertoldo F,
Fracalossi A, Balducci E, Azzarello G, Vinante O and Lo Cascio V:
Amino-bisphosphonates decrease hTERT gene expression in breast
cancer in vitro. Aging Clin Exp Res. 19:91–96. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Valenti MT, Carbonare L Dalle, Bertoldo F,
Donatelli L and Lo Cascio V: The effects on hTERT gene expression
is an additional mechanism of amino-bisphosphonates in prostatic
cancer cells. Eur J Pharmacol. 580:36–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gatta V, Drago D, Fincati K, Valenti MT,
Carbonare L Dalle, Sensi SL and Zatta P: Microarray analysis on
human neuroblastoma cells exposed to aluminum, β(1–42)-amyloid or
the beta (1–42)-amyloid aluminum complex. PloS One. 6:e159652011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Murayama T, Kawasoe Y, Yamashita Y, Ueno
Y, Minami S, Yokouchi M and Komiya S: Efficacy of the
third-generation bisphosphonate risedronate alone and in
combination with anticancer drugs against osteosarcoma cell lines.
Anticancer Res. 28:2147–2154. 2008.PubMed/NCBI
|
|
33
|
Valenti MT, Zanatta M, Donatelli L,
Viviano G, Cavallini C, Scupoli MT and Carbonare L Dalle: Ascorbic
acid induces either differentiation or apoptosis in MG-63
osteosarcoma lineage. Anticancer Res. 34:1617–1627. 2014.PubMed/NCBI
|
|
34
|
Galindo M, Pratap J, Young DW,
Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS and van
Wijnen AJ: The bone-specific expression of Runx2 oscillates during
the cell cycle to support a G1-related antiproliferative function
in osteoblasts. J Biol Chem. 280:20274–20285. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Valenti MT, Carbonare L Dalle, Donatelli
L, Bertoldo F, Giovanazzi B, Caliari F and Lo Cascio V: STEAP mRNA
detection in serum of patients with solid tumours. Cancer Lett.
273:122–126. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Carbonare L Dalle, Gasparetto A, Donatelli
L, Dellantonio A and Valenti MT: Telomerase mRNA detection in serum
of patients with prostate cancer. Urol Oncol. 31:205–210. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pratap J, Lian JB and Stein GS: Metastatic
bone disease: Role of transcription factors and future targets.
Bone. 48:30–36. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li XQ, Lu JT, Tan CC, Wang QS and Feng YM:
RUNX2 promotes breast cancer bone metastasis by increasing integrin
α5-mediated colonization. Cancer Lett. 380:78–86. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang C, Long F, Wan J, Hu Y and He H:
MicroRNA-205 acts as a tumor suppressor in osteosarcoma via
targeting RUNX2. Oncol Rep. 35:3275–3284. 2016.PubMed/NCBI
|
|
41
|
Ge C, Zhao G, Li Y, Li H, Zhao X, Pannone
G, Bufo P, Santoro A, Sanguedolce F, Tortorella S, et al: Role of
Runx2 phosphorylation in prostate cancer and association with
metastatic disease. Oncogene. 35:366–376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Blyth K, Vaillant F, Jenkins A, McDonald
L, Pringle MA, Huser C, Stein T, Neil J and Cameron ER: Runx2 in
normal tissues and cancer cells: A developing story. Blood Cells
Mol Dis. 45:117–123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He Y, de Castro LF, Shin MH, Dubois W,
Yang HH, Jiang S, Mishra PJ, Ren L, Gou H, Lal A, et al: p53 loss
increases the osteogenic differentiation of bone marrow stromal
cells. Stem Cells. 33:1304–1319. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Engelmann D and Pützer BM: Emerging from
the shade of p53 mutants: N-terminally truncated variants of the
p53 family in EMT signaling and cancer progression. Sci Signal.
7:re92014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chimge NO, Baniwal SK, Little GH, Chen YB,
Kahn M, Tripathy D, Borok Z and Frenkel B: Regulation of breast
cancer metastasis by Runx2 and estrogen signaling: The role of
SNAI2. Breast Cancer Res. 13:R1272011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lee JY and Kong G: Roles and epigenetic
regulation of epithelial-mesenchymal transition and its
transcription factors in cancer initiation and progression. Cell
Mol Life Sci. Jul 26–2016.(Epub ahead of print). View Article : Google Scholar
|
|
47
|
Akech J, Wixted JJ, Bedard K, van der Deen
M, Hussain S, Guise TA, van Wijnen AJ, Stein JL, Languino LR,
Altieri DC, et al: Runx2 association with progression of prostate
cancer in patients: Mechanisms mediating bone osteolysis and
osteoblastic metastatic lesions. Oncogene. 29:811–821. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Leong DT, Lim J, Goh X, Pratap J, Pereira
BP, Kwok HS, Nathan SS, Dobson JR, Lian JB, Ito Y, et al:
Cancer-related ectopic expression of the bone-related transcription
factor RUNX2 in non-osseous metastatic tumor cells is linked to
cell proliferation and motility. Breast Cancer Res. 12:R892010.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Baniwal SK, Khalid O, Gabet Y, Shah RR,
Purcell DJ, Mav D, Kohn-Gabet AE, Shi Y, Coetzee GA and Frenkel B:
Runx2 transcriptome of prostate cancer cells: Insights into
invasiveness and bone metastasis. Mol Cancer. 9:2582010. View Article : Google Scholar : PubMed/NCBI
|