Introduction
Colorectal cancer (CRC) represents the third most
common cancer in men and the second most common cancer in women
globally (1–3). With the development of early diagnosis
and treatment modalities, the 5-year survival rate of CRC has been
improved over the past two decades (2,4).
Currently, combination chemotherapy strategy has been recognized as
the standard of care in international guidelines (4,5). However,
even with aggressive intervention, it does not ensure a cure,
particularly for patients with malignant CRC (6,7). Research
has been focused on tumor-suppressor genes, oncogenes and cell
signaling pathways, including their role in the proliferation,
apoptosis and aggressiveness of these tumors (8). The resulting information has led the way
for an increasing interest in potential genetics-based treatments
(9).
MicroRNAs (miRNAs or miRs) are a class of noncoding
RNA molecules that negatively regulate the translation of messenger
(m) RNAs by interacting with complementary sites in the 3′
untranslated region (UTR) (10).
miRNAs regulate various biological processes, including cell
growth, invasion, cell cycle and apoptosis (10). The close association between miRNAs
and tumors is also well studied (11). Several miRNAs have been regarded as
oncomiRs or tumor suppressor genes in specific cancers by directly
targeting oncogenes or tumor suppressor genes (12). Recently, mounting evidence indicates
that miR-217 can function as a tumor suppressor gene or as an
oncogene depending on the cell type (13,14).
miR-217 has been proved to target the oncogenes sirtuin 1 (SirT1)
or KRAS in endothelial and pancreatic ductal adenocarcinoma cells,
although it targets the tumor suppressor gene phosphatase and
tensin homolog (PTEN) in kidney (13–16).
However, its function and expression in CRC are still unclear.
The present study identified that mitogen-activated
protein kinase (MAPK) 1 is a novel miR-217 target, which is
regarded as a critical component of the MAPK signaling pathway
(17). Considering that the MAPK
signaling pathway is important in regulating pathological cell
growth (18), it is considered that
downregulation of MAPK inhibitors would represent an ideal
anti-tumor therapy. It was additionally demonstrated that miR-217
could inhibit tumor proliferation and enhance apoptosis of CRC via
the MAPK pathway.
Materials and methods
Cell culture
The malignant CRC cell lines RKO and SW480 were
obtained from the Chinese Academy of Sciences (Shanghai, China).
All cells were cultured in Dulbecco's modified Eagle medium (Lonza
Inc., Allendale, NJ, USA) supplemented with 0.1% insulin, 50 U/ml
penicillin/streptomycin, 1% non-essential amino acids and 10% fetal
bovine serum (Lonza Inc.) at 37°C in an atmosphere of 5% CO2. All
cell lines were tested periodically for mycoplasma contamination
using MycoAlert Mycoplasma Detection kit (Lonza Inc.).
Transfection and luciferase activity
assay
miRNA transfection and luciferase activity assay
were performed as described previously (13). Briefly, the human CRC cell lines RKO
and SW480 were seeded on 24-well plates and transfected 24 h later
using Lipofectamine 2000 according to the manufacturer's protocol
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The
miR-217 precursor molecules (Ambion; Thermo Fisher Scientific,
Inc.) were co-transfected with 4 ng/well of plasmid pRL-CMV
(Promega Corporation, Madison, WI, USA) and 80 ng/well of
luciferase reporter plasmids (Shanghai GenePharma Co., Ltd.,
Shanghai, China).
Tissue samples
The present study protocol complies with National
Regulations on the Use of Clinical Samples in China (19), and was approved by the by Ethics
Committee of Chinese PLA General Hospital (Beijing, China). CRC
specimens were obtained from patients with CRC at the Chinese PLA
General Hospital from July 2000 to July 2006. Follow-up was
conducted in all patients, and survival time was completed in
December 2009. The follow-up was carried out every 3 months by
e-mail or telephone, and the last follow-up was in January 2010.
The clinicopathological parameters of the patients are shown in
Table I. Written informed consent was
obtained from all patients.
 | Table I.Association between miR-217
expression and clinicopathological variables of colorectal cancer
patients. |
Table I.
Association between miR-217
expression and clinicopathological variables of colorectal cancer
patients.
|
|
| miR-217
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | No. of
patients | Low | High | P-value |
|---|
| No. of
patients | 121 | 53 | 68 |
|
| Age, years |
|
|
| 0.14 |
|
<50 | 71 | 32 | 39 |
|
|
≥50 | 50 | 21 | 29 |
|
| Gender |
|
|
| 0.10 |
|
Male | 69 | 28 | 41 |
|
|
Female | 52 | 25 | 27 |
|
| MTD, cm |
|
|
| 0.08 |
|
<5 | 80 | 38 | 42 |
|
| ≥5 | 41 | 15 | 26 |
|
| Tumor location |
|
|
| 0.11 |
|
Rectum | 57 | 27 | 30 |
|
|
Colon | 64 | 26 | 38 |
|
| Depth of
invasion |
|
|
| 0.14 |
|
T1/T2 | 38 | 18 | 20 |
|
|
T3/T4 | 83 | 35 | 48 |
|
| Survival
status |
|
|
| 0.01 |
|
Alive | 23 | 5 | 18 |
|
|
Dead | 98 | 48 | 50 |
|
| TNM stage |
|
|
| 0.09 |
| II | 49 | 24 | 25 |
|
|
III/IV | 72 | 29 | 43 |
|
Protein extraction and western
blotting
Protein extraction and western blot analysis were
conducted as described previously (20). The primary antibodies used were
anti-MAPK1 (Abcam, Cambridge, UK, ab102930, 1:500), anti-p38 MAPK
(Abcam, ab7952, 1:800), anti-B-cell lymphoma (Bcl)-2
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany, SAB4300339,
1:500), anti-Raf-1 (Abcam, ab32025, 1:800) and anti-β-actin (Santa
Cruz Biotechnology, Inc., sc-8432, 1:500).
Reverse transcription-quantitative
polymerase chain reaction analysis (RT-qPCR)
The RNeasy Mini kit (Qiagen GmbH, Hilden, Germany)
was used to extract total RNA from RKO cells and frozen tissue
specimens. The expression of miR-217 was measured using the
standard TaqMan® MicroRNA Assay (Applied Biosystems;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) as previously
reported (21). The primers were
designed as follows: MAPK1, 5′-ATGACCTCCTATGGCATCGA-3′ (forward)
and 5′-TGATGTTCTGTGCGTGGAGC-3′ (reverse); Bcl-2,
5′-CCGGGAGATCGTGATGAAGT-3′ (forward) and 5′-ATCCCAGCCTCCGTTATCCT-3′
(reverse); Bcl-extra large (xl), 5′-ATGGAGAACAATAAAACCT-3′
(forward) and 5′-CTAGTGATAAAAGTAGAGTTC-3′ (reverse); and KRAS,
5′-GGCCTGCTGAAAATGACTGAAT-3′ (forward) and
5′-ATTGTTGGATCATATTCGTCCAC-3′ (reverse). First-strand complementary
DNA was synthesized using a ReverTra Ace® qPCR RT kit
(Toyobo, Co., Ltd., Osaka, Japan). RT-qPCR was performed using the
ABI PRISM® 7900 HT Sequence Detection System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) under the following
cycling conditions: 95°C for 3 min followed by 40 cycles of 95°C
for 10 sec and 55°C for 45 sec. Melting curve analysis was used to
check the specificity of amplification. Relative gene expression
was calculated by using the comparative Cq method (22).
Apoptosis assay
An Annexin V-fluorescein isothiocyanate (FITC)
apoptosis kit was used to measure apoptosis according to the
manufacturer's instructions (ImmunoChemistry Technologies, LLC,
Bloomington, MN, USA). Cells were analyzed by flow cytometry after
being stained with an anti-Annexin V-FITC antibody (Santa Cruz
Biotechnology, Inc., sc-1929, 1:1,000). For each experiment, at
least 20,000 cells were analyzed.
Colony formation and proliferation
assay
Colony forming capacity was evaluated as previously
described (23). At daily intervals,
the number of viable cells was determined by MTT assay to measure
the cell proliferation. Cell proliferation was analyzed by Cell
Counting kit (Sigma-Aldrich; Merck Millipore, 03285) according to
the manufacturer's instructions. The optical density was measured
by using a microplate reader (Thermo Fisher Scientific, Inc.) at
570 nm.
Statistics
The χ2 test was used to analyze the
miR-217 expression and clinicopathological characteristics. Cox
proportional-hazards regression analysis was used to evaluate
predictors of survival. Overall survival times in CRC patients were
compared using Kaplan-Meier survival analysis. Analyses were
performed using SPSS 10.0 (SPSS, Inc., Chicago, IL, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
miR-217 expression and association
with clinicopathological characteristics in CRC
The expression levels of miR-217 were measured in
121 human CRC specimens and 48 matched adjacent noncancerous
tissues. As shown in Fig. 1A, the
expression levels of miR-217 were markedly higher in normal tissues
compared with those in CRC tissues (mean ± standard deviation:
7.82±1.8 vs. 2.71±0.89, P<0.001). In addition, it was observed
that low-grade CRC [World Health Organization (WHO) grade I,
3.12±1.08] exhibited greater expression of miR-217 compared with
high-grade CRC (WHO grade II–III, 1.98±1.16, P<0.05). Thus,
miR-217 expression is associated with the proliferation of CRC and
with tumor progression.
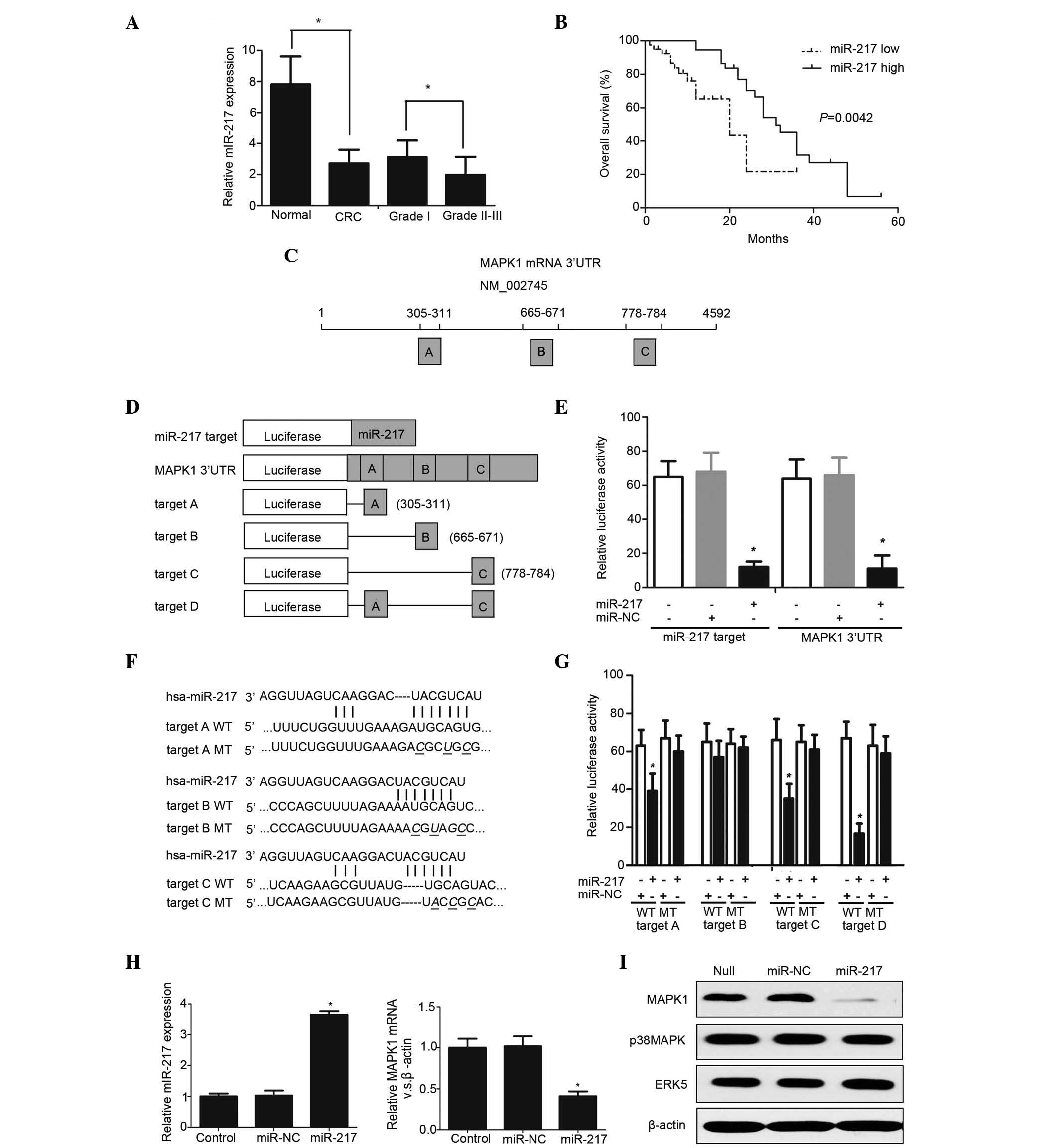 | Figure 1.(A) miR-217 levels of were measured
in CRC specimens by RT-qPCR. *P<0.05. (B) Kaplan-Meier survival
curves according to miR-217 expression. *P=0.0042. (C) Chart of
MAPK1 mRNA sequence. Predicted consequential pairing of MAPK1
region: Region A, positions 395–401 of MAPK1 3′ UTR; region B:
positions 665–671 of MAPK1 3′ UTR; region C, positions 778–784 of
MAPK1 3′ UTR. (D) Schematic diagram of luciferase reporter
constructs transfected into RKO cells. (E) Luciferase reporter
plasmids were co-transfected into RKO cells, and the relative
luciferase activity was determined. *P<0.05. (F) Sequence of
wild type and mutant miR-217 target sites in the MAPK1 3′ UTR. (G)
The contribution of the three miR-217 target sites was measured by
a luciferase reporter assay *P<0.01. (H) The expression level of
miR-217 was measured in stably expressing subclones by
TaqMan® MicroRNA Assay. RT-qPCR demonstrated that MAPK1
mRNA was significantly decreased in RKO cells transfected with
miR-217 compared with mock- and miR-NC-transfected cells.
*P<0.05. (I) Western blot analysis revealed that miR-217
specifically repressed MAPK1 protein expression without influencing
the expression of the other MAP kinase family members (p38 MAPK and
ERK5). miR, microRNA; CRC, colorectal cancer; MAPK,
mitogen-activated protein kinase; mRNA, messenger RNA; UTR,
untranslated region; NC, negative control; hsa, Homo sapiens; WT,
wild type; MT, mutant; ERK, extracellular signal-regulated kinase;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction. |
To explore whether miR-217 expression is correlated
with clinicopathological parameters, the association between
miR-217 expression and clinical characteristics of patients with
CRC was explored. The mean miR-217 expression level of CRC was
2.71, which was utilized to divide high-grade CRC patients into a
high group (≥2.71, n=68) and a low group (<2.71, n=53).
Univariate analysis indicated a significant association between
miR-217 expression and prognosis (Table
II), while no significant associations were identified between
prognosis and other clinical characteristics. The Kaplan-Meier
survival curves revealed that the survival rate probability was
higher in CRC patients exhibiting high miR-217 expression than in
patients with low miR-217 expression (Fig. 1B). Multivariate analysis further
revealed that miR-217 expression is an independent factor for
overall survival (Table II). These
data suggest that miR-217 expression may be a critical prognostic
factor for high-grade CRC.
 | Table II.Univariate and multivariate analyses
of different prognostic parameters in patients with colorectal
cancer by log-rank test and Cox regression analysis. |
Table II.
Univariate and multivariate analyses
of different prognostic parameters in patients with colorectal
cancer by log-rank test and Cox regression analysis.
| Variable | Univariate log-rank
test (P-value) | Cox multivariate
analysis (P-value) | Relative risk |
|---|
| Age | 0.178 | 0.148 | 0.987 |
| Gender | 0.238 | 0.435 | 1.012 |
| MTD | 0.876 | 0.706 | 0.824 |
| Tumor location | 0.762 | 0.604 | 0.987 |
| Depth of
invasion | 0.541 | 0.442 | 1.112 |
| TNM stage | 0.089 | 0.168 | 1.862 |
| miR-217
expression | 0.009 | 0.018 | 6.438 |
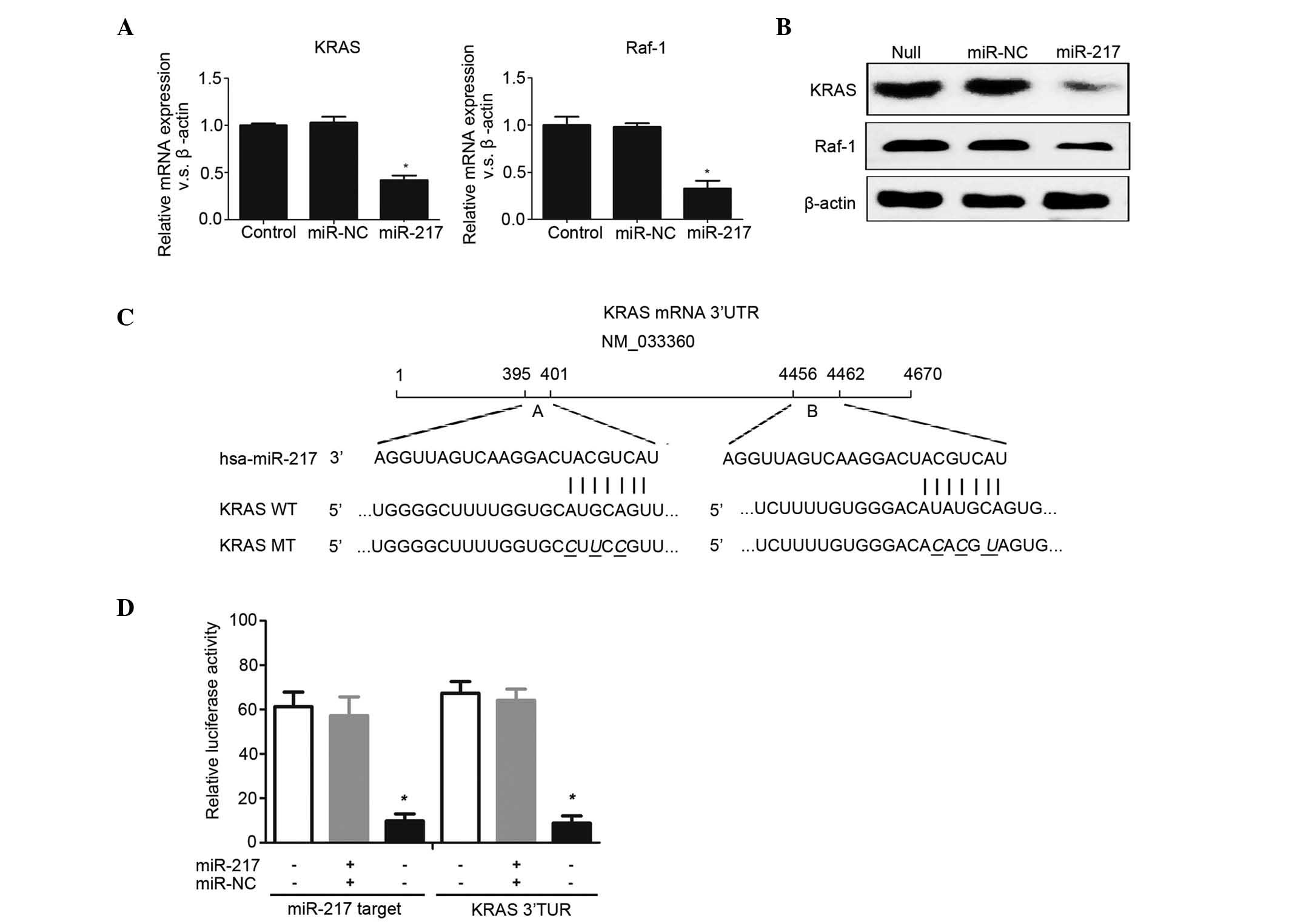
MAPK1 is a potential target of
miR-217
We hypothesized that MAPK1 is a potential target of
miR-217 because it has three putative miR-217 target sites in its
3′ UTR by using TargetScan (www.targetscan.org) (Fig.
1C). Luciferase reporter vectors that are fully complementary
to either the full-length MAPK1 3′ UTR or to the intact mature
miR-7 sequence were used (Fig. 1D).
Next, the mutant sequence of the putative target site or its
relevant target site were cloned into an identical reporter vector
to determine the major targets in RKO malignant CRC cells. By using
a luciferase reporter assay, it was indicated that only when
miR-217 was present, wild-type MAPK1 3′ UTR and miR-217 could
significantly reduce the relative luciferase activity (Fig. 1E and F). Furthermore, it was revealed
that the reporter vectors with putative mir-217 target sites A or C
resulted in 38.2±4.5% or 42.8±3.9% decrease in relative luciferase
activity, respectively, compared with the corresponding mutant
introduced with miR-217 in RKO cells (Fig. 1G). In addition, the relative
luciferase activity was only reduced to 91.2±7.2% when the vector
harbored only the putative target site B. Finally, it was observed
that the relative luciferase activity was decreased to 19.1±1.4% by
a new artificial target D (including putative target sites A and C)
(Fig. 1G). This result was similar to
the wild-type MAPK1 3′ UTR-induced relative luciferase activity in
RKO cells. Transfection of RKO cells with miR-217 resulted in a
~5.6-fold increase in miR-217 expression compared with mock- and
miR-negative control (NC)-transfected cells, while MAPK1 mRNA was
reduced to 24.8% when the RKO CRC cells were transfected with
miR-217 (Fig. 1H). Western blotting
revealed that miR-217 specifically decreased MAPK1 expression at
the protein level (Fig. 1I) without
affecting the expression of the other MAPK family members (p38 MAPK
and extracellular-signal-regulated kinase 5). Thus, these data
suggest that MAPK1 is a novel target of miR-217.
miR-217 represses the survival of CRC
cells in vitro
In light of the above findings, we tested the
hypothesis that miR-217 may repress CRC cell proliferation and
enhance cell apoptosis by inhibiting MAPK1 expression. To verify
our hypothesis, an RKO cell line stably transfected with miR-217,
miR-NC or mock (Null) was established to investigate the effects of
miR-217 expression on CRC cell proliferation. It was observed that
overexpression of miR-217 significantly inhibited tumor cell
proliferation in stable miR-217-transfected RKO and SW480 cells
compared with mock cells and miR-NC-transfected cells, as measured
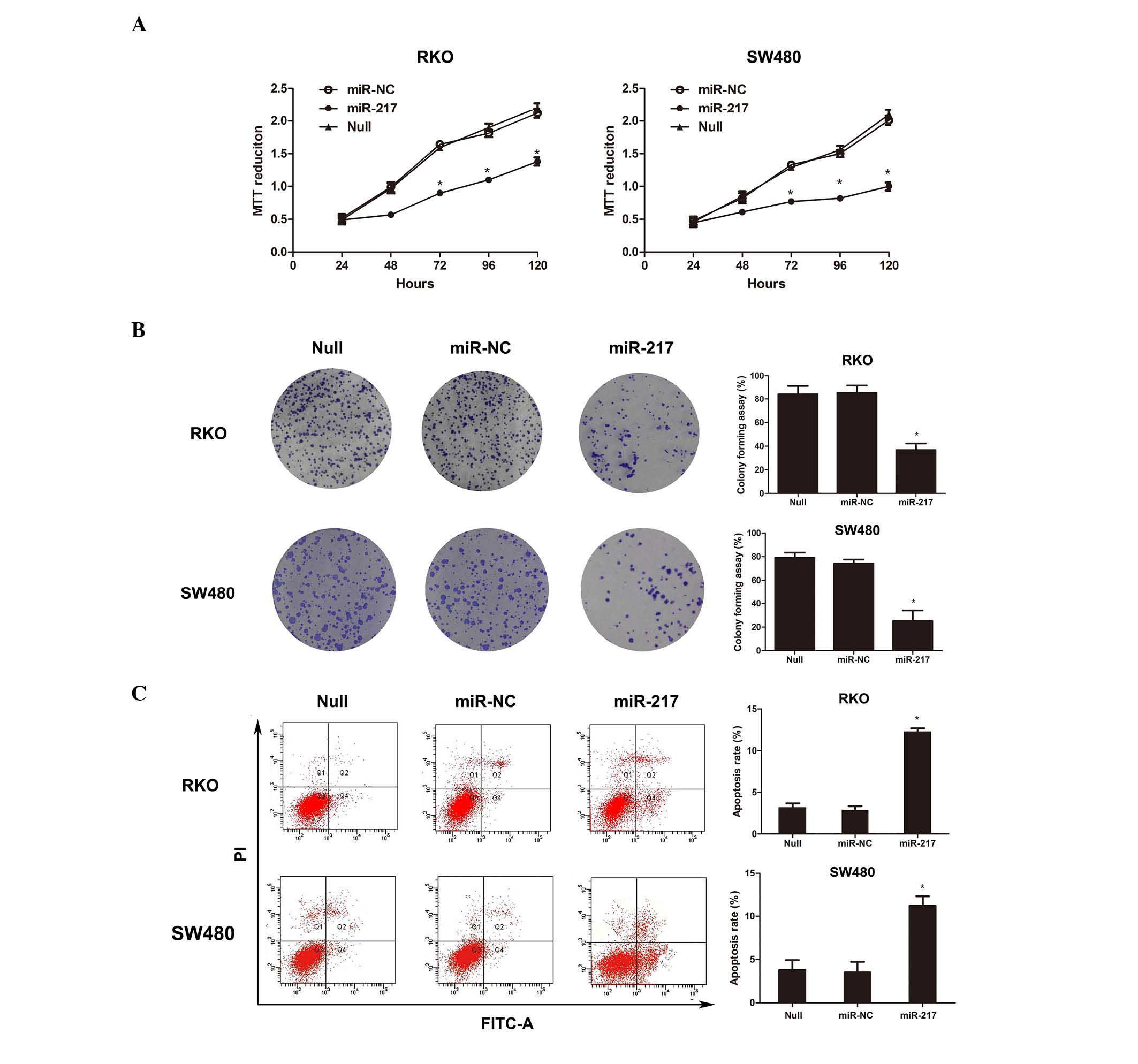
by daily MTT assays (Fig. 2A).
Furthermore, colony formation assays revealed that transfection of
RKO and SW480 cells with miR-217 resulted in significantly lower
anchorage-independent growth compared with cells transfected with
miR-NC (Fig. 2B).
miR-217 enhances the apoptosis of CRC
cells in vitro
Given the effects of miR-217 expression in RKO and
SW480 cell proliferation, we hypothesized that miR-217 could
regulate the apoptosis rate of CRC cells. As predicted, apoptosis
assay indicated that RKO and SW480 cells stably transfected with
miR-217 exhibited a higher apoptosis rate compared with RKO cells
transfected with miR-NC (Fig. 2C).
These results indicate that miR-217 may play a critical role in
regulating cell apoptosis.
To determine the molecular mechanism of cell death
induced by miR-217, the expression of Bcl-2 and Bcl-xl in these CRC
transfectants was initially measured via RT-qPCR and western
blotting. Treatment of RKO and SW480 cells with miR-217
significantly decreased Bcl-xl expression, which was associated
with a reduction in activated Bcl-2 expression (Fig. 3A). Western blot analysis revealed
similar results (Fig. 3B). Taken
together, our results indicate that miR-217 is associated with
apoptosis progression, with a strong correlation between Bcl-2,
Bcl-xl and miR-217 in CRC cells.
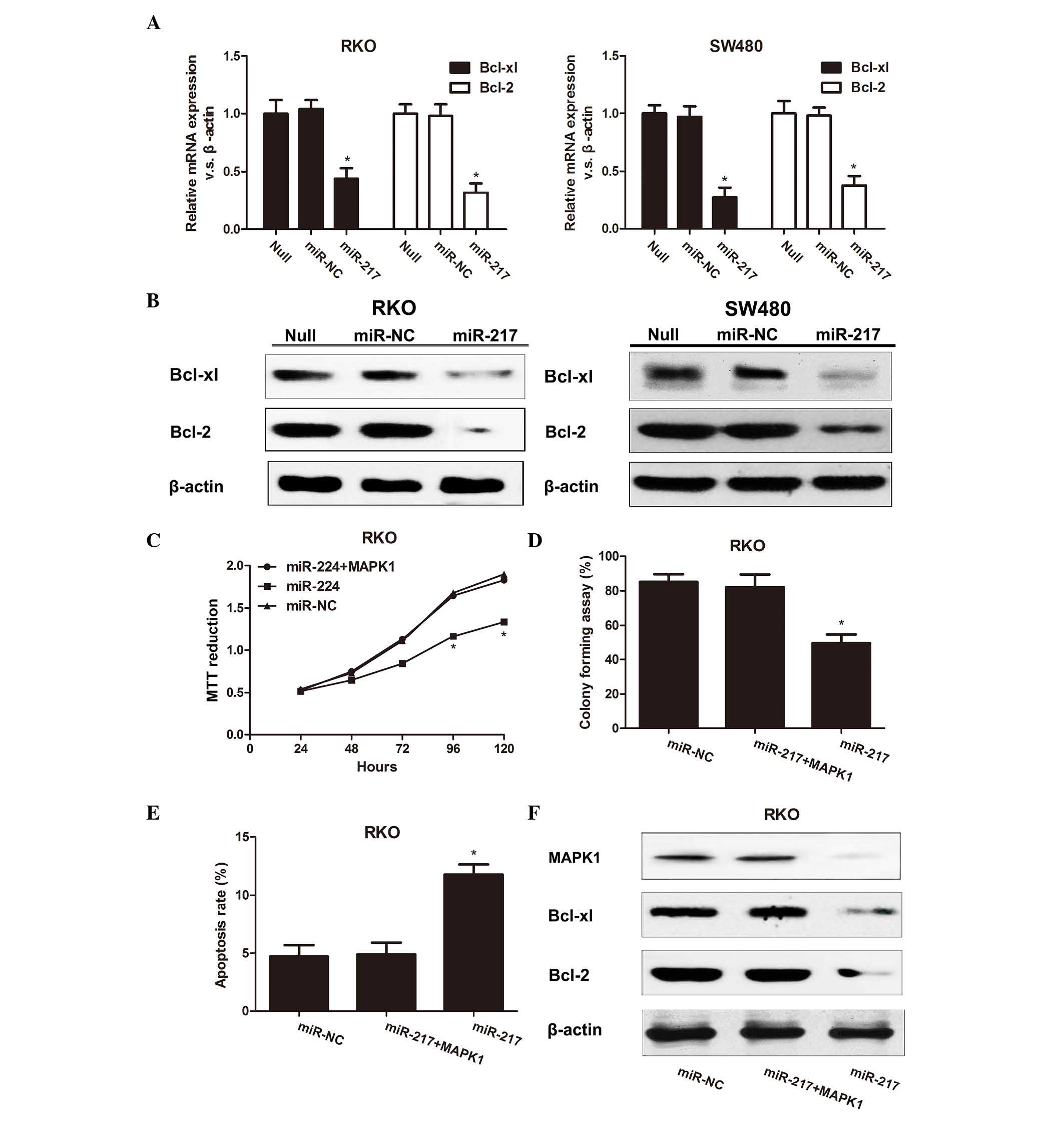 | Figure 3.(A) Bcl-2 and Bcl-xl expression was
measured by reverse transcription-quantitative polymerase chain
reaction. *P<0.05. (B) Western blot analysis revealed similar
results. (C) RKO cells were co-transfected with miR-217 and MAPK1
overexpression plasmid or with a control, and the absorbance
(representative of MTT reduction) was measured at 1, 2, 3, 4 or 5
days after transfection. *P<0.05 compared with the control
groups. (D) Representative results of colony formation assay in RKO
cells co-transfected with miR-217 precursor and MAPK1. *P<0.05
compared with the control groups. (E) Apoptosis assay of RKO cells
transfected with miR-217 precursor and MAPK1 overexpression plasmid
or with a control. *P<0.05 compared with the control groups. (F)
MAPK1, Bcl-2 and Bcl-xl protein expression was measured in RKO
cells co-transfected with miR-217 precursor and MAPK1
overexpression plasmid or with a control. mRNA, messenger RNA; NC,
negative control; miR, microRNA; Bcl, B-cell lymphoma; xl, extra
large; MAPK, mitogen-activated protein kinase. |
The MAPK family member MAPK1 regulates apoptosis and
proliferation in cancer via the Bcl-2 and Bcl-xl apoptotic
signaling pathways (24). To
determine whether these pathways are involved in miR-217-induced
RKO growth and inhibition of apoptosis, the present study first
explored whether changing MAPK1 expression altered the
miR-217-induced effects on apoptosis and growth. Second, the MAPK1,
Bcl-2 and Bcl-xl expression levels in RKO cells overexpressing
miR-217 were investigated.
To examine the function of miR-217-induced MAPK1
signaling in cell proliferation and apoptosis, RKO cells expressing
miR-217 were stably transfected with full-length MAPK1. MTT assay
(Fig. 3C), colony formation assay
(Fig. 3D) and apoptosis assay
(Fig. 3E) demonstrated that
upregulation of MAPK1 in RKO cells overexpressing miR-217 reversed
the increased apoptosis and suppressed the cell proliferation
observed in RKO cells expressing only miR-217, strongly suggesting
that MAPK1 is a critical downstream of miR-217 signaling. In
addition, it was noticed that MAPK1, Bcl-xl and Bcl-2 expression
were significantly lower in RKO cells transfected with miR-217
compared with RKO cells transfected with miR-NC (Figs. 1I, 3A and
3B), suggesting that miR-217 is an important regulator of the
MAPK1-induced apoptotic pathway. Furthermore, upregulation of MAPK1
by expression of full-length MAPK1 substantially increased Bcl-2
and Bcl-xl expression in RKO cells stably expressing miR-217,
compared with cells expressing only miR-217 (Fig. 3F). These results suggest that MAPK1,
Bcl-xl and Bcl-2 are important downstream effectors of
miR-217-induced inhibition of apoptosis and enhanced proliferation
in CRC cells.
miR-217 regulates the RAS-MAPK
signaling pathway
The present study further explored the effects of
miR-217 overexpression in the MAPK signaling pathway. It was
observed that the mRNA levels of KRAS and Raf-1, significant
upstream components of the MAPK pathway (25,26), were
decreased by 0.32- and 0.45-fold, respectively, in
miR-217-transfected RKO cells by using RT-qPCR (Fig. 4A). Western blotting revealed similar
results at the protein level (Fig.
4B). These findings indicated that miR-217 may be a critical
regulator of the RAS-Raf-MAPK pathway. Since miRNAs could regulate
one pathway by affecting its multiple and functionally related
targets (10), the current study
explored whether miR-217 could modulate KRAS and Raf-1. It was
revealed that there are no miR-217 target sites in the Raf-1 3′
UTR, but two putative sites (A and B) were identified in the KRAS
3′ UTR (Fig. 4C). These two target
sites significantly inhibited the luciferase activity, which is
consistent with a previous study reporting that miR-217 targets the
oncogene KRAS in pancreatic ductal adenocarcinoma cells (15) (Fig. 4D).
These data suggest that miR-217 can modulate KRAS and MAPK1
expression via directly binding to its target sites, indicating
that miR-217 can inhibit RKO cell proliferation and enhance
apoptosis by regulating the RAS/Raf/MAPK signaling pathway.
Discussion
miR-217, a novel of regulator of gene expression, is
a special miRNA that can act as an oncogene or as a tumor
suppressor gene depending on the cell type (27). Previous studies have shown that
miR-217 acts as a tumor suppressor by targeting the oncogene SirT1
in endothelial cells (13), while it
also acts as an oncogene by targeting PTEN, a tumor suppressor
gene, in kidney cells and by enhancing the germinal center (GC)
reaction in GC B-cells (16,28). These findings prompted us to explore
its expression and biological function in CRC. The current study
determined the miR-217 expression levels in clinical CRC specimens
and revealed that miR-217 expression was significantly associated
with CRC histopathology. Kaplan-Meier analysis indicated that
patients with CRC expressing high miR-217 levels had significantly
longer overall survival than patients with low miR-217 expression,
suggesting that miR-217 may play a critical role as a negative
regulator of CRC development and progression. Furthermore,
statistical analyses demonstrated that the miR-217 expression level
was an independent prognostic parameter of patient outcomes. Thus,
this is the first study to show that miR-217 expression is a novel
molecular marker for predicting the prognosis of CRC.
The present study demonstrated that miR-217
suppresses tumor growth and enhances apoptosis in RKO CRC cells by
blocking a novel miR-217 target, MAPK1. This protein has been
regarded as an integration point for multiple biochemical signals,
and is associated with various cellular processes, including
proliferation, apoptosis, transcription regulation and development
(29). Furthermore, it was
demonstrated that upregulation of miR-217 markedly increased the
apoptosis of RKO cells. Western blot assay demonstrated that RKO
cells overexpressing miR-217 could decrease the protein expression
levels of Bcl-2 and Bcl-xl, which are important molecules of the
cellular apoptosis pathway (30,31).
The current results revealed a novel mechanism by
which miR-217 suppresses cell growth and enhances apoptosis in RKO
cells. miR-217 mediates MAPK1, Bcl-2 and Bcl-xl expression, which
play critical roles in CRC cell apoptosis (32,33).
Importantly, upregulation of miR-217 in RKO cells enhanced
apoptosis and inhibited growth, an effect that was significantly
reversed by constitutive MAPK1 expression. Furthermore, MAPK1
activation resulted in a significant increase in Bcl-2 and Bcl-xl
expression in RKO cells stably expressing miR-217 compared with
cells expressing only miR-217. MAPK1 is a key regulatory component
of the intrinsic apoptotic pathway that is required for programmed
cell death (34). The intrinsic
pathway of apoptosis regulates the activity of the Bcl-2 family
proteins (Bcl-2 and Bcl-xl) that control the integrity of the
mitochondrial membrane (35). Our
results are consistent with previous studies reporting that MAPK
could suppress the activation of apoptosis by increasing the level
of Bcl-2 and Bcl-xl (36,37). The above finding demonstrates that
miR-217 is required for RKO proliferation and apoptosis via
MAPK1-induced intrinsic apoptosis signaling.
The present study also explored the function of
miR-217 in the MAPK pathway to clarify the critical mechanism
underlying tumorigenesis, proliferation and apoptosis. The MAPK
pathway is regarded as an important cascade of proteins in the
cell, transferring signals to the nucleus from receptors on the
surface of the cell (38). This is a
critical step for the progression of tumors (39,40).
Activated RAS phosphorylates and activates MAPK, and then produces
certain changes in the cell, such as cell proliferation, apoptosis,
invasion and drug resistance (41,42). It
was observed that KRAS, which is an important upstream signal of
the MAPK pathway (43,44), also contained miR-217 target sites in
the 3′ UTR. Since the transcription of KRAS, Raf-1 and MAPK1,
critical members of the MAPK pathway (25,43), was
significantly reduced in RKO cells treated with miR-217, it was
concluded that miR-217 modulates the RAS/Raf/MAPK signaling
pathway. The RAS/Raf/MAPK pathway is well documented to affect the
cellular apoptotic pathway, which is characterized by promoting
cytochrome c release (25). RAS/MAPK
activity has been correlated with the downregulation of
antiapoptotic components such as Bcl-2 and Bcl-xl (25). Indeed, the MAPK-activated Bcl-2
protein family has been shown to directly affect the release of
cytochrome c from mitochondria, offering a possible explanation for
the observed cell apoptosis alteration (25).
In conclusion, the present study provides the first
line of evidence that miR-217 suppresses tumor growth and enhances
apoptosis in CRC, and that these effects are associated with the
downregulation of RAS/Raf/MAPK signaling. Our results suggest that
miR-217 is a promising therapeutic target for the treatment of
CRC.
References
|
1
|
Debarros M and Steele SR: Colorectal
cancer screening in an equal access healthcare system. J Cancer.
4:270–280. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cunningham D, Atkin W, Lenz HJ, Lynch HT,
Minsky B, Nordlinger B and Starling N: Colorectal cancer. Lancet.
375:1030–1047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qaseem A, Denberg TD, Hopkins RH Jr,
Humphrey LL, Levine J, Sweet DE and Shekelle P: Clinical Guidelines
Committee of the American College of Physicians: Screening for
colorectal cancer: A guidance statement from the American College
of Physicians. Ann Intern Med. 156:378–386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ragnhammar P, Hafström L, Nygren P and
Glimelius B: SBU-group. Swedish Council of Technology Assessment in
Health Care: A systematic overview of chemotherapy effects in
colorectal cancer. Acta Oncol. 40:282–308. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bergsland EK: Is more not better?
Combination therapies in colorectal cancer treatment. Hematol Oncol
Clin North Am. 29:85–116. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Saltz LB and Minsky B: Adjuvant therapy of
cancers of the colon and rectum. Surg Clin North Am. 82:1035–1058.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bauer TW and Spitz FR: Adjuvant and
neoadjuvant chemoradiation therapy for primary colorectal cancer.
Surg Oncol. 7:175–181. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Moss JA: Gene therapy review. Radiol
Technol. 86:155–184. 2014.PubMed/NCBI
|
|
9
|
Roth JA and Cristiano RJ: Gene therapy for
cancer: What have we done and where are we going? J Natl Cancer
Inst. 89:21–39. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kalla R, Ventham NT, Kennedy NA, Quintana
JF, Nimmo ER, Buck AH and Satsangi J: MicroRNAs: New players in
IBD. Gut. 64:504–517. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xue J, Niu J, Wu J and Wu ZH: MicroRNAs in
cancer therapeutic response: Friend and foe. World J Clin Oncol.
5:730–743. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: MicroRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Menghini R, Casagrande V, Cardellini M,
Martelli E, Terrinoni A, Amati F, Vasa-Nicotera M, Ippoliti A,
Novelli G, Melino G, et al: MicroRNA 217 modulates endothelial cell
senescence via silent information regulator 1. Circulation.
120:1524–1532. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xia H, Ooi LL and Hui KM:
MicroRNA-216a/217-induced epithelial-mesenchymal transition targets
PTEN and SMAD7 to promote drug resistance and recurrence of liver
cancer. Hepatology. 58:629–641. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM and
Chen J: The miR-217 microRNA functions as a potential tumor
suppressor in pancreatic ductal adenocarcinoma by targeting KRAS.
Carcinogenesis. 31:1726–1733. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kato M, Putta S, Wang M, Yuan H, Lanting
L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, et al:
TGF-beta activates Akt kinase through a microRNA-dependent
amplifying circuit targeting PTEN. Nat Cell Biol. 11:881–889. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Voong LN, Slater AR, Kratovac S and
Cressman DE: Mitogen-activated protein kinase ERK1/2 regulates the
class II transactivator. J Biol Chem. 283:9031–9039. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Giehl K, Skripczynski B, Mansard A, Menke
A and Gierschik P: Growth factor-dependent activation of the
Ras-Raf-MEK-MAPK pathway in the human pancreatic carcinoma cell
line PANC-1 carrying activated K-ras: Implications for cell
proliferation and cell migration. Oncogene. 19:2930–2942. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu Y, Xiao T, Zhang F, Chen Y, Liu Y, Li
Y, Chen YD, Li Z and Guan M: Effect of mitochondrial tRNA(Lys)
mutation on the clinical and biochemical characteristics of Chinese
essential hypertensive subjects. Biochem Biophys Res Commun.
454:500–504. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Q, Qian J, Wang J, Luo C, Chen J, Hu
G and Lu Y: Knockdown of RLIP76 expression by RNA interference
inhibits invasion, induces cell cycle arrest, and increases
chemosensitivity to the anticancer drug temozolomide in glioma
cells. J Neurooncol. 112:73–82. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hui AB, Shi W, Boutros PC, Miller N,
Pintilie M, Fyles T, McCready D, Wong D, Gerster K, Waldron L,
Jurisica I, et al: Robust global micro-RNA profiling with
formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest.
89:597–606. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Q, Wang JY, Zhang XP, Lv ZW, Fu D, Lu
YC, Hu GH, Luo C and Chen JX: RLIP76 is overexpressed in human
glioblastomas and is required for proliferation, tumorigenesis and
suppression of apoptosis. Carcinogenesis. 34:916–926. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Subramanian M and Shaha C: Upregulation of
Bcl-2 through ERK phosphorylation is associated with human
macrophage survival in an estrogen microenvironment. J Immunol.
179:2330–2338. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vitagliano O, Addeo R, D'Angelo V, Indolfi
C, Indolfi P and Casale F: The Bcl-2/Bax and Ras/Raf/MEK/ERK
signaling pathways: Implications in pediatric leukemia pathogenesis
and new prospects for therapeutic approaches. Expert Rev Hematol.
6:587–597. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lemieux E, Cagnol S, Beaudry K, Carrier J
and Rivard N: Oncogenic KRAS signalling promotes the Wnt/β-catenin
pathway through LRP6 in colorectal cancer. Oncogene. 34:4914–4927.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo J, Feng Z, Huang Z, Wang H and Lu W:
MicroRNA-217 functions as a tumor suppressor gene and correlates
with cell resistance to cisplatin in lung cancer. Mol Cells.
37:664–671. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
de Yébenes VG, Bartolomé-Izquierdo N,
Nogales-Cadenas R, Pérez-Durán P, Mur SM, Martínez N, Di Lisio L,
Robbiani DF, Pascual-Montano A, Cañamero M, et al: MiR-217 is an
oncogene that enhances the germinal center reaction. Blood.
124:229–239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Joshi S and Platanias LC: Mnk kinase
pathway: Cellular functions and biological outcomes. World J Biol
Chem. 5:321–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bates DJ and Lewis LD: Manipulating the
apoptotic pathway: Potential therapeutics for cancer patients. Br J
Clin Pharmacol. 76:381–395. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang T and Saghatelian A: Emerging roles
of lipids in BCL-2 family-regulated apoptosis. Biochim Biophys
Acta. 1831:1542–1554. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ye L, Yuan G, Xu F, Sun Y, Chen Z, Chen M,
Li T, Sun P, Li S and Sun J: The small-molecule compound BM-1197
inhibits the antiapoptotic regulators Bcl-2/Bcl-xL and triggers
apoptotic cell death in human colorectal cancer cells. Tumour Biol.
36:3447–3455. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao SL, Hong J, Xie ZQ, Tang JT, Su WY,
Du W, Chen YX, Lu R, Sun DF and Fang JY: TRAPPC4-ERK2 interaction
activates ERK1/2, modulates its nuclear localization and regulates
proliferation and apoptosis of colorectal cancer cells. PLoS One.
6:e232622011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Abdel-Magid AF: ERK2 inhibitors may
provide treatment for cancer. ACS Med Chem Lett. 4:576–577. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Adams JM and Cory S: The Bcl-2 protein
family: Arbiters of cell survival. Science. 281:1322–1326. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang D, Weng Q, Zhang L, He Q and Yang B:
VEGF and Bcl-2 interact via MAPKs signaling pathway in the response
to hypoxia in neuroblastoma. Cell Mol Neurobiol. 29:391–401. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Scheid MP, Schubert KM and Duronio V:
Regulation of bad phosphorylation and association with Bcl-x(L) by
the MAPK/Erk kinase. J Biol Chem. 274:31108–31113. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Orton RJ, Sturm OE, Vyshemirsky V, Calder
M, Gilbert DR and Kolch W: Computational modelling of the
receptor-tyrosine-kinase-activated MAPK pathway. Biochem J.
392:249–261. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cheng Y, Zhang G and Li G: Targeting MAPK
pathway in melanoma therapy. Cancer Metastasis Rev. 32:567–584.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Furukawa K and Hohmann S: Synthetic
biology: Lessons from engineering yeast MAPK signalling pathways.
Mol Microbiol. 88:5–19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pritchard AL and Hayward NK: Molecular
pathways: Mitogen-activated protein kinase pathway mutations and
drug resistance. Clin Cancer Res. 19:2301–2309. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zassadowski F, Rochette-Egly C, Chomienne
C and Cassinat B: Regulation of the transcriptional activity of
nuclear receptors by the MEK/ERK1/2 pathway. Cell Signal.
24:2369–2377. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Collins MA and di Magliano M Pasca: Kras
as a key oncogene and therapeutic target in pancreatic cancer.
Front Physiol. 4:4072013.PubMed/NCBI
|
|
44
|
Sundaram MV: RTK/Ras/MAPK signaling.
WormBook. 1–19. 2006.PubMed/NCBI
|