Introduction
Hepatocellular carcinoma (HCC) is a common malignant
cancer of the digestive system, resulting from interactions between
the environment and the human genome (1). Epidemiological studies have shown that
40–50% of cases of HCC worldwide each year are in China, and it
represents the second leading cause of cancer-associated mortality
(2,3).
The majority of patients with HCC are diagnosed in the middle or
late stages of the disease and have poor overall survival rates.
Although the curative effect of comprehensive treatment for HCC
based on surgery has improved substantially, clinical cure rates
and long-term survival rates for HCC remain low (4,5). In
addition, 60–70% of patients with HCC have recurrence or metastasis
within 5 years following cancer resection (6).
Due to intensive investigations of various types of
cancer, an increasing number of cancer-associated genes have been
found, including polycomb group (PcG) protein family members. PcG
proteins were first identified in the developmental study of
Drosophila (7), with functions
in chromatin modification, gene transcription and carcinogenesis
(8,9).
As a member of the PcG family, Ring1 and YY1 binding protein (RYBP)
is a transcriptional repressor, and has been implicated in
embryonic development, chronic rhinosinusitis, apoptosis and cancer
(10–13). Previous studies have shown that RYBP
can interact with multiple apoptotic proteins to promote tumor
apoptosis (14). RYBP inhibits mouse
double minute 2 homolog-mediated p53 proteasome degradation, which
is important in maintaining p53 stability (14). In addition, RYBP can be induced by a
variety of antitumor drugs and compounds, including etoposide and
LAQ824 (15), to synergistically
facilitate tumor necrosis factor α and induce the apoptosis of
tumor cells (13). A previous study
found that RYBP was downregulated in patients with cervical cancer
due to the lack of chromosome 3p13 (16). Low expression levels of RYBP in
cervical cancer tissues had an effect on drug treatment effect and
patient prognosis (17). In prostate
cancer, abnormal RYBP is involved in transmembrane protease, serine
2-ETS-related gene fusion, and is associated with the prognosis of
patients (18,19). However, the expression and function of
RYBP in HCC remains to be fully elucidated.
Invasion and metastasis are important biological
characteristics of HCC. As a critical process in the development of
malignant tumor cells from epithelial cells, epithelial-mesenchymal
transition (EMT) is a well-known early marker of tumor invasion and
metastasis (20,21). The predominant features of EMT include
loss of the E-cadherin/catenin complex, keratin cytoskeleton
transformation for vimentin and the morphological characteristics
of mesenchymal cells. Through the EMT process, epithelial cells
lose polarity, obtain the ability to invade, inhibit apoptosis and
degrade extracellular matrix (22).
The expression and function of EMT-associated transcription factors
are important for further understanding the role of EMT in
regulating the malignant biological behavior of HCC. The Zinc
finger E-box binding homeobox (ZEB) family is found in the early
embryonic developmental process, and its family members include
ZEB1 and ZEB2. Studies have shown that ZEB1 is important in the
development of colon cancer, prostate cancer, lung cancer,
endometrial cancer and other types of invasive cancer (23,24). ZEB2
is similar to ZEB1, and high expression levels of ZEB2 can promote
the expression of mesenchymal proteins to obtain a mesenchymal
phenotype, inducing the occurrence of tumor EMT (25). However, whether RYBP is involved in
the EMT process in HCC via an association with ZEB1 or ZEB2 remains
to be elucidated.
The aim of the present study was to investigate the
possible role of RYBP in HCC carcinogenesis. The results
demonstrated that RYBP was downregulated in HCC and affected the
survival rates of patients with HCC via an association with the
EMT-associated factors, ZEB1 and ZEB2.
Materials and methods
Patients and specimens
The present study was approved by the ethics
committee of Guilin Medical University (Guilin, China), and written
informed consent was obtained from each patient involved in the
study. A total of 20 paired cancerous and matched adjacent normal
tissues were collected from patients with HCC undergoing
hepatectomy at the Affiliated Hospital of Guilin Medical University
between 2012 and 2014. The tissues were snap-frozen in liquid
nitrogen and stored at −80°C following surgery for reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blot analyses. Another 216 paired paraffin-embedded HCC
samples for use in immunohistochemical analysis, were collected
between 2010 and 2014 and obtained from the Affiliated Hospital of
Guilin Medical University and Zhengzhou People's Hospital
(Zhengzhou, China). The tissues were prepared into a tissue
microarray chip by Guilin Fanpu Biological Technology Co., Ltd.
(Guilin, China). The survival rates were calculated from the date
of surgery to the date the patient succumbed to morality or the
last follow-up. Medical details, including age, tumor size and
serum level of α-fetoprotein, were collected from the medical
records of each patient. Tumor staging was performed according to
the World Health Organization standards (26), and histological tumor grading was
based on Edmondson-Steiner classification (27).
RT-qRCR analysis
Fozen tissue samples were pulverized by mortar and
pestle in liquid nitrogen. Then, ice-cold TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) was added to the
powdered tissues, which were subsequently transferred to Eppendorf
tubes (Eppendorf, Hamburg, Germany) on ice for RNA extraction.
Total RNA was extracted and purified from 20 pairs of fresh frozen
HCC tissues and corresponding noncancerous tissues. The RT step was
carried out using PrimeScript™ II 1st strand cDNA Synthesis kit
(Takara Biotechnology Co., Ltd., Dalian, China). The levels of
messenger RNA (mRNA) were quantitated using SYBR® Green
Realtime PCR Master Mix (Toyobo Co., Ltd., Osaka, Japan) and
analyzed in a ViiA™ 7 Real-Time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The primers used for RT-qPCR
analysis were purchased from Invitrogen (Thermo Fisher Scientific,
Inc.) and were as follows: RYBP, forward 5′-TCGCAACTTCCATTGATT-3′
and reverse 5′-TCACACTCAGTCATACCT-3′; GAPDH, forward
5′-TCGCAACTTCCATTGATT-3′ and reverse 5′-TCACACTCAGTCATACCT-3′. The
amplification conditions consisted of the following: 30 min at 42°C
for reverse trancsription and 2 min at 94°C for Taq activation,
followed by 35 cycles of 94°C for 20 sec, 58°C for 20 sec and
elongation at 72°C for 30 sec. GAPDH was used as the internal
control for determining the mRNA expression of RYBP. Fluorescent
data were converted into quantification cycle (Cq). ΔCq values of
each sample were calculated as Cq gene of interest - Cq GAPDH. A
ΔCq value of 3.33 corresponds to a magnitude lower of gene
expression compared with that of GAPDH. The experiment was
performed in triplicate. The results were normalized with
respective internal controls.
Western blot analysis
The homogenized HCC samples were lysed in
radioimmunoprecipitation assay lysis buffer, containing 150 mmol/l
NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS and 50 mmol/l
Tris (pH 7.4), and the lysates were harvested by centrifugation at
16,000 × g at 4°C for 30 min. The concentration of protein
was determined by BCA Protein Assay kit (Beyotime Institute of
Biotechnology, Haimen, China). Subsequently, 20 µg of protein was
separated by electrophoresis on a 12% sodium dodecyl sulfate
polyacrylamide gel and transferred onto a polyvinylidene fluoride
membrane. Following blocking of nonspecific binding sites for 60
min with 5% nonfat milk, the membranes were incubated overnight at
4°C with anti-rabbit polyclonal antibody against RYBP (GR104527-2;
Abcam, Cambridge, MA, USA) at a 1:1,000 dilution. The membranes
were then washed three times with Tris-buffered saline with
Tween-20 (TBST) for 10 min and were probed with an anti-rabbit
immunoglobulin G (IgG) antibody (GGHL-15PXSPP; Immunology
Consultants Laboratory, Portland, OR, USA) at a 1:1,000 dilution at
room temperature for 1 h. Following three washes with TBST, the
membranes were developed using an enhanced chemiluminescence system
(Cell Signaling Technology, Inc., Danvers, MA, USA). The band
intensity was measured by densitometry using Quantity One software
version 4.62 (Bio-Rad Laboratories Inc., Hercules, CA, USA). The
protein level of RYBP was normalized to the level of β-actin,
detected using a mouse monoclonal antibody against β-actin (KC5A08;
Kangchen Biotech, Shanghai, China) overnight at 4°C at a 1:10,000
dilution.
Immunohistochemical analysis and
scoring
Formalin-fixed, paraffin-embedded tissue blocks were
used for immunohistochemical analysis to detect the expression of
RYBP, ZEB1 and ZEB2 using standard methods. The tissues were first
fixed with 10% formalin at 4°C for 24 h. The paraffin-embedded
tissue sections (6 µm thick) previously constructed into a tissue
microarray chip were deparaffinized and quenched for endogenous
peroxidase activity with methanol and 3% hydrogen peroxide for 15
min. The sections were then processed in 10 mmol/l citrate buffer
(pH 6.0) and heated at 120°C for 5 min to retrieve the antigen. The
sections were incubated for at 37°C 1 h with anti-RYBP (goat
anti-rabbit polyclonal antibody; 1:100 dilution), anti-ZEB1 (goat
anti-rabbit polyclonal antibody; ab124512; 1:100 dilution; Abcam)
and anti-ZEB2 antibodies (goat anti-rabbit polyclonal antibody;
1:100 dilution; ab138222; Abcam) diluted 1:100 in 1% bovine serum
albumin (BSA) (Beyotime Institute of Biotechnology). As a negative
control, sections were incubated with 1% BSA/PBS without primary
antibody. The sections were then washed with PBS for 5 min and
incubated with the anti-rabbit IgG antibody (1:1,000 dilution;
Immunology Consultants Laboratory) at 37°C for 15 min. Following
rinsing in PBS, the reaction was visualized under a light
microscope by incubating the sections with diaminobenzidine
solution for 15 min, following which the sections were weakly
counterstained with hematoxylin. All the immunostained sections
were evaluated in a blinded-manner with no knowledge of the
clinicopathological information. For the assessment of RYBP, ZEB1
and ZEB2, five fields in each specimen were randomly selected and
>500 cells were counted under a microscope (Olympus Corporation,
Tokyo, Japan) to determine the mean percentage of immunostained
cells relative to the total number of cells. The positive cell
staining percentages were scored into four categories: 0 for 0%, 1
for 1–33%, 2 for 34–66% and 3 for 67–100% staining. The
immunohistochemical staining intensities were also scored into four
grades (0, 1, 2 and 3), according to the brown color intensity of
the cells: 0 for no color, 1 for light color, 2 for medium color
and 3 for dark brown color. The sum of the percentage and intensity
scores was used as the final RYBP, ZEB1 and ZEB2 staining score.
The staining scores were defined as low expression for scores of
0–2 and high expression for scores of 3–6.
Statistical analysis
All statistical analyses were performed using SPSS
version 19.0 (IMB SPSS, Armonk, NY, USA). The paired-samples-test
and one-way analysis of variance were used to compare the mRNA
expression of RYBP normalized to GAPDH between the HCC and
corresponding adjacent non-tumor tissues, whereas the χ2
test was applied for the comparison of dichotomous variables. The
Kaplan-Meier estimate was used for survival analysis, and the
log-rank test was selected to compare the cumulative survival
durations in the patients. P<0.05 was considered to indicate a
statistically significant difference.
Results
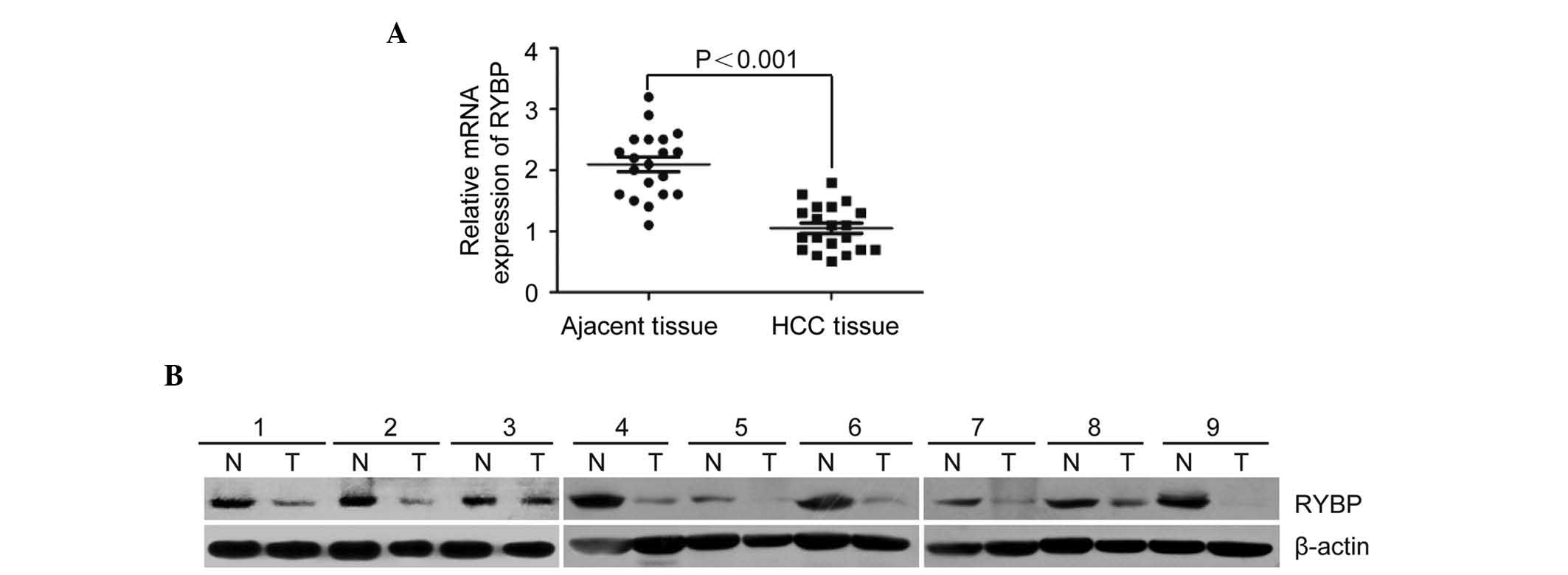
Expression of RYBP is low in HCC
tissues
To investigate the expression profile of RYBP in
HCC, the present study performed RT-qPCR and western blot analyses
in 20 pairs of HCC tissues and matched adjacent non-tumor tissues.
The mRNA expression levels of RYBP was markedly lower in the HCC
samples, compared with the high levels of expression in the matched
adjacent tissues (P=0.012; Fig.
1A).
Consistent with the RT-qPCR data, the protein
expression of RYBP was also low in 16 of the 20 (80.0%) paired
tissue samples, determined using western blot analysis (Fig. 1B). The protein level of RYBP was
significantly lower in the HCC tissues, compared with that in the
adjacent non-tumor tissues (P<0.01).
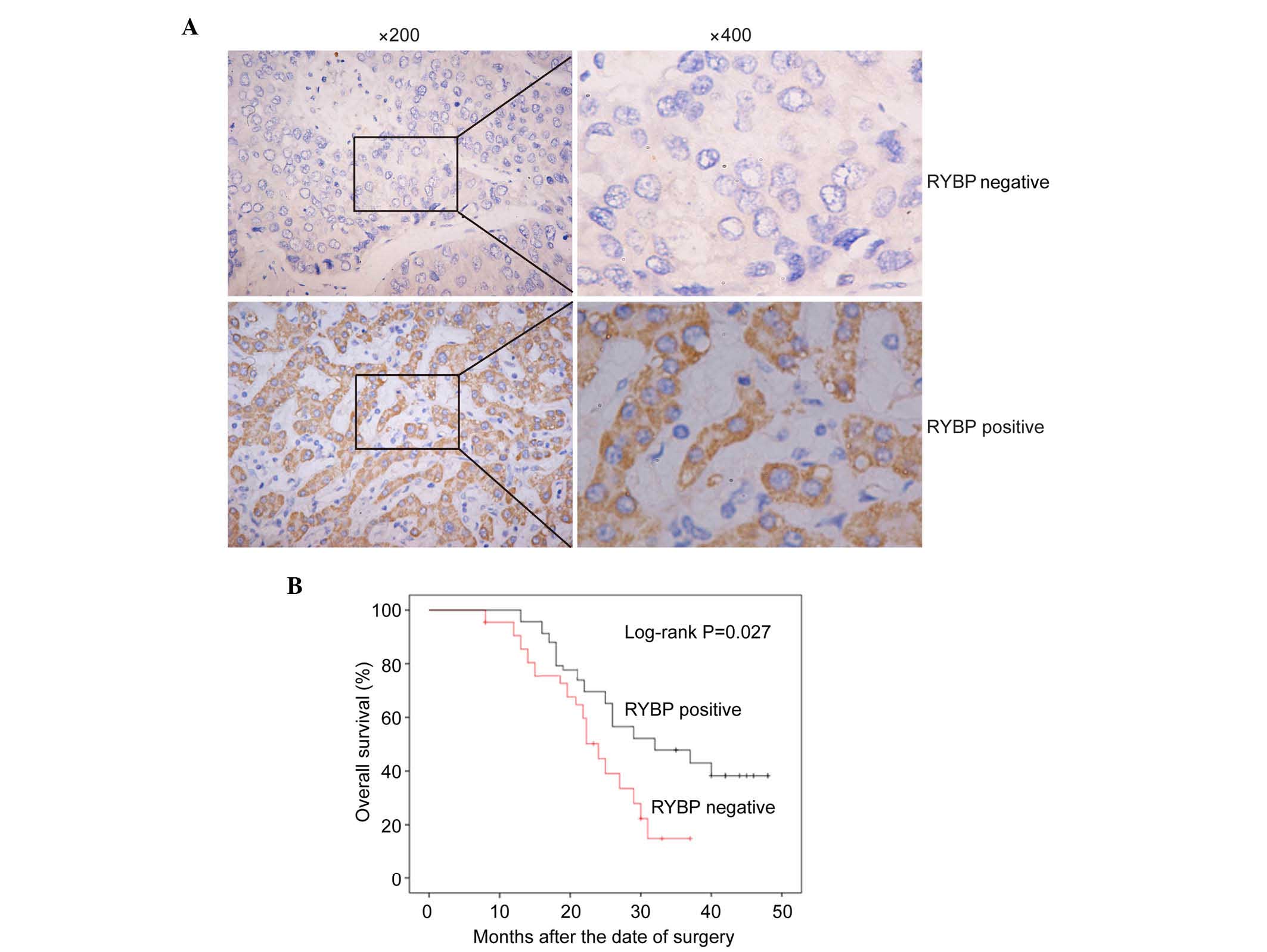
Association between low protein
expression levels of RYBP and the clinicopathological
characteristics and prognosis of HCC
Immunohistochemical analysis was then performed in
all 216 archival paraffin-embedded HCC samples. In total, 60 of the
216 (27.8%) cases were positive for the expression of RYBP in the
cancerous tissues, whereas 156 of the 216 (72.2%) cases were
negative for the expression of RYBP (Fig.
2A; P<0.01). The association between the protein expression
levels of RYBP and the clinicopathological characteristics of RYBP
was determined using the χ2 test. As shown in Table I, the negative expression of RYBP was
significantly associated with tumor size (P=0.046) and metastasis
(P=0.028), which suggested that the expression of RYBP was
correlated with the diagnosis and prognosis of HCC.
 | Table I.Association between the expression of
RYBP and clinicopathological features of hepatocellular
carcinoma. |
Table I.
Association between the expression of
RYBP and clinicopathological features of hepatocellular
carcinoma.
|
| Expression of RYBP
(n) |
|
|
|---|
|
|
|
|
|
|---|
| Variable | Positive | Negative | χ2
value | P-value |
|---|
| Gender |
|
|
| 0.311 |
|
Male | 43 | 122 | 1.027 |
|
|
Female | 17 | 34 |
|
|
| Age (years) |
|
|
| 0.380 |
|
≥50 | 33 | 96 | 0.770 |
|
|
<50 | 27 | 60 |
|
|
| Tumor size
(cm) |
|
|
| 0.046 |
| ≤5 | 42 | 87 | 3.648 |
|
|
>5 | 18 | 69 |
|
|
| Tumor grade |
|
|
| 0.262 |
| I | 23 | 73 | 1.257 |
|
|
II+III | 37 | 83 |
|
|
| Tumor stage |
|
|
| 0.303 |
|
I+II | 32 | 71 | 1.062 |
|
|
III+IV | 28 | 85 |
|
|
| Tumor number |
|
|
| 0.076 |
| 1 | 21 | 81 | 4.979 |
|
| ≥2 | 39 | 75 |
|
|
| Metastasis |
|
|
| 0.028 |
|
Yes | 25 | 91 | 4.841 |
|
| No | 35 | 65 |
|
|
| α-fetoprotein
(ng/ml) |
|
|
| 0.073 |
|
≥400 | 27 | 94 | 4.094 |
|
|
<400 | 33 | 62 |
|
|
To determine the prognostic value of RYBP in HCC,
the present study also assessed the association between the
expression of RYBP and survival rates using Kaplan-Meier analysis
with a log-rank test. As shown in Fig.
2B, the survival rates of the patients with HCC were
significantly different between cases positive for the expression
of RYBP and cases negative for the expression of RYBP (P=0.027). In
patients with HCC, a low expression level of RYBP indicated a
poorer prognosis, compared with those with a high expression level
of RYBP.
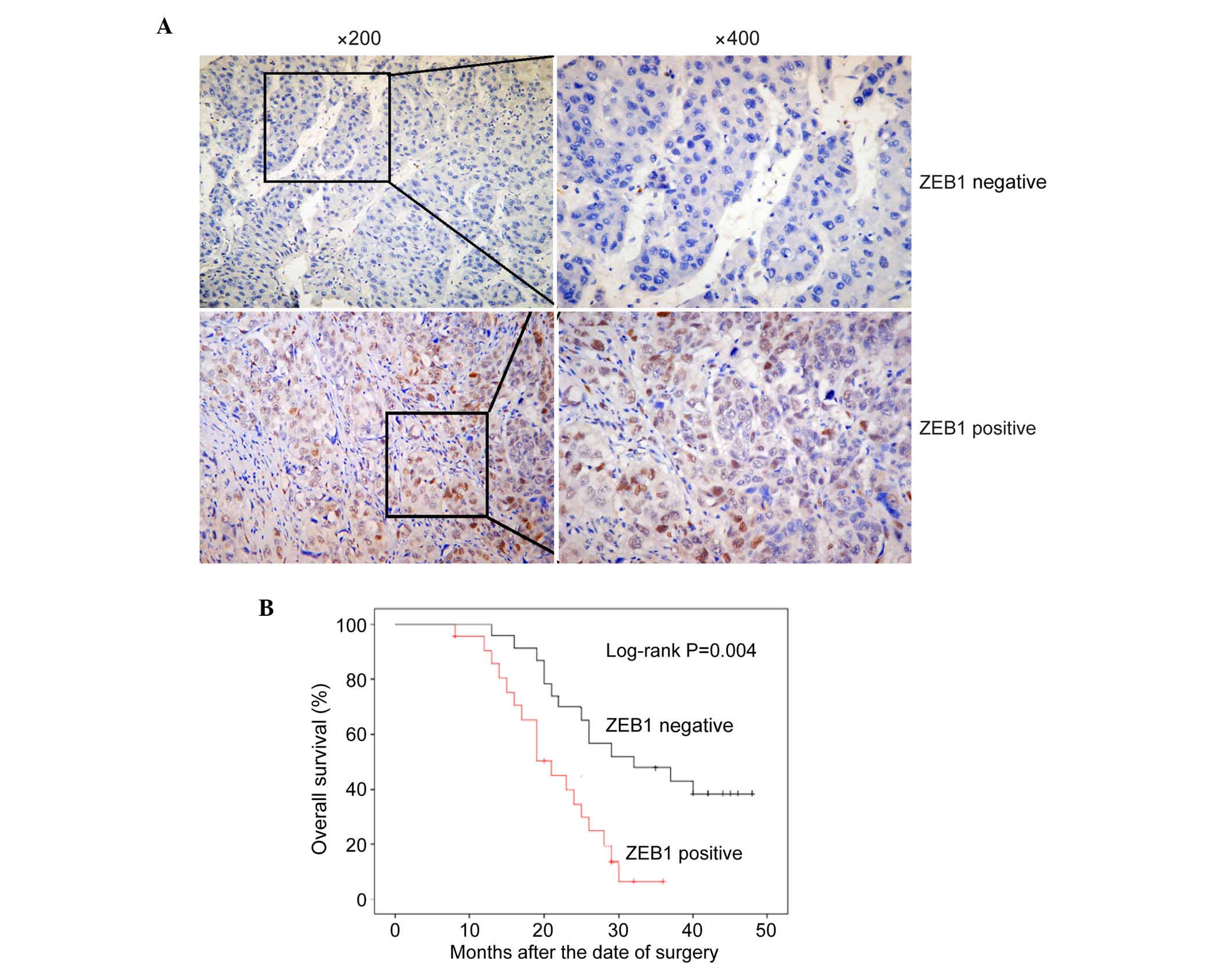
Association between the protein
expression of ZEB1 and the clinicopathological features of HCC
As there was a significant correlation between RYBP
and HCC metastasis, the present study aimed to investigate the role
of RYBP in EMT. A number of well-known EMT markers were selected
and their expression was detected in HCC tissues using
immunohistochemistry. ZEB1 was the first EMT marker assessed, which
represses the E-cadherin promoter and induces EMT by recruiting
SMARCA4/BRG1 (28). In all 216
archival paraffin-embedded HCC samples, 159 of 216 (73.6%) cases
were positive for the expression of ZEB1, whereas 57 of 216 (26.4%)
cases were negative for the expression of ZEB1 (Fig. 3A; P<0.01).
To determine the correlation between ZEB1 and the
clinicopathological characteristics and prognosis of HCC, a
χ2 test and Kaplan-Meier analysis with a log-rank test
were used. As shown in Table II, the
expression of ZEB1 was significantly associated with clinical stage
(P=0.035) and metastasis (P=0.008). As shown in Fig. 3B, the survival rates of patients with
HCC were significantly different between those positive for the
expression of ZEB1 and those negative for the expression of ZEB1
(P<0.001). In patients with HCC, the group with high expression
levels of ZEB1 had shorter survival rates, compared with the group
expressing low levels of ZEB1. These results indicated that ZEB1 is
a suitable EMT marker for HCC.
 | Table II.Association between the expression of
ZEB1 and clinicopathological features of hepatocellular
carcinoma. |
Table II.
Association between the expression of
ZEB1 and clinicopathological features of hepatocellular
carcinoma.
|
| Expression of ZEB1
(n) |
|
|
|---|
|
|
|
|
|
|---|
| Variable | Positive | Negative | χ2
value | P-value |
|---|
| Gender |
|
|
| 0.198 |
|
Male | 125 | 40 | 1.657 |
|
|
Female | 34 | 17 |
|
|
| Age (years) |
|
|
| 0.203 |
|
≥50 | 99 | 30 | 1.618 |
|
|
<50 | 60 | 27 |
|
|
| Tumor size
(cm) |
|
|
| 0.113 |
| ≤5 | 100 | 29 | 2.518 |
|
|
>5 | 59 | 28 |
|
|
| Tumor grade |
|
|
| 0.098 |
| I | 76 | 20 | 2.746 |
|
|
II+III | 83 | 37 |
|
|
| Tumor stage |
|
|
| 0.035 |
|
I+II | 69 | 34 | 4.443 |
|
|
III+IV | 90 | 23 |
|
|
| Tumor number |
|
|
| 0.116 |
| 1 | 70 | 32 | 2.471 |
|
| ≥2 | 89 | 25 |
|
|
| Metastasis |
|
|
| 0.008 |
|
Yes | 94 | 22 | 7.108 |
|
| No | 65 | 35 |
|
|
| α-fetoprotein
(ng/ml) |
|
|
| 0.065 |
|
≥400 | 95 | 26 | 3.402 |
|
|
<400 | 64 | 31 |
|
|
Statistical analysis was also performed to examine
the association between the expression of RYBP and ZEB1 in the HCC
tissues. As shown in Table III, the
expression of RYBP was negatively correlated with the expression of
ZEB1 in HCC tissues (r=−0.473; P<0.001). These results suggested
that a low level of RYBP may promote EMT in HCC.
 | Table III.Correlation between the expression of
RYBP and ZEB1 in hepatocellular carcinoma tissues. |
Table III.
Correlation between the expression of
RYBP and ZEB1 in hepatocellular carcinoma tissues.
|
| ZEB1 expression
(n) |
|
|
|
|---|
|
|
|
|
|
|
|---|
| RYBP
expression | Low | High | χ2
value | r-value | P-value |
|---|
| Low | 21 | 135 | 48.315 | −0.473 |
<0.001 |
| High | 36 | 24 |
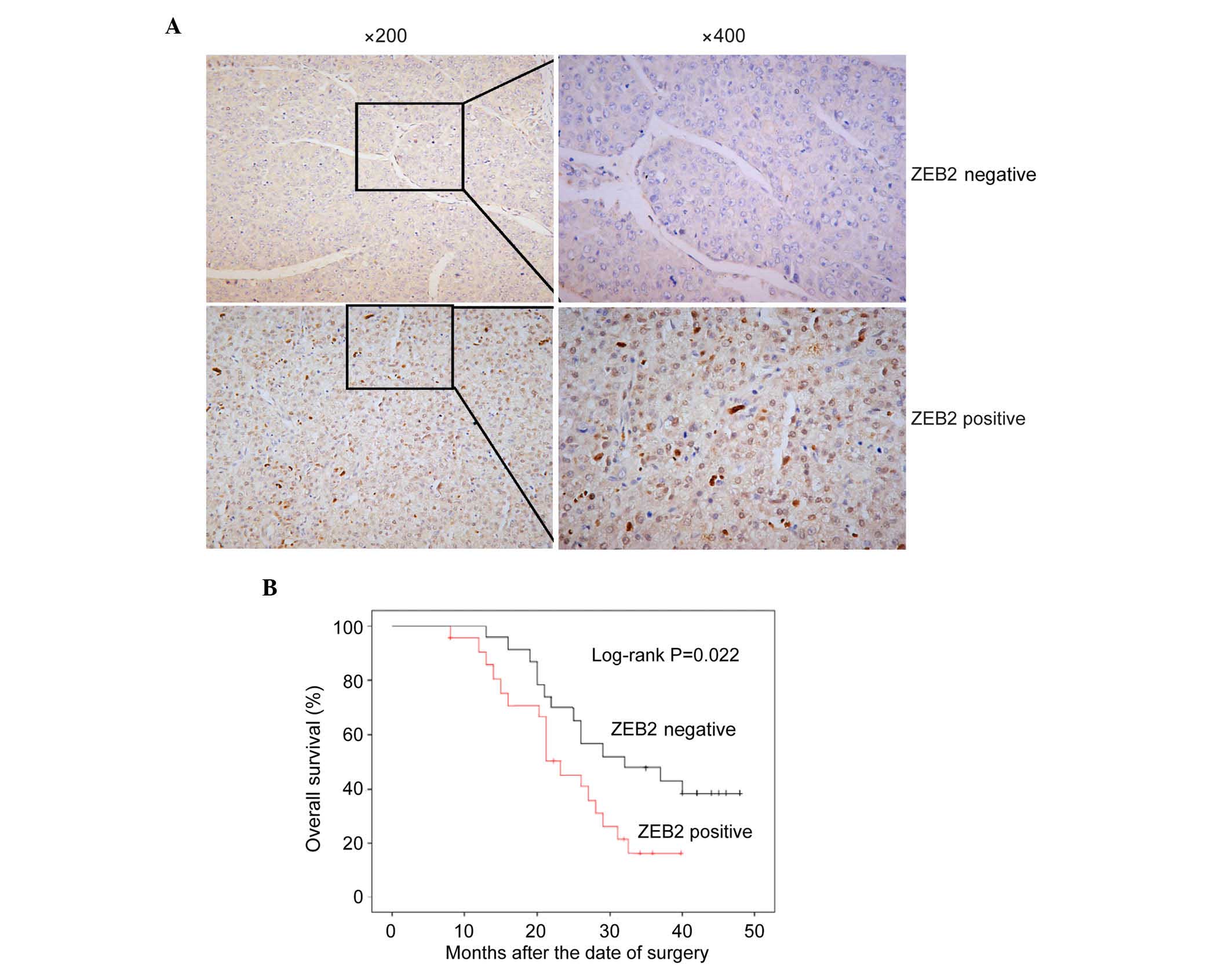
Association between the protein
expression of ZEB2 with the clinicopathological features of
HCC
As with ZEB1, ZEB2 is a transcription inhibitor of
E-cadherin implicated in gastric cancer (29,30). In
the 216 archival paraffin-embedded HCC samples, 135 of the 216
(62.5%) cases were positive for the expression of ZEB2 in cancerous
tissues, whereas 81 of the 216 (37.5%) cases showed low expression
levels of ZEB2 (Fig. 4A; P<0.01).
The correlation between the expression of ZEB2 and the
clinicopathological characteristics and prognosis of HCC were
examined using a χ2 test and Kaplan-Meier analysis. As
shown in Table IV, the positive
expression of ZEB2 was significantly associated with metastasis
(P=0.008). The log-rank test showed that the survival rate of
patients with HCC in the ZEB2-positive expression group was
significantly shorter, compared with that in the ZEB2-negative
expression group (Fig. 4B;
P<0.001). As with ZEB1, a significant correlation was found
between the expression of ZEB2 and the metastasis and prognosis of
HCC.
 | Table IV.Association between the expression of
ZEB2 and clinicopathological features of hepatocellular
carcinoma. |
Table IV.
Association between the expression of
ZEB2 and clinicopathological features of hepatocellular
carcinoma.
|
| ZEB2 (n) |
|
|
|---|
|
|
|
|
|
|---|
| Variable | Positive | Negative | χ2
value | P-value |
|---|
| Gender |
|
|
| 0.172 |
|
Male | 99 | 66 | 1.864 |
|
|
Female | 36 | 15 |
|
|
| Age (years) |
|
|
| 0.210 |
|
≥50 | 85 | 44 | 1.572 |
|
|
<50 | 50 | 37 |
|
|
| Tumor size
(cm) |
|
|
| 0.076 |
| ≤5 | 89 | 40 | 5.760 |
|
|
>5 | 46 | 41 |
|
|
| Tumor grade |
|
|
| 0.258 |
| I | 56 | 40 | 1.280 |
|
|
II+III | 79 | 41 |
|
|
| Tumor stage |
|
|
| 0.062 |
|
I+II | 71 | 32 | 3.475 |
|
|
III+IV | 64 | 49 |
|
|
| Tumor number |
|
|
| 0.139 |
| 1 | 69 | 33 | 2.185 |
|
| ≥2 | 66 | 48 |
|
|
| Metastasis |
|
|
| 0.007 |
|
Yes | 82 | 34 | 7.170 |
|
| No | 53 | 47 |
|
|
| α-fetoprotein
(ng/ml) |
|
|
| 0.071 |
|
≥400 | 82 | 39 | 3.258 |
|
|
<400 | 53 | 42 |
|
|
Statistical analysis of the association between the
expression of RYBP and ZEB2 also showed a negative correlation
(r=−0.416; P<0.001; Table V).
Taken together, RYBP was negatively correlated with ZEB1 and ZEB2,
suggesting the importance of RYBP in the EMT process in HCC.
 | Table V.Correlation between the expression of
RYBP and the expression of ZEB2 in hepatocellular carcinoma
tissues. |
Table V.
Correlation between the expression of
RYBP and the expression of ZEB2 in hepatocellular carcinoma
tissues.
|
| ZEB2
expression |
|
|
|
|---|
|
|
|
|
|
|
|---|
| RYBP | Low | High | χ2
value | r value | P-value |
|---|
| Low | 39 | 117 | 37.44 | −0.416 |
<0.001 |
| High | 42 | 18 |
Discussion
RYBP is a conserved alkaline protein, which is
composed of 228 amino acid residues, and contains a zinc finger
structure at the amino terminal, a lysine-rich middle region and a
serine/threonine-rich carboxyl terminal. When interacting with DNA
or with other proteins, the conformational structure of RYBP is
altered and, due to this specific structure, RYBP combines and
regulates other members of the PcG family, including Ring1 (Ring1A
and Ring1B), YY1 and M33 (31).
Several studies have indicated that RYBP is closely
associated with various types of cancer, however, the expression of
RYBP in cancer remains controversial. In cervical cancer and
prostate cancer, the expression of RYBP was found to decrease
following 3p13 deletion (16,18), where the coding gene of human RYBP
protein is located. The downregulation of RYBP led to a decrease in
the survival rates of patients with cervical cancer and prostate
cancer. However, an abnormal increase in the expression level of
RYBP has been reported in acute leukemia (32). In addition, the expression levels of
RYBP in paired tumor and non-tumor samples have been investigated
using immunohistochemical methods, and 10% of cancer cases were
found positive for RYBP, predominantly in oligodendroglial tumors,
pituitary adenoma, Hodgkin's lymphoma and T cell lymphoma (33). Wang et al (34) found that the overexpression of RYBP
inhibited tumor cell growth and migration, induced apoptosis and
increased the chemical sensitivity of cells, whereas the knockout
of RYBP led to the opposite result (34). In the present study, it was shown that
the expression of RYBP was low in HCC, in accordance with previous
studies in liver and lung cancer (34). The present study also analyzed the
correlation between the expression of RYBP and the prognosis of
patients with HCC, and found that the negative expression of RYBP
indicated a poor prognosis in patients with HCC. The results of
previous studies and those of the present study suggested that RYBP
can be used to predict the prognosis of patients with HCC and
provide an effective means of treatment.
In addition to RYBP, the present study detected that
the EMT-associated factors, ZEB1 and ZEB2, were overexpressed in
HCC tissues. ZEB1 and ZEB2 are reported to inhibit the
transcription of the epithelial marker E-cadherin to mediate the
EMT process in tumors (35–37). ZEB1 is detected in a variety of
tissues, and is important in the formation and differentiation of
skeletal muscle and T lymphocytes (38,39). The
knockdown of ZEB1 not only restores the expression of E-cadherin in
dedifferentiated and metastatic tumors, but also causes the
reconstruction of epithelial function, including tight junctions
(40). In addition, mutation of ZEB1
has been shown to lead to loss of the mesenchymal marker vimentin
in mouse mesenchymal cells, resulting in a variety of abnormal
functions in mouse embryos (41). It
has been shown that ZEB1 is involved in the invasion and metastasis
of tumor cells. ZEB1 was found to be expressed at high levels in
lung squamous cell carcinoma, particularly in patients positive for
lymph node and distant metastases. When ZEB1 was silenced, the
invasive and metastatic ability of the tumor cells was
significantly inhibited, suggesting that ZEB1 promoted invasion and
metastasis in lung squamous cell carcinoma (42). In addition, ZEB1 has been found to be
upregulated in cervical cancer and breast cancer, and is correlated
with clinical staging, lymph node metastasis and tumor
differentiation (35,43), which indicates it is an important
biological indicator for predicting the invasion and metastasis of
various types of cancer.
As with ZEB1, several studies have demonstrated that
ZEB2 is also important in the regulation of EMT. The overexpression
of ZEB2 combined to the E2 box of the E-cadherin promoter reduces
the function of E-cadherin on tumor epithelial cells, and induces
cells more susceptible to the formation of invasive and metastatic
behavior (44,45). The expression of ZEB2 in breast cancer
is associated with poor prognosis, indicating that ZEB2 may be a
marker for EMT and myoepithelial loss in breast cancer (46). In addition, the overexpression of ZEB2
in HCC increases the RNA level of matrix metalloproteinase-2
(47), which can reduce the level of
type IV collagen. Therefore, ZEB2 can be considered to be closely
associated with tumor invasion and metastasis.
In the present study, ZEB1 and ZEB2 were found to be
associated with the occurrence of distant metastasis in patients
with HCC, as with RYBP. Therefore, the associations between the
expression level of RYBP and the expression levels of ZEB1 and ZEB2
were examined in HCC tissues. The results confirmed that the
expression of RYBP in HCC was negatively correlated with the
expression of ZEB1 and ZEB2; when the expression of RYBP was
downregulated, the expression of ZEB1 and ZEB2 increased,
suggesting the involvement of RYBP in the regulation of the tumor
EMT process.
Taken together, the results of the present study
showed that the negative expression of RYBP promoted the invasion
and metastasis of HCC. Furthermore, the expression of RYBP in HCC
was associated with the presence of ZEB1 and ZEB2, suggesting that
RYBP may be involved in the process of EMT. Therefore, RYBP offers
potential as a biomarker for the diagnosis and prognosis of HCC in
the future.
Acknowledgements
This study was supported by the National Nature
Science Foundation of China (grant nos. 81460515 and 81160359) and
the Scientific Research Project of Guangxi Universities and
Colleges (grant no. KY2015ZD088).
References
|
1
|
Su Y, Zhao B, Guo F, Bin Z, Yang Y, Liu S,
Han Y, Niu J, Ke X, Wang N, et al: Interaction of benzo[a]pyrene
with other risk factors in hepatocellular carcinoma: A case-control
study in Xiamen, China. Ann Epidemiol. 24:98–103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen M, Therneau T, Orsini LS and Qiao YL:
Design and rationale of the hcc bridge study in China: A
longitudinal, multicenter cohort trial in hepatocellular carcinoma.
BMC Gastroenterol. 11:532011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu Q, Li N, Zeng X, Han Q, Li F, Yang C,
Lv Y, Zhou Z and Liu Z: Hepatocellular carcinoma in a large medical
center of China over a 10-year period: Evolving therapeutic option
and improving survival. Oncotarget. 6:4440–4450. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu L, Miao R, Yang H, Lu X, Zhao Y, Mao
Y, Zhong S, Huang J, Sang X and Zhao H: Prognostic factors after
liver resection for hepatocellular carcinoma: A single-center
experience from China. Am J Surg. 203:741–750. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee SC, Tan HT and Chung MC: Prognostic
biomarkers for prediction of recurrence of hepatocellular
carcinoma: Current status and future prospects. World J
Gastroenterol. 20:3112–3124. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Francis NJ and Kingston RE: Mechanisms of
transcriptional memory. Nat Rev Mol Cell Biol. 2:409–421. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Koppens M and van Lohuizen M:
Context-dependent actions of polycomb repressors in cancer.
Oncogene. 35:1341–1352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khan AA, Lee AJ and Roh TY: Polycomb group
protein-mediated histone modifications during cell differentiation.
Epigenomics. 7:75–84. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bejarano F, González I, Vidal M and
Busturia A: The drosophila RYBP gene functions as a
polycomb-dependent transcriptional repressor. Mech Dev.
122:1118–1129. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pirity MK, Locker J and Schreiber-Agus N:
Rybp/DEDAF is required for early postimplantation and for central
nervous system development. Mol Cell Biol. 25:7193–7202. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Endam LM, Filali-Mouhim A, Zhao
L, Desrosiers M, Han D and Zhang L: Polymorphisms in RYBP and AOAH
genes are associated with chronic rhinosinusitis in a Chinese
population: A replication study. PLoS One. 7:e392472012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Novak RL and Phillips AC:
Adenoviral-mediated RYBP expression promotes tumor cell-specific
apoptosis. Cancer Gene Ther. 15:713–722. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen D, Zhang J, Li M, Rayburn ER, Wang H
and Zhang R: RYBP stabilizes p53 by modulating MDM2. EMBO Rep.
10:166–172. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Scott GK, Mattie MD, Berger CE, Benz SC
and Benz CC: Rapid alteration of microRNA levels by histone
deacetylase inhibition. Cancer Res. 66:1277–1281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lando M, Wilting SM, Snipstad K, Clancy T,
Bierkens M, Aarnes EK, Holden M, Stokke T, Sundfør K, Holm R, et
al: Identification of eight candidate target genes of the recurrent
3p12-p14 loss in cervical cancer by integrative genomic profiling.
J Pathol. 230:59–69. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lando M, Holden M, Bergersen LC, Svendsrud
DH, Stokke T, Sundfør K, Glad IK, Kristensen GB and Lyng H: Gene
dosage, expression, and ontology analysis identifies driver genes
in the carcinogenesis and chemoradioresistance of cervical cancer.
PLoS Genet. 5:e10007192009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Krohn A, Seidel A, Burkhardt L, Bachmann
F, Mader M, Grupp K, Eichenauer T, Becker A, Adam M, Graefen M, et
al: Recurrent deletion of 3p13 targets multiple tumour suppressor
genes and defines a distinct subgroup of aggressive ERG
fusion-positive prostate cancers. J Pathol. 231:130–141. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Feik E, Schweifer N, Baierl A,
Sommergruber W, Haslinger C, Hofer P, Maj-Hes A, Madersbacher S and
Gsur A: Integrative analysis of prostate cancer aggressiveness.
Prostate. 73:1413–1426. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lindsey S and Langhans SA: Crosstalk of
oncogenic signaling pathways during epithelial-mesenchymal
transition. Front Oncol. 4:3582014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mitra A, Mishra L and Li S: EMT, CTCs and
CSCs in tumor relapse and drug-resistance. Oncotarget.
6:10697–10711. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gregory PA, Bracken CP, Smith E, Bert AG,
Wright JA, Roslan S, Morris M, Wyatt L, Farshid G, Lim YY, et al:
An autocrine TGF-beta/ZEB/miR-200 signaling network regulates
establishment and maintenance of epithelial-mesenchymal transition.
Mol Biol Cell. 22:1686–1698. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Z, Sun B, Qi L, Li H, Gao J and Leng
X: Zinc finger E-box binding homeobox 1 promotes vasculogenic
mimicry in colorectal cancer through induction of
epithelial-to-mesenchymal transition. Cancer Sci. 103:813–820.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kurahara H, Takao S, Maemura K, Mataki Y,
Kuwahata T, Maeda K, Ding Q, Sakoda M, Iino S, Ishigami S, et al:
Epithelial-mesenchymal transition and mesenchymal-epithelial
transition via regulation of ZEB-1 and ZEB-2 expression in
pancreatic cancer. J Surg Oncol. 105:655–661. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fritz A April, Percy C, Jack A,
Shanmugaratnam K, Sobin L, Parkin DM and Whelan S: World Health
Organization: International Classification of Diseases for
Oncology. 3rd. WHO Press; Geneva: 2000
|
|
27
|
Edmondson HA and Steiner PE: Primary
carcinoma of the liver: A study of 100 cases among 48,900
necropsies. Cancer. 7:462–503. 1954. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sánchez-Tilló E, Lázaro A, Torrent R,
Cuatrecasas M, Vaquero EC, Castells A, Engel P and Postigo A: ZEB1
represses E-cadherin and induces an EMT by recruiting the SWI/SNF
chromatin-remodeling protein BRG1. Oncogene. 29:3490–3500. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cong N, Du P, Zhang A, Shen F, Su J, Pu P,
Wang T, Zjang J, Kang C and Zhang Q: Downregulated microRNA-200a
promotes EMT and tumor growth through the wnt/β-catenin pathway by
targeting the E-cadherin repressors ZEB1/ZEB2 in gastric
adenocarcinoma. Oncol Rep. 29:1579–1587. 2013.PubMed/NCBI
|
|
30
|
Vandewalle C, Comijn J, De Craene B,
Vermassen P, Bruyneel E, Andersen H, Tulchinsky E, Van Roy F and
Berx G: SIP1/ZEB2 induces EMT by repressing genes of different
epithelial cell-cell junctions. Nucleic Acids Res. 33:6566–6578.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Garcia E, Marcos-Gutiérrez C, del Mar
Lorente M, Moreno JC and Vidal M: RYBP, a new repressor protein
that interacts with components of the mammalian polycomb complex,
and with the transcription factor YY1. EMBO J. 18:3404–3418. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sánchez-Beato M, Sánchez E, Garcia JF,
Pérez-Rosado A, Montoya MC, Fraga M, Artiga MJ, Navarrete M,
Abraira V, Morente M, et al: Abnormal PcG protein expression in
hodgkin's lymphoma. Relation with E2F6 and NFkappaB transcription
factors. J Pathol. 204:528–537. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sánchez-Beato M, Sánchez E,
González-Carreró J, Morente M, Diez A, Sánchez-Verde L, Martin MC,
Cigudosa JC, Vidal M and Piris MA: Variability in the expression of
polycomb proteins in different normal and tumoral tissues. A pilot
study using tissue microarrays. Mod Pathol. 19:684–694. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang W, Cheng J, Qin JJ, Voruganti S, Nag
S, Fan J, Gao Q and Zhang R: RYBP expression is associated with
better survival of patients with hepatocellular carcinoma (HCC) and
responsiveness to chemotherapy of HCC cells in vitro and in vivo.
Oncotarget. 5:11604–11619. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu JM, Sun W, Hua F, Xie J, Lin H, Zhou DD
and Hu ZW: BCL6 induces EMT by promoting the ZEB1-mediated
transcription repression of E-cadherin in breast cancer cells.
Cancer Lett. 365:190–200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Galván JA, Zlobec I, Wartenberg M, Lugli
A, Gloor B, Perren A and Karamitopoulou E: Expression of E-cadherin
repressors SNAIL, ZEB1 and ZEB2 by tumour and stromal cells
influences tumour-budding phenotype and suggests heterogeneity of
stromal cells in pancreatic cancer. Br J Cancer. 112:1944–1950.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cai MY, Luo RZ, Chen JW, Pei XQ, Lu JB,
Hou JH and Yun JP: Overexpression of ZEB2 in peritumoral liver
tissue correlates with favorable survival after curative resection
of hepatocellular carcinoma. PLoS One. 7:e328382012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Siles L, Sánchez-Tilló E, Lim JW, Darling
DS, Kroll KL and Postigo A: ZEB1 imposes a temporary
stage-dependent inhibition of muscle gene expression and
differentiation via CtBP-mediated transcriptional repression. Mol
Cell Biol. 33:1368–1382. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang J, Lee S, Teh CE, Bunting K, Ma L and
Shannon MF: The transcription repressor, ZEB1, cooperates with
CtBP2 and HDAC1 to suppress IL-2 gene activation in T cells. Int
Immunol. 21:227–235. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Elsum IA, Martin C and Humbert PO:
Scribble regulates an EMT polarity pathway through modulation of
MAPK-ERK signaling to mediate junction formation. J Cell Sci.
126:3990–3999. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu Y, El-Naggar S, Darling DS, Higashi Y
and Dean DC: Zeb1 links epithelial-mesenchymal transition and
cellular senescence. Development. 135:579–588. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang J, Lu C, Zhang J, Kang J, Cao C and
Li M: Involvement of ZEB1 and E-cadherin in the invasion of lung
squamous cell carcinoma. Mol Biol Rep. 40:949–956. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ran J, Lin DL, Wu RF, Chen QH, Huang HP,
Qiu NX and Quan S: ZEB1 promotes epithelial-mesenchymal transition
in cervical cancer metastasis. Fertil Steril. 103:1606–1614.e1-e2.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Koopmansch B, Berx G, Foidart JM, Gilles C
and Winkler R: Interplay between KLF4 and ZEB2/SIP1 in the
regulation of E-cadherin expression. Biochem Biophys Res Commun.
431:652–657. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dai YH, Tang YP, Zhu HY, Lv L, Chu Y, Zhou
YQ and Huo JR: ZEB2 promotes the metastasis of gastric cancer and
modulates epithelial mesenchymal transition of gastric cancer
cells. Dig Dis Sci. 57:1253–1260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Si W, Huang W, Zheng Y, Yang Y, Liu X,
Shan L, Zhou X, Wang Y, Su D, Gao J, et al: Dysfunction of the
reciprocal feedback loop between GATA3- and ZEB2-nucleated
repression programs contributes to breast cancer metastasis. Cancer
Cell. 27:822–836. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang Z, Sun B, Li Y, Zhao X, Zhao X, Gu Q,
An J, Dong X, Liu F and Wang Y: ZEB2 promotes vasculogenic mimicry
by TGF-β1 induced epithelial-to-mesenchymal transition in
hepatocellular carcinoma. Exp Mol Pathol. 98:352–359. 2015.
View Article : Google Scholar : PubMed/NCBI
|