Introduction
Bladder cancer is the most common urological
malignancy among human urothelial cell carcinomas (UCCs) and
accounts for ~90% of all bladder cancers (1). The prognosis of patients with
non-invasive bladder cancer is generally favorable, whereas
patients with invasive bladder cancer typically show postoperative
distant metastasis or local recurrence following a radical
cystectomy (2). Feline
sarcoma-related protein (Fer) is a 94-kDa non-receptor protein
tyrosine kinase, which was shown to reside in the cytoplasm and
nucleus of mammalian cells (3).
Previous studies have reported Fer activation or upregulation in
numerous cancers, including renal (4), hepatic (5), prostate (6), and triple negative breast cancer
(7). Furthermore, it has been
reported that Fer is able to modulate cell migration and invasion
in numerous cell types (8). However,
the biological role of Fer in bladder UCC has yet to be defined,
and the molecular mechanisms underlying Fer-mediated cell migration
and invasion remain unclear.
Metastasis is commonly associated with the
progression of malignancy, and the invasive nature of tumor cells
is a major prerequisite to cancer metastasis (9). Cell migration has a critical role in
cancer cell invasion and metastasis, and is initiated via
activation of the epithelial-mesenchymal transition (EMT) in tumor
cells. Molecular alterations in the epithelial marker, E-cadherin,
and the mesenchymal markers, β-catenin, N-cadherin and vimentin,
may result in dysfunctional cell-cell adhesion and loss of
cell-cell junctions, which are associated with cell phenotype
transformation (10). Multitudinous
transcription factors, including the Snail-family members, snail
family transcriptional repressor 1 (Snail), snail family
transcriptional repressor 2 (Slug) and twist family bHLH
transcription factor 1 (Twist1), induce the EMT by repressing
E-cadherin expression (11,12). The EMT also involves a series of
complex changes in numerous signaling pathways, including the Wnt,
Notch and mitogen-activated protein kinase (MAPK) signaling
pathways (13).
The MAPK signaling pathway not only promotes cell
differentiation, proliferation and survival, but also mediates
oncogenesis (14). In a previous
study, the MAPK signaling pathway regulated the expression of the
activator protein 1 (AP-1) transcription factor, which regulates
the expression of members of the proto-oncogene Jun protein (c-Jun,
JunB and JunD) and Fos protein (c-Fos, FosB, Fra-1 and Fra-2)
families (15).
Extracellular-signal-regulated kinases (ERKs) are key components of
the MAPK signaling pathway and abnormal activation of the ERK
cascade is associated with metastasis in numerous human cancers
(16).
In the present study, Fer expression in bladder UCC
tissues and cell lines was assessed, and its prognostic value for
survival in patients with bladder UCC was evaluated. In addition,
the regulatory effect of Fer on T24 cell migration and invasion,
and the EMT process, was investigated and was shown to be mediated
via the ERK/AP-1 signaling pathway.
Materials and methods
Tissue samples
This study was approved by the Second Affiliated
Hospital of Anhui Medical University (Hefei, China). Tumor samples
from resected specimens were collected from two cohorts of patients
with primary bladder UCC that underwent transurethral resection,
partial cystectomy or radical cystectomy at the Second Affiliated
Hospital of Anhui Medical University between December 2008 and
October 2013. Cohort A consisted of 12 patients (10 females, 2
males; median age, 55 years; age range, 45–78 years), from whom
fresh tumor samples and adjacent histologically normal bladder
tissues were obtained for analysis of Fer mRNA and protein
expression. Cohort B comprised 78 patients (68 females, 10 males;
median age, 65 years; age range, 45–78 years), whose
paraffin-embedded specimens were used for immunohistochemical
analysis. A further 20 paraffin-embedded normal bladder mucosal
samples adjacent to the neoplastic bladder tissue were obtained
from the same UCC patients, and were used as a control. None of the
patients had received preoperative treatment. The tumors were
stratified as non-muscle invasive bladder cancer or muscle invasive
bladder cancer, according to the 2002 Union for International
Cancer Control TNM classification of tumor stage (17). The tumor grade was assigned using the
2004 World Health Organization/International Society of Urological
Pathology classification system (18). This study was conducted according to
the guidelines in the Declaration of Helsinki. Written informed
consent was obtained from all participants.
Cell line culture and maintenance
Bladder cancer cell lines, T24, 5637 and BIU-87, and
an immortalized normal human urothelial cell line, SV-HUC-1, were
purchased from Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China). T24, 5637 and BIU-87 cell lines were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS;
Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) at 37°C in 5%
CO2 and at 95% humidity. SV-HUC-1 cells were cultured in
F12k medium (Wisent, Inc., St. Bruno, QC, Canada) containing 10%
FBS, 100 units/ml penicillin and 100 µg/ml streptomycin in an
atmosphere containing 5% CO2 at 37°C. The activator,
epidermal growth factor (EGF; 0.1 ng/ml), was obtained from
Sigma-Aldrich (Merck Millipore, Darmstadt, Germany). At 70%
confluence, T24 cells transfected with Fer-siRNA were serum-starved
overnight, followed by incubation with EGF at 37°C for 48 h.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the cells using RNAiso
Plus (Takara Bio, Inc., Otsu, Japan), according to the
manufacturer's protocol. Total RNA (5 µg) was reverse transcribed
into cDNA using the M-MLV First-Strand Synthesis system (Promega
Corporation, Madison, WI, USA). cDNA was analyzed in triplicate
using the MJ Real-Time PCR system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The primers used are shown in Table I. qPCR was performed using the Power
SYBR Green Master Mix (Takara Bio, Inc.) and the ABI 7300 Real-Time
PCR detection system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). PCR conditions were as follows: Initial denaturation step at
95°C for 15 sec, followed by 40 cycles of amplification and
quantification at 95°C for 10 sec, 60°C for 30 sec and 72°C for 30
sec. Fold changes in expression of each gene were calculated using
the comparative quantification method (19).
 | Table I.Oligonucleotide primer sequences used
in RT-qPCR.. |
Table I.
Oligonucleotide primer sequences used
in RT-qPCR..
| Gene | Sequence |
|---|
| Fer | F:
5′-TTCGAGGGCACTGGGTTTTC-3′ |
|
| R:
5′-TTCCCTTGCCCAGTAATTCTCC-3′ |
| MMP-2 | F:
5′-TACAGGATCATTGGCTACACACC-3′ |
|
| R:
5′-GGTCACATCGCTCCAGACT-3′ |
| MMP-9 | F:
5′-TGTACCGCTATGGTTACACTCG-3′ |
|
| R:
5′-RGGCAGGGACAGTTGCTTCT-3′ |
| N-cadherin | F:
5′-TGCCAGTCACTTGCTAACAAAAG-3′ |
|
| R:
5′-GTGTGCGCTGGGAGAATAAAG-3′ |
| β-catenin | F:
5′-AAAGCGGCTGTTAGTCACTGG-3′ |
|
| R:
5′-CGAGTCATTGCATACTGTCCAT-3′ |
| E-cadherin | F:
5′-GACCGAGAGAGTTTCCCTACG-3′ |
|
| R:
5′-TCAGGCACCTGACCCTTGTA-3′ |
| Vimentin | F:
5′-ATGACCGCTTCGCCAACTAC-3′ |
|
| R:
5′-CGGGCTTTGTCGTTGGTTAG-3′ |
| Slug | F:
5′-ATACCACAACCAGAGATCCTCA-3′ |
|
| R:
5′-GACTCACTCGCCCCAAAGATG-3′ |
| Snail | F:
5′-TGAGGCCAAGGATCTCCAGG-3′ |
|
| R:
5′-GGGCAGGTATGGAGAGGAAG-3′ |
| Twist1 | F:
5′-GGGAGTCCGCAGTCTTACGA-3′ |
|
| R:
5′-AGACCGAGAAGGCGTAGCTG-3′ |
| GAPDH | F:
5′-CATGACCACAGTCCATGCCAT- 3′ |
|
| R:
5′-AAGGC-CATGCCAGTGAGCTTC-3′ |
Western blot assay
Cell lysate was prepared by extracting proteins
using RIPA buffer (Fermentas Inc., Burlington, ON, Canada)
supplemented with 1% protease inhibitors (Sigma-Aldrich; Merck
Millipore). Protein concentrations were measured using the
bicinchoninic acid protein assay (Pierce; Thermo Fisher Scientific,
Inc.), after which proteins were diluted to equal concentrations,
boiled for 5 min and separated by 7.5–10% SDS-PAGE, followed by
transblotting onto an Immun-Blot® polyvinylidene
fluoride membrane (Bio-Rad Laboratories, Inc.). The membranes were
blocked with 5% non-fat milk in TBST and subsequently probed with
primary antibodies overnight at 4°C. The primary antibodies
included: Anti-Fer (cat. no. ab52479; 1:500; Abcam, Cambridge, UK),
anti-matrix metalloproteinase (MMP)-2 (cat. no. 4022), anti-MMP-9
(cat. no. 3852S), anti-E-cadherin (cat. no. 4065), anti-vimentin
(cat. no. 3295S), anti-N-cadherin (cat. no. 4061S), anti-β-catenin
(cat. no. 9562), anti-Slug (cat. no. 9585P), anti-Snail (cat. no.
3879S), anti-Twist1 (cat. no. 4119S), anti-phospho-ERK1/2 (cat. no.
9911S), anti-p-c-jun (cat. no. 8221S) and anti-p-c-fos (cat. no.
5348S) (all 1:500; Cell Signaling Technology, Inc., Danvers, MA,
USA). Rabbit anti-GAPDH polyclonal antibody (cat. no. sc-25778;
1:500; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) was used
as an internal control. Primary antibodies were detected by
incubating the membranes with a horseradish peroxidase
(HRP)-conjugated secondary antibody (cat. no. 7071; 1:1,000; Cell
Signaling Technology, Inc.) for 1 h at room temperature. The blots
were subsequently developed using an enhanced chemiluminescence
detection kit (GE Healthcare Life Sciences) and exposure to
film.
Immunohistochemical analysis
Immunohistochemical staining was performed using a
Dako Envision System (Dako North America, Inc., Carpinteria, CA,
USA), according to the manufacturer's protocol. Briefly, 4-µm
paraffin-embedded tissue sections were heated for 1 h at 65°C,
deparaffinized with xylene and rehydrated using a graded series of
ethanol/distilled water. Subsequently, the tissue sections were
submerged in EDTA buffer (pH 8.0), heated in a microwave for
antigen retrieval, treated with 0.3% H2O2 for 15 min to
block endogenous peroxidase activity and incubated overnight with a
rabbit anti-Fer monoclonal antibody (1:50; Abcam) at 4°C. Next, the
tissue sections were washed, incubated with HRP-conjugated antibody
at 4°C for 30 min and visualized with diaminobenzidine.
RNA interference (RNAi)
siRNAs targeting Fer and negative control siRNA were
purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). The
sequences of the three siRNAs targeting Fer were as follows:
siRNA1, 5′-AAAGAAATTTATGGCCCTGAG-3′; siRNA2,
5′-CAGATAGATCCTAGTACAGAA-3′; and siRNA3,
5′-AACTACGGTTGCTGGAGACAG-3′). The sequence of the negative control
was as follows: 5′-UUCUCCGAACGUGUCACGU-3′. The sequences of the
siRNAs were designed using an RNAi algorithm (https://rnaidesigner.thermofisher.com/rnaiexpress/sort.do).
For siRNA transfection, the cells were seeded onto 6-well plates
and, after reaching 40–50% confluency, were transfected with
negative control or one of the Fer-specific siRNAs using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Cells were harvested for RNA extraction after 48 h and protein
extraction after 72 h of transfection.
Cell wound healing assay
T24 cells in 6-well plates were transfected with
control or Fer-siRNA. Upon reaching 90–95% confluence after 24 h,
wounds were generated by scratching the surface of the plates with
a 0–20 µl pipette tip. Wound closure was monitored at various time
points (0, 12 and 24 h) by observation under an inverted
microscope, and the degree of cell migration was quantified by the
ratio of gap distance at 24 h to that at 0 h. The experiment was
performed in triplicate.
Matrigel invasion assay
Cell invasion assays were performed using 24-well
Transwell chambers with a pore size of 8 µm (Costar; Corning, Inc.,
New York, NY, USA). The inserts were coated with 100 µl Matrigel
(dilution, 1:8; BD Biosciences, Franklin Lakes, NJ, USA). T24 cells
were trypsinized following transfection with control or Fer-siRNA
for 48 h and transferred to the upper Matrigel chamber in 100 µl
serum-free medium for 24 h. The lower chamber was filled with
medium containing 10% FBS as a chemoattractant. Following
incubation, the non-invaded cells on the upper membrane were
removed using a cotton tip, and the invaded cells on the bottom
membrane were evaluated by light microscopy. The cells were stained
with crystal violet and the optical density (OD) of the crystal
violet solution removed from the cells using 300 µl glacial acetic
acid (33%) was measured at 570 nm. All experiments were performed
in triplicate.
Statistical analysis
Statistical analyses were performed using SPSS
software, version 17.0 (SPSS Inc., Chicago, IL, USA). A
paired-samples t-test was used to compare the mRNA and protein
expression of Fer in the UCC tissues with that in the paired
adjacent normal tissue samples. The relationship between Fer
protein expression and clinicopathological features was analyzed
using χ2 tests. Overall survival curves were constructed
using the Kaplan-Meier method and were analyzed using the log-rank
test. Paired t-tests and Student's t-tests were used to analyze the
findings of the in vitro cell assay. P<0.05 was
considered to indicate a statistically significant difference.
Results
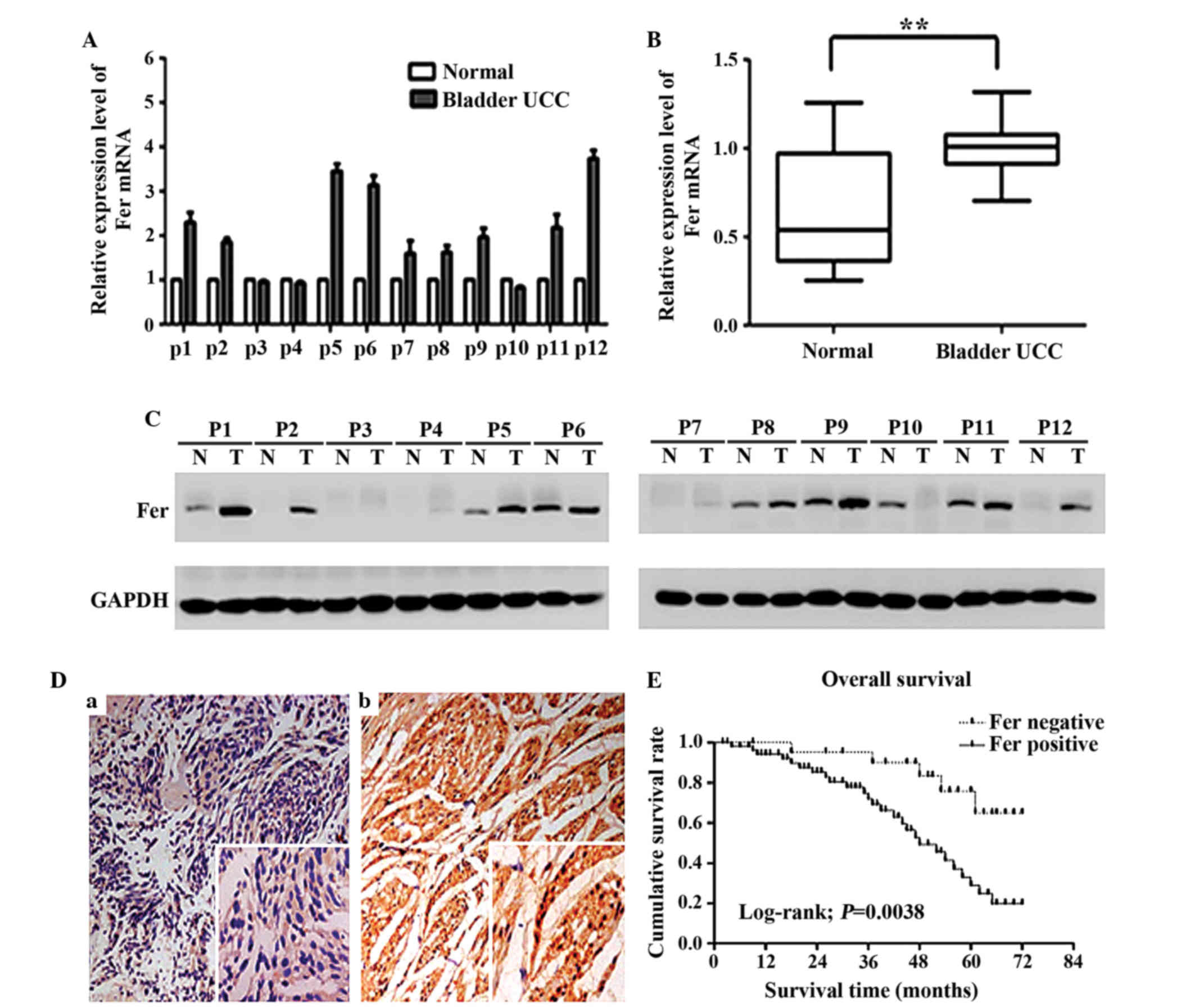
Fer is significantly upregulated in
bladder UCC tissues and is correlated with clinicopathological
parameters
To investigate the role of Fer in bladder UCC
development, the mRNA and protein expression of Fer in 12 bladder
UCC tissue samples and adjacent normal bladder tissues were
detected by RT-qPCR and western blotting, respectively. As shown in
Fig. 1A and B, the relative mRNA
expression level of Fer in bladder UCC tissues was significantly
higher than that in adjacent normal bladder tissues (P<0.01),
which was consistent with the results of the western blot (Fig. 1C). Immunohistochemical analysis was
performed to further analyze the expression of Fer in 78 bladder
UCC tissues, as compared with 20 paired adjacent normal tissues. As
shown in Fig. 1D, Fer staining was
negligible in the normal bladder tissues. Conversely, Fer was
positively expressed in both the cytoplasm and nucleus of 55
(70.5%) cancer tissues. Furthermore, it was observed that Fer
protein expression significantly correlated with the tumor stage
(P=0.042), histological grade (P=0.023) and lymph node status
(P=0.014), but was not associated with age (P=0.459), gender
(P=0.246) and tumor multiplicity (P=0.803) (Table II). The prognostic value of Fer for
overall survival in bladder UCC patients was evaluated by comparing
the patients with positive and negative Fer expression. According
to the Kaplan-Meier survival analysis, bladder UCC patients with
positive Fer expression had markedly lower overall survival rates
than patients with negative Fer expression (log-rank value=8.390;
P=0.0038; Fig. 1E). These results
suggest that the Fer expression status may be useful for predicting
the overall survival of patients with bladder UCC.
 | Table II.Relationship between Fer protein
expression and various clinicopathological parameters in 78 bladder
UCC tissues. |
Table II.
Relationship between Fer protein
expression and various clinicopathological parameters in 78 bladder
UCC tissues.
|
|
| Fer expression |
|
|---|
|
|
|
|
|
|---|
| Variable | n | Negative | Positive | P-value |
|---|
| Age, years (median
65) |
|
|
| 0.459 |
|
<65 | 38 | 13 | 25 |
|
|
≥65 | 40 | 10 | 30 |
|
| Gender |
|
|
| 0.246 |
|
Male | 68 | 19 | 49 |
|
|
Female | 10 | 4 | 6 |
|
| Tumor stage |
|
|
| 0.042 |
| Ta,
T1 | 46 | 18 | 28 |
|
| T2, T3,
T4 | 32 | 5 | 27 |
|
| Histological
grade |
|
|
| 0.023 |
| G1 | 19 | 10 | 9 |
|
| G2 | 28 | 8 | 20 |
|
| G3 | 31 | 5 | 26 |
|
| Tumor
multiplicity |
|
|
| 0.803 |
|
Unifocal | 52 | 16 | 36 |
|
|
Multifocal | 26 | 7 | 19 |
|
| Lymph node
status |
|
|
| 0.014 |
| N0 | 56 | 21 | 35 |
|
| N1,
N2 | 22 | 2 | 20 |
|
Knockdown of the Fer gene using siRNA
inhibits the migration of T24 cells
To determine the optimum cell model for
investigating the role of Fer in bladder UCC, the mRNA and protein
expression levels of Fer in various bladder UCC cells lines were
evaluated. The protein and mRNA expression of Fer was upregulated
in three bladder UCC cell lines (BIU-87, T24 and 5637), as compared
with the normal bladder epithelium cell line, SV-HUC-1.
Furthermore, high levels of Fer expression were observed in T24
cells compared with 5637 and BIU-87 cells (Fig. 2A and B). Therefore, T24 cells were
selected to assess the effects of Fer silencing on bladder UCC
cells by transfecting the cells with three positive Fer-siRNAs in
order to obtain efficient and specific Fer depletion. As shown in
Fig. 2C, the relative mRNA expression
levels of Fer were significantly decreased by 72% in T24 cells
transfected with siRNA1, as compared with the cells transfected
with normal control siRNA (P=0.003), and were significantly lower
than those cells transfected with siRNA2 and siRNA3. This result
was also observed for the protein expresion levels (Fig. 2D). Therefore, Fer-siRNA1 was selected
for the further analyses, as it demonstrated the most effective
silencing effects on Fer in T24 cells. As shown in Fig. 2E and F, in monolayer wound healing
assays, it was demonstrated that the cells transfected with
Fer-siRNA showed a significantly reduced migration distance
(0.358±0.030 mm), as compared with the cells transfected with the
normal control siRNA group (0.551±0.033 mm) (P<0.05).
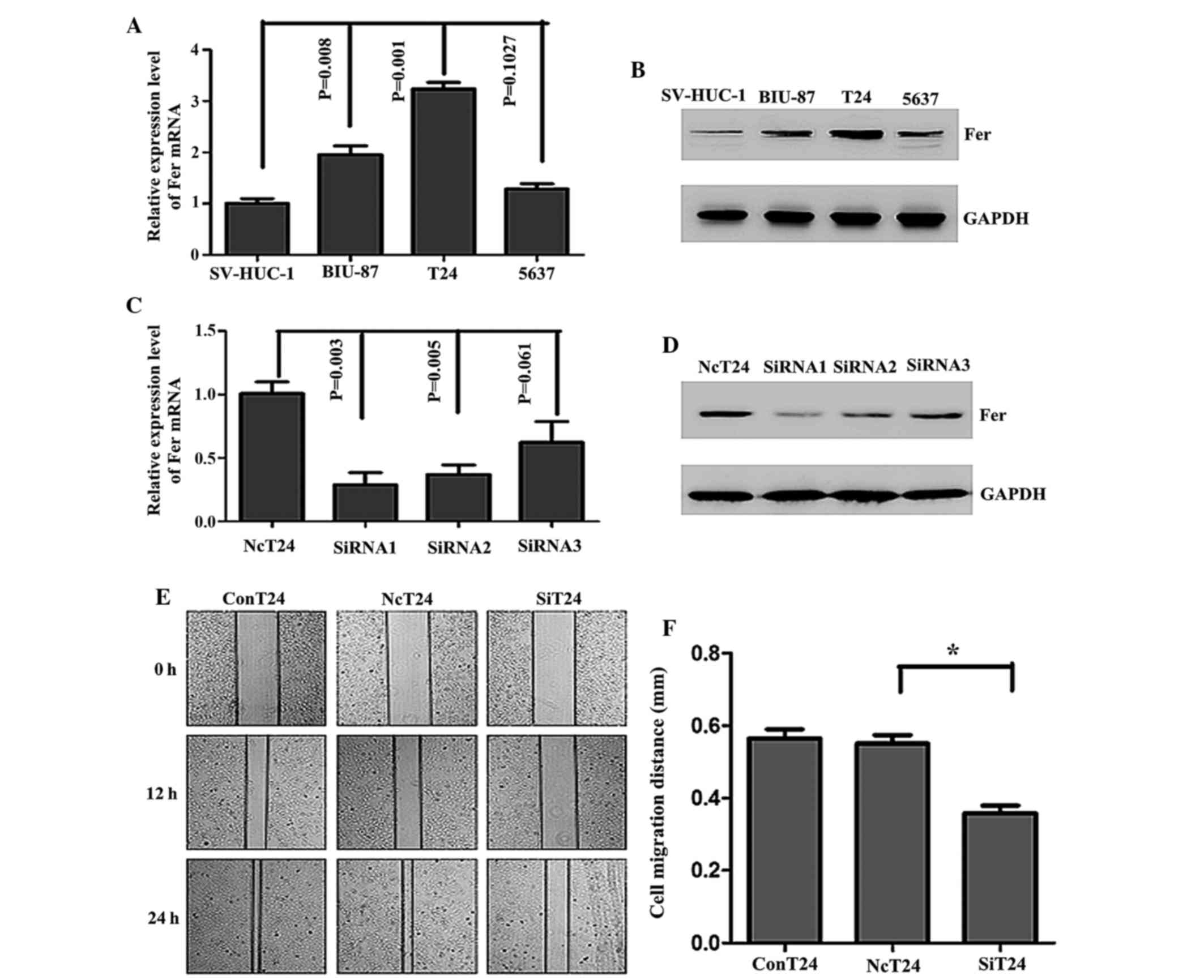 | Figure 2.mRNA and protein expression of Fer was
upregulated in human bladder cancer cell lines and Fer knockdown
using Fer-specific siRNA inhibited the migration of T24 cells. The
(A) mRNA and (B) protein expression of Fer in human bladder cancer
cell lines and an immortalized normal urothelial cell line,
SV-HUC-1, were detected by RT-qPCR and western blotting,
respectively. GAPDH was used as an internal control. Data are
presented as the mean ± standard deviation. The transfection
efficiency following transfection of T24 cells with Fer-specific
siRNAs (siRNA1, siRNA2 and siRNA3) or normal control siRNA for 48
and 72 h was assessed by (C) RT-qPCR and (D) western blotting. (E)
Wound healing assays were conducted to assess the capacity of cell
migration in the three groups of cells at 0, 12 and 24 h. (F) The
relative migration distances were calculated. *P<0.05. The
experiments were repeated in triplicate. Fer, feline
sarcoma-related protein; siRNA, small interfering RNA; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction;
NcT24, T24 cells transfected with normal control siRNA; SiT24, T24
cells transfected with Fer-specific siRNAs; ConT24, untreated T24
cells. |
Fer gene silencing inhibits cell
invasion and suppresses MMP family protein expression in T24
cells
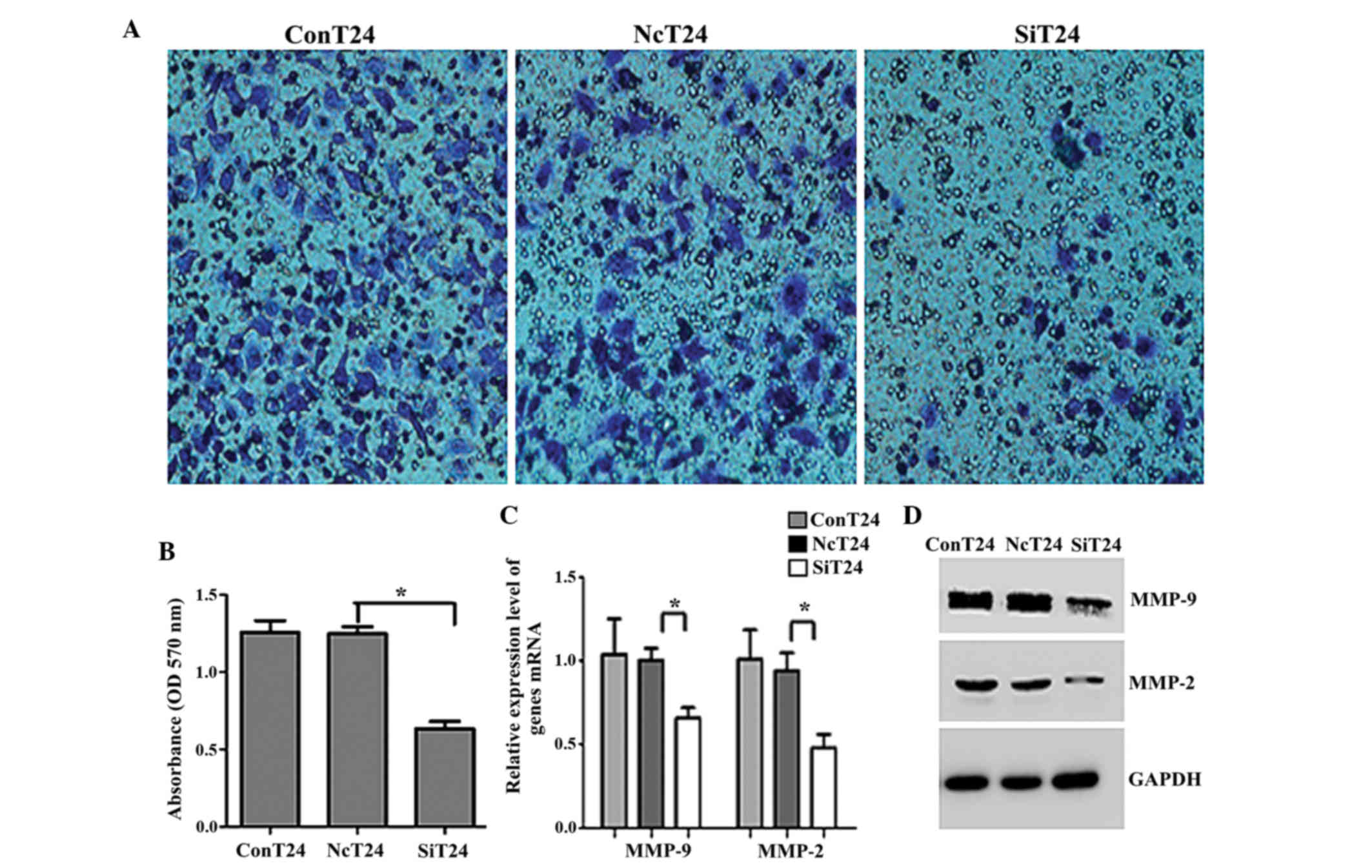
To functionally confirm the role of Fer in
aggressive bladder cancers, matrigel invasion assays were
performed. The OD of the Fer-siRNA-transfected cells was
significantly reduced compared with the OD of the T24 cells
transfected with normal control siRNA (0.635±0.066 vs. 1.249±0.062,
respectively; P<0.05; Fig. 3A and
B). Previous studies have reported that MMPs are the principal
mediators of alterations in the cancer microenvironment during
cancer metastasis (20). Therefore,
the present study investigated whether the MMP protein family was
involved in bladder UCC invasion and metastasis. As shown in
Fig. 3C and D, the expression levels
of MMP-2 and MMP-9 were significantly downregulated in
Fer-siRNA-transfected T24 cells, as compared with the cells
transfected with normal control siRNA (P<0.05).
Fer gene silencing induces changes in
the morphology and markers of EMT in T24 cells via the ERK/AP-1
pathway
The morphology of T24 cells transfected with
Fer-siRNA or normal control siRNA was analyzed by light microscopy.
T24 cells transfected with normal control siRNA had an irregular
fibroblastoid morphology, tight intercellular structure and a clear
contour, while the majority of the Fer-siRNA-transfected T24 cells
displayed a rounded shape, typical of an epithelial cobblestone
appearance (Fig. 4A). These changes
from a mesenchymal morphology to an epithelial morphology suggested
that Fer silencing was able to reverse the EMT of T24 cells. As
shown in Fig. 4B and C, silencing of
Fer in T24 cells increased the expression of E-cadherin and
decreased the expression of β-catenin, N-cadherin and vimentin. In
addition, the expression levels of the EMT-regulating transcription
factors, Slug, Snail and Twist1, were downregulated in
Fer-siRNA-transfected cells. Furthermore, there was a marked
decrease in the expression of phospho-ERK1/2, p-c-jun and p-c-fos
following Fer-knockdown in the T24 cells, but not total ERK/AP-1
(Fig. 4D). These findings suggest
that inhibition of Fer by siRNA blocked the phosphorylation of
ERK/AP-1. To further investigate whether the ERK/AP-1 pathway is
involved in the Fer-induced EMT in T24 cells, an activator (EGF) of
the ERK pathway was used. As shown in Fig. 4E, the EMT was promoted and the AP-1
levels were upregulated in siRNA-transfected cells incubated with
EGF for 48 h. These results suggest that the ERK/AP-1 pathway has
an important role in Fer-induced EMT.
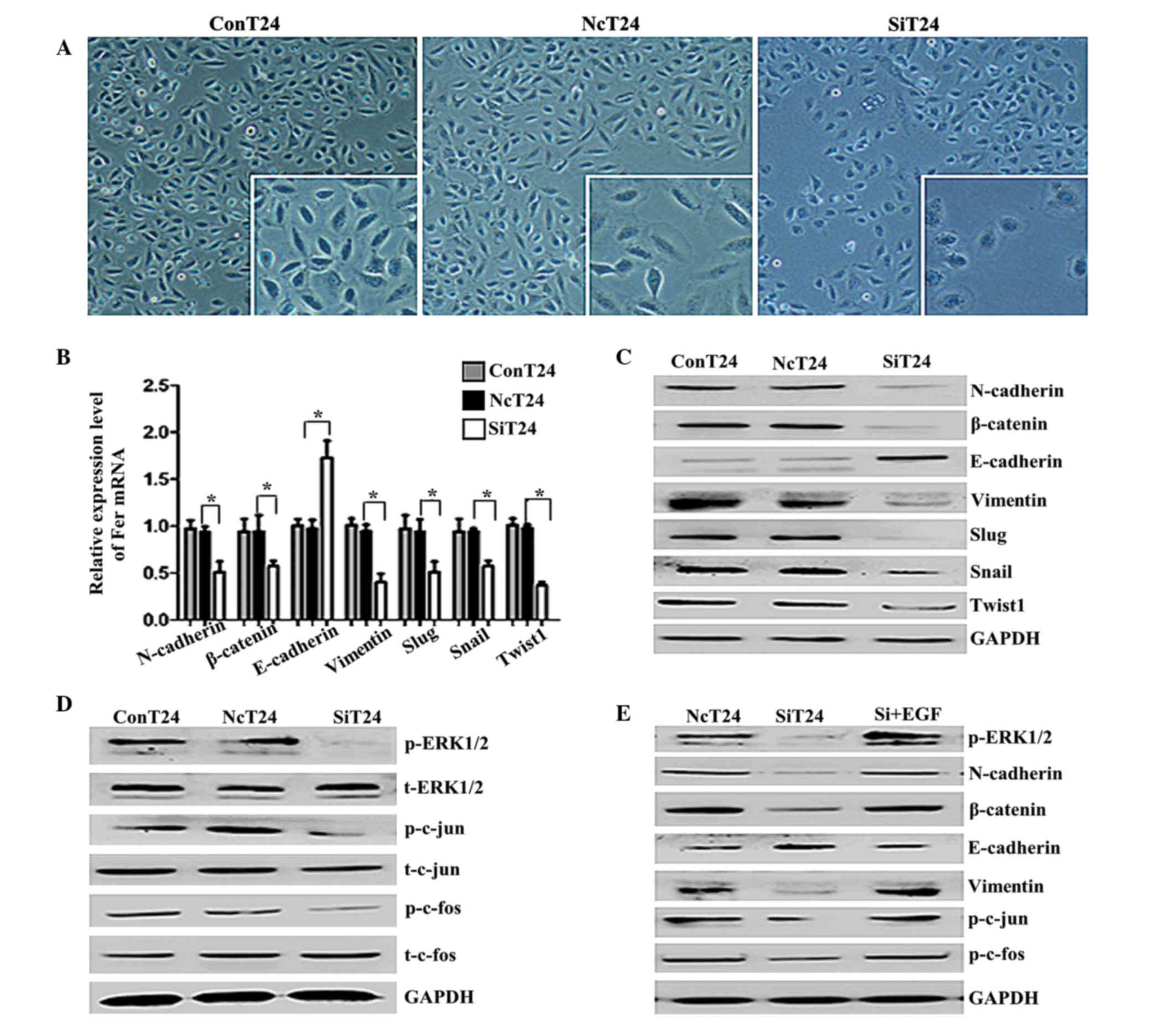 | Figure 4.Knockdown of Fer induced a
mesenchymal-epithelial transition-like phenotype and altered the
expression of EMT-associated genes via the ERK/AP-1 pathway in T24
cells. (A) Light microscopy of T24 cells transfected with Fer-siRNA
(magnification, ×100; framed in white, ×200). At 72 h following
transfection, the majority of Fer-siRNA-transfected T24 cells were
rounded, which is typical of an epithelial cobblestone appearance.
Knockdown of Fer differentially regulated the (B) mRNA and (C)
protein expression of EMT-associated proteins and transcription
factors in T24 cells. GAPDH was used as an internal control. Data
are presented as the mean ± standard deviation. *P<0.05. (D) The
phosphorylation levels of ERK1/2 and AP-1 in T24 cells were
decreased by silencing of Fer expression, as demonstrated in a
western blot analysis. (E) T24 cells transfected with Fer-siRNA
were treated with EGF (0.1 ng/ml) for 48 h, and the effect of EGF
on the expression of EMT-associated proteins and the
phosphorylation levels of ERK1/2 and AP-1 were detected by western
blotting. Fer, feline sarcoma-related protein; EMT,
epithelial-mesenchymal transition; ERK/AP-1, extracellular
signal-regulated kinase/activator protein-1; siRNA, small
interfering RNA; NcT24, T24 cells transfected with normal control
siRNA; SiT24, T24 cells transfected with Fer-specific siRNAs;
ConT24, untreated T24 cells; EGF, epidermal growth factor; Snail,
snail family transcriptional repressor 1; Slug, snail family
transcriptional repressor 2; Twist1, twist family bHLH
transcription factor 1. |
Discussion
It has been reported that Fer is extensively
expressed in numerous mammalian cells, and is associated with tumor
progression (21). However, the
expression pattern and biological significance of Fer in bladder
UCC are unclear. The present study demonstrated that Fer was
significantly upregulated in bladder UCC tissues and cell lines, as
compared with adjacent normal bladder tissues and the SV-HUC-1
normal human urothelial cell line, respectively. These findings
indicate that Fer is involved in the progression of bladder UCC. In
addition, an immunohistochemical analysis of bladder UCC specimens
suggested that Fer expression was significantly associated with
tumor stage, histological grade and lymph node status. Notably, Fer
expression was an independent prognostic factor for a poor
prognosis in patients with bladder UCC patients.
Several studies have reported that Fer is involved
in tumor invasion and metastasis (5,22,23); however, the role and cellular
mechanisms of Fer in bladder UCC are not well-known. In the present
study, siRNA was used to knockdown Fer expression in T24 cells, and
the ability of the cells to migrate and invade was investigated.
Notably, T24 cell migration and invasion was significantly reduced
in cells transfected with Fer-siRNA. Increasingly it has been
suggested that MMPs, which degrade the extracellular matrix and
cell adhesion molecules, enhance cancer cell metastasis (24,25). The
present study demonstrated that knockdown of Fer expression was
able to downregulate the expression of MMP-9 and MMP-2; thus
suggesting that Fer may have an important role in the migration and
invasion of T24 cells via the regulation of MMP gene
expression.
During the process of metastasis, cancer cells often
initiate the EMT, which is a dynamic cellular process thought to
promote the acquisition of migratory and invasive abilities
(26). During the EMT, morphological
changes from the epithelial polarized morphology to the mesenchymal
fibroblastoid morphology occur (27).
Previous studies reported that Fer is involved in
integrin/E-cadherin-mediated signaling pathways and has a role in
regulating the cross-talk between cadherin-catenin complexes via
focal adhesions (28–30). Furthermore, it was revealed that
multiple small G protein/MAPK cascades were involved in the
downstream signal transduction by Fps/Fes tyrosine kinases
(31). The initiation of the EMT is
dependent on the concomitant activity of MAPK pathways to induce
the morphogenic process of the EMT (16).
In the present study, transfection of T24 cells with
Fer-siRNA resulted in alterations in the morphology of T24 cells
from a mesenchymal to an epithelial phenotype. In addition,
silencing of Fer expression was shown to increase the expression of
epithelial junction proteins (E-cadherin) and decrease the
expression of mesenchymal markers (N-cadherin, vimentin and
β-catenin). Furthermore, Fer-siRNA decreased the expression of
Snail, Slug and Twist1 in T24 cells, which are known to potently
modulate epithelial cell plasticity and induce the EMT phenotype
(12,32,33).
Another significant finding was that Fer inhibition disrupted the
ERK/AP-1 signaling pathway by suppressing the phosphorylation of
ERK/AP-1. These results suggested that Fer silencing inhibited the
EMT via suppression of the ERK/AP-1 signaling pathway.
In summary, the present study demonstrated that Fer
was upregulated in patients with bladder UCC and that the Fer
expression status was associated with tumor development and
prognosis in these patients. In addition, it was shown that
knockdown of Fer reduced the migration and invasion of T24 cells,
and reversed the EMT by blocking the ERK/AP-1 signaling pathway.
However, further studies are required to explore whether Fer-siRNA
affected other upstream or downstream signaling molecules. These
results suggested that targeting Fer signaling using novel
approaches may be useful for reversing the EMT phenotype, which
would likely result in the reversal of neoplasm recurrence and
elimination of bladder UCC.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81373005, 81072330
and 81202194) and by the Priority Academic Program Development of
Jiangsu Higher Education Institutions.
References
|
1
|
Bo J, Yang G, Huo K, Jiang H, Zhang L, Liu
D and Huang Y: microRNA-203 suppresses bladder cancer development
by repressing bcl-w expression. FEBS J. 278:786–792. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ben-Dor I, Bern O, Tennenbaum T and Nir U:
Cell cycle-dependent nuclear accumulation of the p94fer tyrosine
kinase is regulated by its NH2 terminus and is affected by kinase
domain integrity and ATP binding. Cell Growth Differ. 10:113–129.
1999.PubMed/NCBI
|
|
4
|
Miyata Y, Kanda S, Sakai H and Greer PA:
Feline sarcoma-related protein expression correlates with malignant
aggressiveness and poor prognosis in renal cell carcinoma. Cancer
Sci. 104:681–686. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li H, Ren Z, Kang X, Zhang L, Li X, Wang
Y, Xue T, Shen Y and Liu Y: Identification of
tyrosine-phosphorylated proteins associated with metastasis and
functional analysis of FER in human hepatocellular carcinoma cells.
BMC Cancer. 9:3662009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zoubeidi A, Rocha J, Zouanat FZ, Hamel L,
Scarlata E, Aprikian AG and Chevalier S: The Fer tyrosine kinase
cooperates with interleukin-6 to activate signal transducer and
activator of transcription 3 and promote human prostate cancer cell
growth. Mol Cancer Res. 7:142–155. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Albeck JG and Brugge JS: Uncovering a
tumor suppressor for triple-negative breast cancers. Cell.
144:638–640. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Allard P, Zoubeidi A, Nguyen LT, Tessier
S, Tanguay S, Chevrette M, Aprikian A and Chevalier S: Links
between Fer tyrosine kinase expression levels and prostate cell
proliferation. Mol Cell Endocrinol. 159:63–77. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stetler-Stevenson WG and Yu AE: Proteases
in invasion: Matrix metalloproteinases. Semin Cancer Biol.
11:143–152. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Batlle E, Sancho E, Francí C, Domínguez D,
Monfar M, Baulida J and De Herreros A García: The transcription
factor snail is a repressor of E-cadherin gene expression in
epithelial tumour cells. Nat Cell Biol. 2:84–99. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bolós V, Peinado H, Pérez-Moreno MA, Fraga
MF, Esteller M and Cano A: The transcription factor slug represses
E-cadherin expression and induces epithelial to mesenchymal
transitions: A comparison with Snail and E47 repressors. J Cell
Sci. 116:499–511. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sebolt-Leopold JS and Herrera R: Targeting
the mitogen-activated protein kinase cascade to treat cancer. Nat
Rev Cancer. 4:937–947. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Angel P and Karin M: The role of Jun, Fos
and the AP-1 complex in cell-proliferation and transformation.
Biochim Biophys Acta. 1072:129–157. 1991.PubMed/NCBI
|
|
16
|
Raman M, Chen W and Cobb MH: Differential
regulation and properties of MAPKs. Oncogene. 26:3100–3112. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sobin DH and Wittekind C: TNM
Classification of Malignant Tumors. 6th. Wiley-Liss; New York, NY:
2003, View Article : Google Scholar
|
|
18
|
Miyamoto H, Miller JS, Fajardo DA, Lee TK,
Netto GJ and Epstein JI: Non-invasive papillary urothelial
neoplasms: The 2004 WHO/ISUP classification system. Pathol Int.
60:1–8. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Groblewska M, Siewko M, Mroczko B and
Szmitkowski M: The role of matrix metalloproteinases (MMPs) and
their inhibitors (TIMPs) in the development of esophageal cancer.
Folia Histochem Cytobiol. 50:12–19. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hao QL, Heisterkamp N and Groffen J:
Isolation and sequence analysis of a novel human tyrosine kinase
gene. Mol Cell Biol. 9:1587–1593. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Orlovsky K, Theodor L, Malovani H, Chowers
Y and Nir U: Gamma interferon down-regulates Fer and induces its
association with inactive Stat3 in colon carcinoma cells. Oncogene.
21:4997–5001. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ahn J, Truesdell P, Meens J, Kadish C,
Yang X, Boag AH and Craig AW: Fer protein-tyrosine kinase promotes
lung adenocarcinoma cell invasion and tumor metastasis. Mol Cancer
Res. 11:952–963. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Deryugina EI and Quigley JP: Matrix
metalloproteinases and tumor metastasis. Cancer Metastasis Rev.
25:9–34. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Moss LA Shuman, Jensen-Taubman S and
Stetler-Stevenson WG: Matrix metalloproteinases: Changing roles in
tumor progression and metastasis. Am J Pathol. 181:1895–1899. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Scheel C, Onder T, Karnoub A and Weinberg
RA: Adaptation versus selection: The origins of metastatic
behavior. Cancer Res. 67:11476–11479; discussion 11479–11480. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Craig AW and Greer PA: Fer kinase is
required for sustained p38 kinase activation and maximal chemotaxis
of activated mast cells. Mol Cell Biol. 22:6363–6374. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kogata N, Masuda M, Kamioka Y, Yamagishi
A, Endo A, Okada M and Mochizuki N: Identification of Fer tyrosine
kinase localized on microtubules as a platelet endothelial cell
adhesion molecule-1 phosphorylating kinase in vascular endothelial
cells. Mol Biol Cell. 14:3553–3564. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Piedra J, Miravet S, Castaño J, Pálmer HG,
Heisterkamp N, de Herreros A García and Duñach M: p120
Catenin-associated Fer and Fyn tyrosine kinases regulate
beta-catenin Tyr-142 phosphorylation and beta-catenin-alpha-catenin
interaction. Mol Cell Biol. 23:2287–2297. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Senis YA, Sangrar W, Zirngibl RA, Craig
AW, Lee DH and Greer PA: Fps/Fes and Fer non-receptor
protein-tyrosine kinases regulate collagen- and ADP-induced
platelet aggregation. J Thromb Haemost. 1:1062–1070. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cano A, Pérez-Moreno MA, Rodrigo I,
Locascio A, Blanco MJ, del Barrio MG, Portillo F and Nieto MA: The
transcription factor snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu AN, Zhu ZH, Chang SJ and Hang XS:
Twist expression associated with the epithelial-mesenchymal
transition in gastric cancer. Mol Cell Biochem. 367:195–203. 2012.
View Article : Google Scholar : PubMed/NCBI
|