Introduction
Colorectal carcinoma (CRC) is the third most
frequent malignant cancer worldwide (1) and has rapidly increased in incidence in
recent years (2). In the United
States of America, CRC is the leading cause of cancer-associated
morbidity and mortality (3). In
China, CRC ranks as the fourth most frequently occurring cancer and
the fifth leading cause of cancer-associated mortality (4). Chemotherapy is important in the
comprehensive treatment of CRC, and remains the primary treatment
for resectable and advanced CRC (5).
However, certain patients with CRC who are treated with
chemotherapeutics exhibit a poor initial response or gradually
develop resistance, a phenomenon known as multidrug resistance
(MDR) (6,7). The emergence of MDR is an obstacle to
the successful treatment of CRC, leading to poor prognosis, relapse
and metastasis of the tumor (8). The
underlying molecular mechanisms of chemoresistance are complex and
multifactorial, and remain to be completely elucidated. Multiple
processes participate in the development of MDR, including those
involved in drug transport and metabolism, DNA damage repair and
apoptotic regulation (9,10).
Evidence increasingly suggests that microRNAs (miRs)
serve an essential role in MDR (11–14). miRs
are evolutionarily conserved, small non-coding single-stranded RNA
molecules of between 19 and 24 nucleotides in length (15,16). The
primary function of miRs is to negatively regulate gene expression
at the post-transcriptional level, through binding to the 3′
untranslated region of their target mRNA, in part or in full, and
subsequently mediating its degradation or inhibiting its
translation (17,18). miRs are involved in a number of
biological processes, including metabolism, differentiation,
proliferation, cell-cycle control and apoptosis (18–20).
Furthermore, increasing research indicates that miRs function as
oncogenes and tumor suppressors, and that the dysregulation of miRs
is associated with the development of MDR in various types of
cancer (21–23). For example, miR-17-5p was demonstrated
to be upregulated in chemoresistant CRC cells, which promoted cell
invasiveness (24). In addition,
miR-508-5p was observed to be downregulated in drug-resistant
gastric cancer cells, where its overexpression was demonstrated to
reverse drug resistance (25).
Various miRs are known to be involved in the regulation of MDR in
cancer, including miR-625-3p, miR-1915, miR-200c and miR-203
(26–29).
miR-93 belongs to the miR-106b-25 cluster and is
located at the human chromosome locus 7q22.1 (30). miR-93 appears to be frequently
dysregulated in various types of human cancer, including gastric,
ovarian, breast and, in particular, CRC (31–35).
Furthermore, miR-93 has been demonstrated to participate in the
development of MDR in ovarian cancer cells and prolactinomas
(36,37). Zhou et al (33) reported that the expression levels of
miR-93 were regulated by chemotherapeutics in human colon cancer
cells and, thus, may be associated with treatment-resistance in
colon cancer. However, to the best of our knowledge, the exact role
of miR-93 in the development of MDR in CRC has not yet been
investigated and verified. Therefore, in the present study, the
role of miR-93-5p in the modulation of drug-resistance in human CRC
was examined using HCT-8 and multidrug-resistant HCT-8/vincristine
(VCR) cell lines. The results of the current study suggest that
miR-93-5p overexpression in CRC serves an essential role in the
development of MDR, via miR-93-5p negatively regulating the
expression of its target gene, cyclin-dependent kinase inhibitor 1A
(CDKN1A).
Materials and methods
Cell lines and cultures
The HCT-8 human ileocecal colorectal adenocarcinoma
cell line was obtained from the Kunming Cell Bank of the Chinese
Academy of Sciences (Kunming, China), while the HCT-8/VCR
multidrug-resistant variant was purchased from the Modern Analysis
and Testing Center of Central South University (Changsha, China).
Cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a
humidified atmosphere containing 5% CO2. A total of 1
µg/ml VCR was added to HCT-8/VCR cell cultures to maintain their
MDR phenotype.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) of miR-93-5p
Total RNA (40 ng) was extracted from the cultured
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Polyadenylation was subsequently carried out, in
which first strand cDNA synthesis was performed using the miRcute
miRNA First-strand cDNA Synthesis kit (Tiangen Biotech Co., Ltd.,
Beijing, China), according to the manufacturer's protocol. The
miRcute miRNA qPCR Detection kit (Tiangen Biotech Co., Ltd.) was
used for the following RT-qPCR analysis. Briefly, the 20 µl PCR
reaction volume included 10 µl 2X miRcute miRNA Premix, 2 µl first
strand cDNA, 7.2 µl RNase-free ddH2O, 0.4 µl forward
primer (final concentration 200 nM) and 0.4 µl reverse primer
(final concentration 200 nM). The reactions were incubated in a
96-well optical plate at 94°C for 2 min, followed by 40 cycles at
94°C for 20 sec and 60°C for 34 sec. The universally expressed U6
small nuclear RNA (snRNA) was used as an internal control for
normalization. Primers for miR-93-5p (cat. no. CD201-0041) and U6
(cat. no., CD201-0145) were acquired from Tiangen Biotech Co., Ltd.
The 2−ΔΔCq method (38)
was used to calculate the relative expression levels of miR-93-5p
for each cell line. The treated group included HVT-8 cells, whereas
the control group was HCT-8/VCR cells. ∆Cq was calculated as
follows: Average miR-93-5p Cq-average U6 snRNA Cq. ∆∆Cq was
calculated as the following: ∆Cq of the treated group-∆Cq of the
control group. The results are presented as the fold change in
expression in HCT-8 cells relative to HCT-8/VCR cells.
miR transfection
The miR-93-5p mimic (cat. no. B01001) and inhibitor
(cat. no. B03001), and the corresponding negative control (NC; cat.
no. B04001), were designed and chemically synthesized by GenePharma
Co., Ltd. (Shanghai, China). HCT-8/VCR and HCT-8 cells were seeded
into 6-well plates at a density of 2×105 cells/well and
cultured at 37°C for 24 h. A total of 100 pmol miR-93-5p mimic,
miR-93-5p inhibitor or NC was transfected into target cells using 5
µl Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.)
and Opti-MEM® I reduced serum medium (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
protocol.
Bioinformatics method
The following online miRNA target prediction
algorithms were used to evaluate the potential target genes of
miR-93-5p: PicTar (http://www.pictar.org/), TargetScan (http://www.targetscan.org/vert_71/) and miRanda
(http://www.microrna.org/microrna/home.do).
RT-qPCR of MDR protein 1 (MDR1) and
CDKN1A
Total RNA (500 ng) was isolated from the cells using
TRIzol® reagent, according to the manufacturer's
protocol. cDNA was synthesized using a PrimeScript™ RT reagent kit
(cat. no., RR037A; Takara Biotechnology Co., Ltd., Dalian, China).
The PCR reaction mix contained 12.5 µl FastStart Universal SYBR
Green Master (Roche Diagnostics GmbH, Mannheim, Germany), 0.25 µl
forward and reverse primers (final concentration, 300 nM) and 2.5
µl cDNA, made up to a final volume of 25 µl with RNase-free
ddH2O. Primers for MDR1, CDKN1A and GAPDH were acquired
from GenePharma Co. Ltd. MDR1 (GeneBank accession number,
NM_000927.4): 5′-GTTGCTGCTTACATTCAGGTTTC-3′ (sense) and
5′-ACCAGCCTATCTCCTGTCGC-3′ (antisense); CDKN1A (GeneBank accession
number, NM_000389.4): 5′-CCTTCCTCATCCACCCCATC-3′ (sense) and
5′-CCCTGTCCATAGCCTCTACTGC-3′ (antisense); GAPDH (GeneBank accession
number, NM_001256799.2): 5′-GTCAGCCGCATCTTCTTT-3′ (sense) and
5′-CGCCCAATACGACCAAAT-3′ (antisense). The ABI 7500 real time PCR
system with 7500 software version 2.0.6 (Applied Biosystems; Thermo
Fisher Scientific, Inc.) was utilized to quantify the expression
levels of RNA. The thermocycling conditions for the RT-qPCR were as
follows: 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for
15 sec and 60°C for 1 min. The expression levels of MDR1, CDKN1A
and GAPDH were assessed using the 2−ΔΔCq method, as
aforementioned, while GAPDH was used as an internal control.
Western blot analysis
At 48 h following transfection, the cells were
washed with 1X PBS and then lysed using radioimmunoprecipitation
assay buffer (Beyotime Institute of Biotechnology, Shanghai, China)
for 30 min at 4°C. The protein concentration was measured using a
bicinchoninic acid protein assay kit (Beijing Solarbio Science
& Technology Co., Ltd., Beijing, China), following the
manufacturer's protocol, and 30 µg protein was separated by 10%
SDS-PAGE for western blot analysis. Once the proteins were
transferred to a polyvinylidene difluoride membrane (Merck
Millipore, Darmstadt, Germany), the membrane was incubated in
bovine serum albumin blocking buffer (Beijing Solarbio Science
& Technology Co., Ltd.) at room temperature for 1 h and washed
with Tris-buffered saline and Tween-20 (TBST; Beijing Solarbio
Science & Technology Co., Ltd.) three times (5 min/time).
Monoclonal antibodies specific to MDR1 (cat. no. 13342; dilution,
1:1,000), CDKN1A (cat. no. 2947; dilution, 1:1,000) or β-actin
(cat. no. 12620; dilution, 1:1,000; Cell Signaling Technology,
Inc., Danvers, MA, USA) were utilized and the membranes were
incubated overnight at 4°C. β-actin was used as the internal
control. Following washing three times with TBST (5 min/time), the
membrane was probed with horseradish peroxidase-conjugated goat
anti-rabbit immunoglobulin G (cat. no. KC-RB-035; dilution,
1:5,000; KangChen Bio-tech, Shanghai, China) for 1 h at room
temperature. Subsequently, the membranes were washed with TBST
three times (5 min/time), ECL Plus Western Blotting Substrate
(Pierce; Thermo Fisher Scientific, Inc.) was used to identify
protein bands and ImageJ software (National Institutes of Health,
Bethesda, MD, USA; version 1.48) was used to quantify their
intensity.
Rhodamine 123 (Rh-123) was used to
measure MDR1 efflux
Rh-123 is a fluorescent dye (Dojindo Molecular
Technologies, Inc., Rockville, MD, USA) that binds to MDR1, which
is an established tool to examine the transport activity of MDR1
(39). A total of 5×105
cells were incubated with 20 µM Rh-123 for 1 h at 37°C. Following
incubation, cells were washed three times with PBS to remove free
Rh-123 from the medium. Cells were subsequently kept in Rh-123-free
medium at 37°C. The intracellular fluorescence intensity of the
remaining Rh-123 was examined using fluorescence-activated cell
sorting with a flow cytometer (FC500; Beckman Coulter, Inc., Brea,
CA, USA).
In vitro drug sensitivity assay
A total of 24 h following miR transfection, the
cells were seeded into 96-well plates at a density of
3×103 cells/well. Following cellular adhesion, VCR was
added to provide the following final concentrations: 0.001, 0.01,
0.1, 1, 10 and 100 times the peak human plasma concentration. The
peak serum concentration of VCR was taken to be 0.5 µg/ml, as
previously determined by Zhu et al (40). The Cell Counting kit-8 (CCK-8; Dojindo
Molecular Technologies, Inc., Rockville, MD, USA) assay was
performed to assess cell viability at 72 h following miR
transfection. The absorbance of each well at 450 nm was detected
using the ELx808™ Absorbance Reader with Gen5 software version
2.00.20 (BioTek Instruments, Inc., Winooski, VT, USA). Furthermore,
the half-maximal inhibitory concentration (IC50) of VCR
on cell viability was determined by calculating the dose-response
curve.
Cell cycle assay
A total of 72 h following miR transfection,
HCT-8/VCR cells were harvested by centrifugation at 300 × g
for 10 min at room temperature. Cells were washed three times with
cold PBS and fixed in ice-cold 75% ethanol at 4°C overnight. Fixed
cells were subsequently rehydrated in 200 µl ice-cold PBS and
treated with 20 µl RNase (Yeasen Biotech Co., Ltd., Shanghai,
China) for 30 min. Subsequently, the cell samples were treated with
400 µl propidium iodide staining solution, followed by incubation
at 4°C for 30 min in the dark. The cell cycle distribution for each
sample was estimated using FlowJo software (version 7.6.1; FlowJo,
LLC, Ashland, OR, USA), according to the manufacturer's
protocol.
Statistical analysis
The data are presented as the mean ± standard
deviation of triplicate results. The difference between means was
statistically analyzed using a two-tailed Student's t-test
when 2 groups were compared, or one-way analysis of variance
(ANOVA) when >2 groups were compared. In ANOVA, post hoc tests
were used for multiple comparisons as follows: The Fisher's least
significant difference and Students-Newman-Keuls methods were
applied if equal variances were assumed; the Games-Howell method
was applied if equal variances were not assumed. All statistical
analyses were performed using SPSS software (version 17.0; SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-93-5p and MDR1 in
HCT-8 and HCT-8/VCR CRC cells
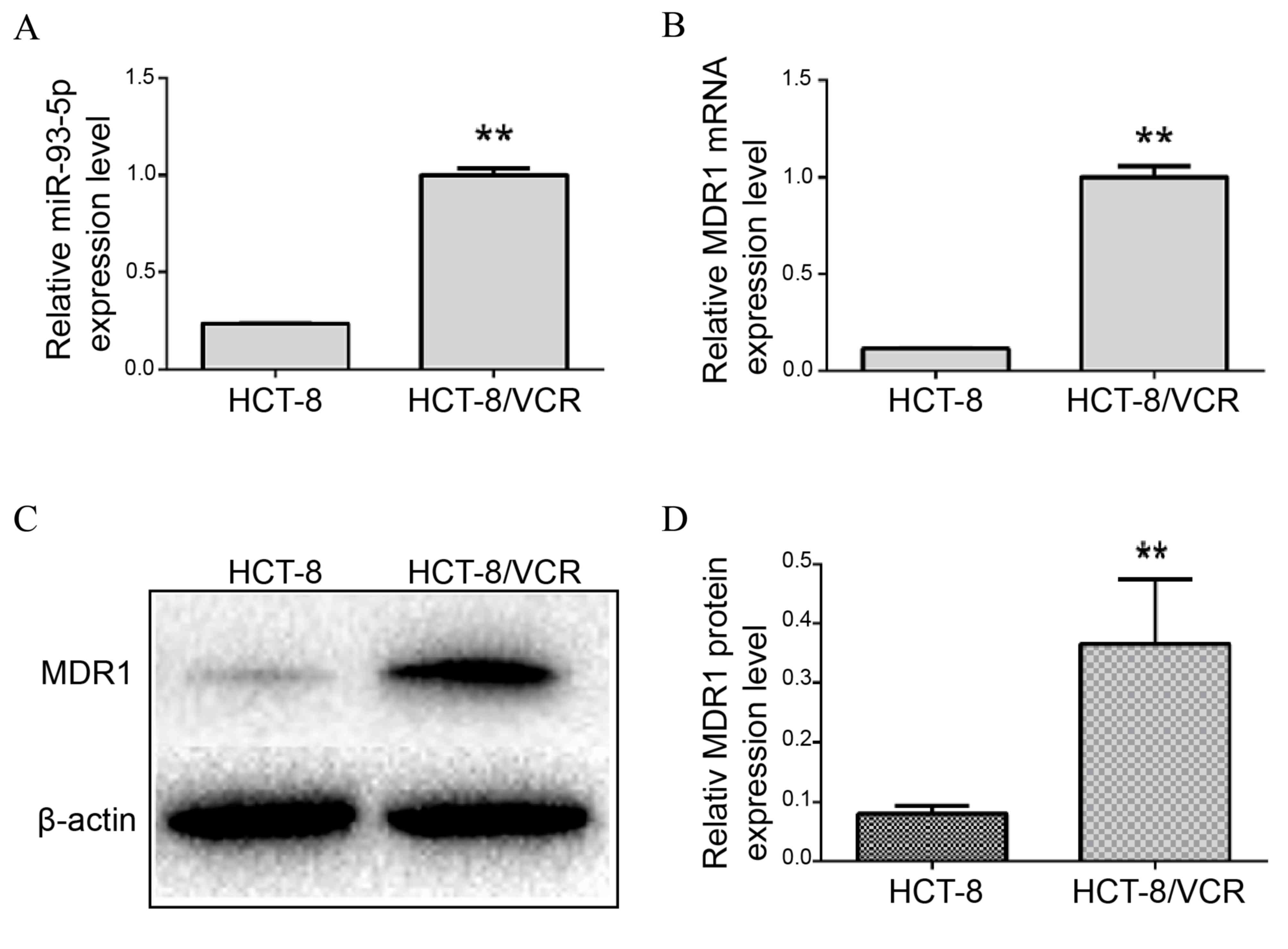
miR-93-5p and MDR1 mRNA expression levels were
examined in HCT-8 and HCT-8/VCR cell lines using RT-qPCR, and MDR1
protein levels were evaluated using western blotting. The results
of the present study demonstrated that the relative expression of
miR-93-5p was significantly increased in HCT-8/VCR cells, compared
with HCT-8 cells (P<0.001; Fig.
1A). In addition, the relative expression levels of MDR1 mRNA
were significantly increased in HCT-8/VCR cells, compared with the
parental HCT-8 cells (P=0.001; Fig.
1B). Similarly, MDR1 protein levels were increased in HCT-8/VCR
cell lines, compared with HCT-8 cell lines (P=0.011; Fig. 1C and D).
miR-93-5p regulates the expression of
MDR1
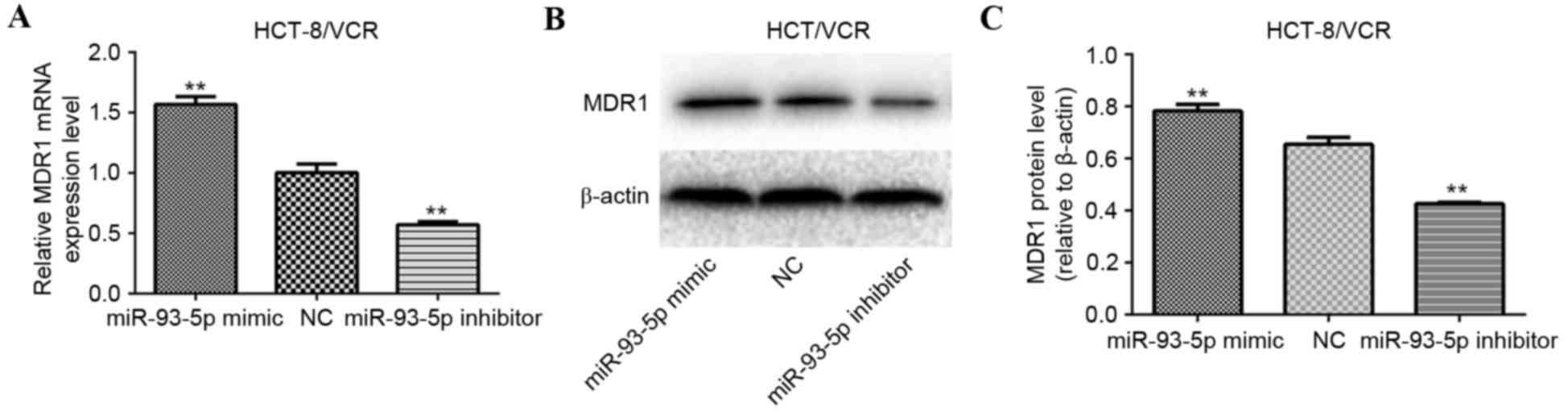
To investigate whether miR-93-5p is involved in the
regulation of MDR1 gene expression, HCT-8/VCR cells were
transfected with the miR-93-5p mimic, miR-93-5p inhibitor or the
NC. MDR1 mRNA and protein expression levels were evaluated using
RT-qPCR and western blotting, respectively. The results indicated
that MDR1 mRNA (P<0.001; Fig. 2A)
and protein (P<0.001; Fig. 2B and
C) expression levels were significantly decreased in HCT-8/VCR
cells transfected with the miR-93-5p inhibitor, compared with the
NC-transfected cells. Conversely, MDR1 mRNA (P<0.001; Fig. 2A) and protein (P<0.001; Fig. 2B and C) expression levels were
significantly increased in HCT-8/VCR cells transfected with the
miR-93-5p mimic, compared with the NC-transfected cells.
Dysregulation of miR-93-5p is
associated with the transport activity of MDR1
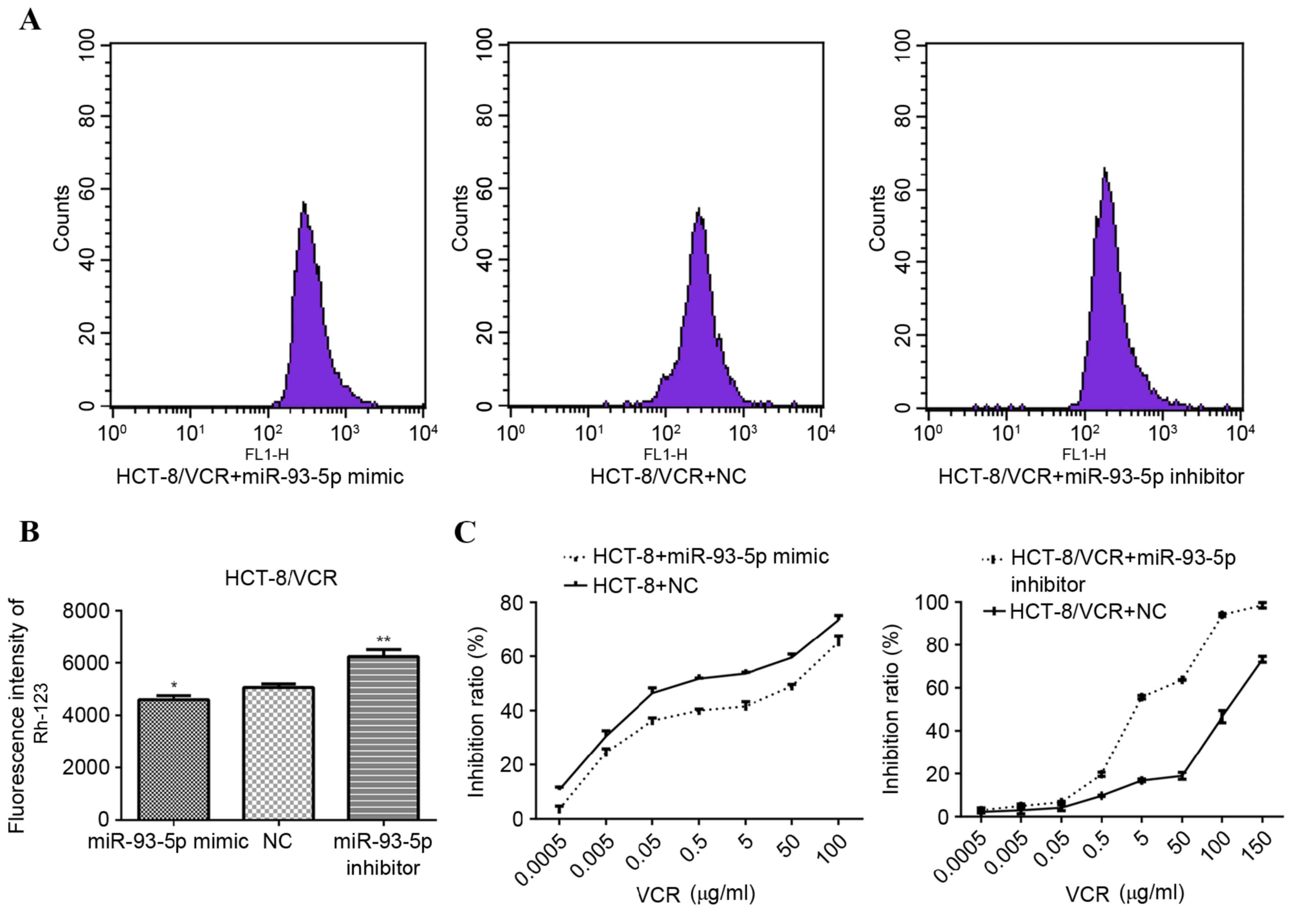
In order to verify the role of miR-93-5p in the
regulation of MDR1 expression, MDR1 transport activity was assessed
by measuring the intracellular accumulation of Rh-123, whereby
increased MDR1 transport activity is indicated by decreased
intracellular Rh-123 fluorescence. As presented in Fig. 3A and B, the fluorescence intensity of
Rh-123 was significantly increased in HCT-8/VCR cells transfected
with the miR-93-5p inhibitor, compared with cells transfected with
the NC (P<0.001). Conversely, HCT-8/VCR cells transfected with
the miR-93-5p mimic exhibited significantly decreased Rh-123
fluorescence intensity, compared with the cells transfected with
the NC (P=0.034; Fig. 3A and B).
These results were consistent with those of the RT-qPCR and western
blot analyses.
miR-93-5p modulates the sensitivity of
human CRC cells to anti-cancer drugs
To determine whether miR-93-5p is directly involved
in the development of drug resistance in human CRC cells, the
sensitivity of miR-93-5p mimic-transfected HCT-8 and miR-93-5p
inhibitor-transfected HCT-8/VCR cells to increasing concentrations
of VCR was examined using a CCK-8 assay. miR-93-5p
inhibitor-transfected HCT-8/VCR cells exhibited enhanced
sensitivity to VCR (IC50, 4.472 µM), compared with the
NC-transfected cells (IC50, 100.7 µM; Fig. 3C). Conversely, the sensitivity of
miR-93-5p mimic-transfected HCT-8 cells to VCR (IC50,
11.37 µM) was decreased, compared with the NC-transfected cells
(IC50, 0.8884 µM; Fig.
3C). The results of the present study indicate that miR-93-5p
has an important role in the regulation of MDR phenotypes in human
CRC cells.
CDKN1A as a target of miR-93-5p
repression
CDKN1A, also known as p21Cip1, is a key
member of the Cip/Kip family of cyclin kinase inhibitors and acts
as a regulator of cell cycle progression in G1 (41,42).
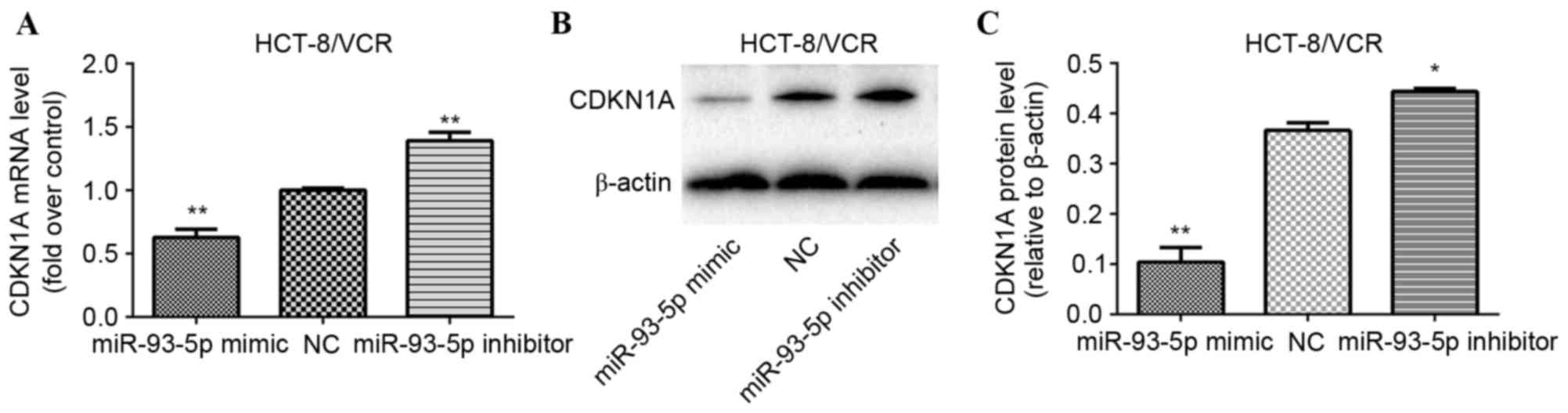
CDKN1A was identified as a potential target of miR-93-5p using the
miR target analysis tools PicTar, TargetScan and miRanda (43,44). In
order to evaluate whether miR-93-5p modulates MDR through the
downregulation of CDKN1A gene expression in human CRC cells,
HCT-8/VCR cells were transfected with a miR-93-5p mimic, miR-93-5p
inhibitor or an NC. A total of 72 h following transfection, RT-qPCR
and western blot analyses were performed to evaluate the expression
levels of CDKN1A mRNA and protein, respectively. Significant
upregulation of CDKN1A mRNA expression levels was observed in the
miR-93-5p inhibitor-transfected cells (P<0.001), and significant
downregulation of CDKN1A mRNA expression levels was observed in the
miR-93-5p mimic-transfected cells (P<0.0001), compared with
NC-transfected cells (Fig. 4A).
Furthermore, as presented in Fig. 4B and
C, transfection with the miR-93-5p mimic significantly reduced
HCT-8/VCR cell CDKN1A protein expression levels (P<0.001), while
transfection with the miR-93-5p inhibitor significantly increased
CDKN1A protein expression levels (P<0.001), compared with
transfection of the NC siRNA.
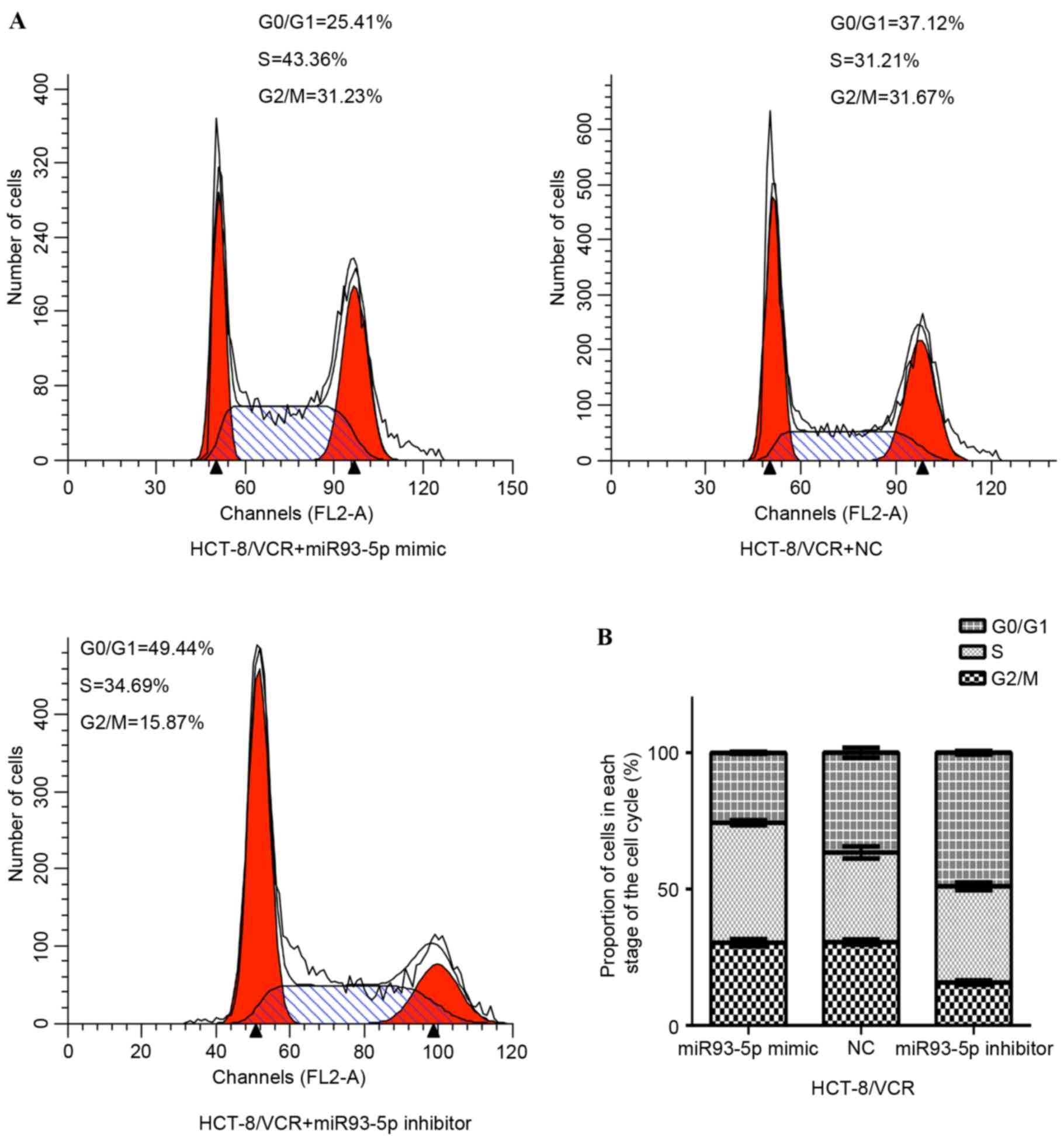
Inhibition of miR-93-5p induces
G1 cell cycle arrest in HCT-8/VCR cells
A total of 72 h following transfection with the
miR-93-5p mimic or inhibitor, the cell cycle distribution of
HCT-8/VCR cells was analyzed using flow cytometry. As presented in
Fig. 5A, the percentage of cells in
G1 was significantly increased in the inhibitor-treated
group, compared with the NC group (48.94±0.64% vs. 36.57±1.81%;
P<0.001). Conversely, the percentage of cells in G2/M
and S was significantly decreased in the inhibitor-treated group,
compared with the NC group (51.06±0.65% vs. 63.43±1.81%;
P<0.001). Furthermore, transfection with the miR-93-5p mimic
significantly decreased the proportion of cells in G1
phase, compared with the NC group (25.63±0.39% vs. 36.57±1.81%;
P<0.001). By contrast, the proportion of cells in the
G2/M and S phase significantly increased, compared with
the NC (74.37±0.39% vs. 63.43±1.81%; P<0.001). The results of
the present study suggest that repression of miR-93-5p induces
G1 arrest, and inhibits cell proliferation and
G1/S transition.
Discussion
MDR involves simultaneous resistance to multiple
anti-cancer drugs and is, therefore, a frequent cause of
chemotherapy failure, particularly in CRC (7,45).
Increasing evidence has revealed that aberrant miR expression
contributes to the development of MDR in CRC (46). Valeri et al (47) reported that CRC cells with upregulated
miR-21 expression exhibited significantly reduced sensitivity to
therapeutics. In addition, Zhang et al (48) demonstrated that miR-153 overexpression
led to oxaliplatin and cisplatin resistance in CRC. Furthermore, Xu
et al (27) observed that
miR-1915 was downregulated in drug-resistant HCT-116 and L-OHP cell
lines, and that the upregulation of miR-1915 sensitized cells to
anticancer drugs by reducing B-cell lymphoma-2 protein expression.
However, the underlying mechanisms of drug resistance in CRC remain
unclear. Additionally, a previous study indicated that miR-93 is
dysregulated in human colon cancer and correlates with treatment
resistance (33). In the present
study, the role of miR-93-5p was further examined and observed to
modulate MDR in human CRC cells by downregulating CDKN1A mRNA and
protein expression.
Recently, miR mimic technology has proven to be a
powerful tool in the study of specific miR functions, due to its
stability and lack of off-target effects (49). In the present study, a synthetic miR
mimic and inhibitor were used to investigate the role of miR-93-5p
in chemoresistance in CRC. The results indicated that the
expression of miR-93-5p was significantly increased in the
drug-resistant HCT-8/VCR cell line, compared with the parental
HCT-8 cell line, and that this was accompanied by significantly
increased MDR1 expression. Further investigation demonstrated that
transfection of HCT-8/VCR cells with an miR-93-5p inhibitor
significantly downregulated MDR1 expression, increased Rh-123
fluorescence intensity, enhanced sensitivity to VCR and induced
G1 cell cycle arrest, compared with the miR-93-5p mimic
and NC-transfected cells. Conversely, transfection of HCT-8 cells
with an miR-93-5p mimic reduced sensitivity to VCR compared with
the NC-transfected cells. The results of the present study suggest
that the role of miR-93-5p in MDR is indirectly associated with
modulation of MDR1 expression and cell cycle control.
At present, the most commonly studied underlying
mechanism of MDR is the overexpression of efflux transporters,
particularly MDR1 (13). MDR1 is a
member of the ATP binding cassette transporter superfamily, which
may reduce the accumulation of intracellular chemotherapeutics in
tumor cells through the efflux of anti-cancer drugs (13). A number of miRs have been demonstrated
to participate in the modulation of drug resistance by regulating
the expression of MDR1, including miR-27a, miR-451, miR-298, miR-9
and miR-122 (50–53). In addition to MDR1 overexpression,
regulation of the cell cycle in tumors has been indicated to be
involved in the development of MDR (54). Huang et al (55) demonstrated that cell cycle regulation
has an important role in chemoresistance in human ovarian cancer.
The present study investigated the role of MDR1 and
miR-93-5p-mediated cell cycle modulation in CRC
chemoresistance.
In order to further elucidate the underlying
mechanisms of MDR, factors involved in the direct regulation of
MDR1 and miR-93-5p-mediated cell cycle progression were examined.
Using PicTar, TargetScan and miRanda, CDKN1A was identified as a
potential target of miR-93-5p. CDKN1A is a cyclin-dependent kinase
inhibitor that belongs to the Cip/Kip family of cyclin kinase
inhibitors and negatively modulates cell cycle progression
(41). CDKN1A inhibits the activity
of cyclin/cyclin-dependent kinase complexes, thus regulating
G1/S cell cycle progression (42). CDKN1A causes G1 cell cycle
arrest when overexpressed in cells (56). Furthermore, CDKN1A has been
demonstrated to contribute to the chemotherapeutic response of
cancer cells. Ding et al (57)
observed that overexpression of CDKN1A sensitized human
osteosarcoma cells to cisplatin. Qin et al (58) reported that upregulated expression of
CDKN1A significantly increased the chemosensitivity of hepatoma
cells to cisplatin. Abukhdeir et al (59) concluded that loss of CDKN1A function
was associated with poor treatment response to tamoxifen in human
breast cancer cells. In addition, Ziller et al (60) demonstrated that CDKN1A was involved in
drug resistance in ovarian carcinoma cells.
The present study aimed to validate the modulation
of CDKN1A by miR-93-5p. Significantly elevated CDKN1A mRNA and
protein expression was observed in miR-93-5p inhibitor-transfected
HCT-8/VCR cells, compared with NC-transfected cells. Concurrently,
transfection of HCT-8/VCR cells with an miR-93-5p inhibitor induced
G1 cell cycle arrest and increased sensitivity to VCR.
The results of the present study suggest that CDKN1A is a direct
target of miR-93-5p and that repression of miR-93-5p restores
CDKN1A function. Furthermore, Wu et al (37) demonstrated that aberrant expression of
miR-93 was negatively associated with CDKN1A protein expression. In
addition, luciferase reporter assays have identified CDKN1A to be a
direct target of miR-93 (37,61,62). The
results of the present study indicate that miR-93-5p participates
in the development of MDR by downregulating the expression of
CDKN1A, subsequently modulating MDR1 expression and cell cycle
progression.
In conclusion, the results of the present study
demonstrate that miR-93-5p is upregulated in drug-resistant CRC
cells and that downregulation of miR-93-5p results in increased
chemotherapeutic sensitivity. This suggests that miR-93-5p
modulates MDR in CRC. Inhibition of miR-93-5p was found to
downregulate MDR1 expression, increase intracellular
chemotherapeutic drug concentration and increase the proportion of
cells in G1, through upregulating CDKN1A mRNA and
protein expression. Therefore, the present study indicates that
miR-93-5p is a novel therapeutic target for the treatment of CRC
with MDR.
Acknowledgements
The present study was supported by The National
Natural Science Foundation of China (grant nos. 81260316 and
81260335). Furthermore, the authors thank Professor Lian-Ying Ge
(Affiliated Tumor Hospital of Guangxi Medical University, Nanning,
China) for her advice, technical assistance and critical reading of
this manuscript.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung JJ, Lau JY, Goh KL and Leung WK: Asia
Pacific Working Group on Colorectal Cancer: Increasing incidence of
colorectal cancer in Asia: Implications for screening. Lancet
Oncol. 6:871–876. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li M and Gu J: Changing patterns of
colorectal cancer in China over a period of 20 years. World J
Gastroenterol. 11:4685–4688. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Henaine AM, Chahine G, Salameh P, Elias E,
Massoud M, Hartmann D, Aulagner G and Armoiry X: MANAGEMENT OF
METASTATIC COLORECTAL CANCER: Current Treatments and New Therapies.
J Med Liban. 63:218–227. 2015.PubMed/NCBI
|
|
6
|
Kozovska Z, Gabrisova V and Kucerova L:
Colon cancer: Cancer stem cells markers, drug resistance and
treatment. Biomed Pharmacother. 68:911–916. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Temraz S, Mukherji D, Alameddine R and
Shamseddine A: Methods of overcoming treatment resistance in
colorectal cancer. Crit Rev Oncol Hematol. 89:217–230. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kelland L: The resurgence of
platinum-based cancer chemotherapy. Nat Rev Cancer. 7:573–584.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dai Z, Huang Y and Sadee W: Growth factor
signaling and resistance to cancer chemotherapy. Curr Top Med Chem.
4:1347–1356. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schickel R, Boyerinas B, Park SM and Peter
ME: MicroRNAs: Key players in the immune system, differentiation,
tumorigenesis and cell death. Oncogene. 27:5959–5974. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu W, Zhu D, Lu S, Wang T, Wang J, Jiang
B, Shu Y and Liu P: miR-497 modulates multidrug resistance of human
cancer cell lines by targeting BCL2. Med Oncol. 29:384–391. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang T, Zheng ZM, Li XN, Li ZF, Wang Y,
Geng YF, Bai L and Zhang XB: MiR-223 modulates multidrug resistance
via downregulation of ABCB1 in hepatocellular carcinoma cells. Exp
Biol Med (Maywood). 238:1024–1032. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xia H and Hui KM: Mechanism of cancer drug
resistance and the involvement of noncoding RNAs. Curr Med Chem.
21:3029–3041. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zong C, Wang J and Shi TM: MicroRNA 130b
enhances drug resistance in human ovarian cancer cells. Tumour
Biol. 35:12151–12156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Barbarotto E, Schmittgen TD and Calin GA:
MicroRNAs and cancer: Profile, profile, profile. Int J Cancer.
122:969–977. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Osada H and Takahashi T: MicroRNAs in
biological processes and carcinogenesis. Carcinogenesis. 28:2–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cho WC: OncomiRs: The discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xue J, Niu J, Wu J and Wu ZH: MicroRNAs in
cancer therapeutic response: Friend and foe. World J Clin Oncol.
5:730–743. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fang L, Li H, Wang L, Hu J, Jin T, Wang J
and Yang BB: MicroRNA-17-5p promotes chemotherapeutic drug
resistance and tumour metastasis of colorectal cancer by repressing
PTEN expression. Oncotarget. 5:2974–2987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shang Y, Zhang Z, Liu Z, Feng B, Ren G, Li
K, Zhou L, Sun Y, Li M, Zhou J, et al: miR-508-5p regulates
multidrug resistance of gastric cancer by targeting ABCB1 and
ZNRD1. Oncogene. 33:3267–3276. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rasmussen MH, Jensen NF, Tarpgaard LS,
Qvortrup C, Rømer MU, Stenvang J, Hansen TP, Christensen LL,
Lindebjerg J, Hansen F, et al: High expression of microRNA-625-3p
is associated with poor response to first-line oxaliplatin based
treatment of metastatic colorectal cancer. Mol Oncol. 7:637–646.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu K, Liang X, Cui D, Wu Y, Shi W and Liu
J: miR-1915 inhibits Bcl-2 to modulate multidrug resistance by
increasing drug-sensitivity in human colorectal carcinoma cells.
Mol Carcinog. 52:70–78. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sui H, Cai GX, Pan SF, Deng WL, Wang YW,
Chen ZS, Cai SJ, Zhu HR and Li Q: miR200c attenuates P-gp-mediated
MDR and metastasis by targeting JNK2/c-Jun signaling pathway in
colorectal cancer. Mol Cancer Ther. 13:3137–3151. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liao H, Bai Y, Qiu S, Zheng L, Huang L,
Liu T, Wang X, Liu Y, Xu N, Yan X and Guo H: MiR-203 downregulation
is responsible for chemoresistance in human glioblastoma by
promoting epithelial-mesenchymal transition via SNAI2. Oncotarget.
6:8914–8928. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Z, Liu M, Zhu H, Zhang W, He S, Hu C,
Quan L, Bai J and Xu N: Suppression of p21 by c-Myc through members
of miR-17 family at the post-transcriptional level. Int J Oncol.
37:1315–1321. 2010.PubMed/NCBI
|
|
31
|
Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim
JH, Kim JW and Kim S: MicroRNA expression profiles in serous
ovarian carcinoma. Clin Cancer Res. 14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim YK, Yu J, Han TS, Park SY, Namkoong B,
Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK and Kim VN: Functional links
between clustered microRNAs: Suppression of cell-cycle inhibitors
by microRNA clusters in gastric cancer. Nucleic Acids Res.
37:1672–1681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou J, Zhou Y, Yin B, Hao W, Zhao L, Ju W
and Bai C: 5-Fluorouracil and oxaliplatin modify the expression
profiles of microRNAs in human colon cancer cells in vitro. Oncol
Rep. 23:121–128. 2010.PubMed/NCBI
|
|
34
|
Yu XF, Zou J, Bao ZJ and Dong J: miR-93
suppresses proliferation and colony formation of human colon cancer
stem cells. World J Gastroenterol. 17:4711–4717. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu S, Patel SH, Ginestier C, Ibarra I,
MartinTrevino R, Bai S, McDermott SP, Shang L, Ke J, Ou SJ, et al:
MicroRNA93 regulates proliferation and differentiation of normal
and malignant breast stem cells. PLoS Genet. 8:e10027512012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fu X, Tian J, Zhang L, Chen Y and Hao Q:
Involvement of microRNA-93, a new regulator of PTEN/Akt signaling
pathway, in regulation of chemotherapeutic drug cisplatin
chemosensitivity in ovarian cancer cells. FEBS Lett. 586:1279–1286.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu ZB, Li WQ, Lin SJ, Wang CD, Cai L, Lu
JL, Chen YX, Su ZP, Shang HB, Yang WL, et al: MicroRNA expression
profile of bromocriptine-resistant prolactinomas. Mol Cell
Endocrinol. 395:10–18. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu H, Cai C and Chen J: Suppression of
P-glycoprotein gene expression in Hs578T/Dox by the overexpression
of caveolin-1. FEBS Lett. 576:369–374. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhu W, Xu H, Zhu D, Zhi H, Wang T, Wang J,
Jiang B, Shu Y and Liu P: miR-200bc/429 cluster modulates multidrug
resistance of human cancer cell lines by targeting BCL2 and XIAP.
Cancer Chemother Pharmacol. 69:723–731. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Harper JW, Adami GR, Wei N, Keyomarsi K
and Elledge SJ: The p21 Cdk-interacting protein Cip1 is a potent
inhibitor of G1 cyclin-dependent kinases. Cell. 75:805–816. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiong Y, Hannon GJ, Zhang H, Casso D,
Kobayashi R and Beach D: p21 is a universal inhibitor of cyclin
kinases. Nature. 366:701–704. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tang Q, Zou Z, Zou C, Zhang Q, Huang R,
Guan X, Li Q, Han Z, Wang D, Wei H, et al: MicroRNA-93 suppress
colorectal cancer development via Wnt/β-catenin pathway
downregulating. Tumour Biol. 36:1701–1710. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chai H, Liu M, Tian R, Li X and Tang H:
miR-20a targets BNIP2 and contributes chemotherapeutic resistance
in colorectal adenocarcinoma SW480 and SW620 cell lines. Acta
Biochim Biophys Sin (Shanghai). 43:217–225. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Baguley BC: Multiple drug resistance
mechanisms in cancer. Mol Biotechnol. 46:308–316. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yu X, Li Z, Yu J, Chan MT and Wu WK:
MicroRNAs predict and modulate responses to chemotherapy in
colorectal cancer. Cell Prolif. 48:503–510. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Valeri N, Gasparini P, Braconi C, Paone A,
Lovat F, Fabbri M, Sumani KM, Alder H, Amadori D, Patel T, et al:
MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating
human DNA MutS homolog 2 (hMSH2). Proc Natl Acad Sci USA.
107:21098–21103. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang L, Pickard K, Jenei V, Bullock MD,
Bruce A, Mitter R, Kelly G, Paraskeva C, Strefford J, Primrose J,
et al: miR-153 supports colorectal cancer progression via
pleiotropic effects that enhance invasion and chemotherapeutic
resistance. Cancer Res. 73:6435–6447. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang Z: The guideline of the design and
validation of miRNA mimics. Methods Mol Biol. 676:211–223. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhu H, Wu H, Liu X, Evans BR, Medina DJ,
Liu CG and Yang JM: Role of MicroRNA miR-27a and miR-451 in the
regulation of MDR1/P-glycoprotein expression in human cancer cells.
Biochem Pharmacol. 76:582–588. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z,
Liu J, Cui Y, Bian X, Bie P and Qian C: MicroRNA-122 sensitizes HCC
cancer cells to adriamycin and vincristine through modulating
expression of MDR and inducing cell cycle arrest. Cancer Lett.
310:160–169. 2011.PubMed/NCBI
|
|
52
|
Bao L, Hazari S, Mehra S, Kaushal D, Moroz
K and Dash S: Increased expression of P-glycoprotein and
doxorubicin chemoresistance of metastatic breast cancer is
regulated by miR-298. Am J Pathol. 180:2490–2503. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Munoz JL, Bliss SA, Greco SJ, Ramkissoon
SH, Ligon KL and Rameshwar P: Delivery of functional anti-miR-9 by
mesenchymal stem cell-derived exosomes to glioblastoma multiforme
cells conferred chemosensitivity. Mol Ther Nucleic Acids.
2:e1262013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Tomida A and Tsuruo T: Drug resistance
mediated by cellular stress response to the microenvironment of
solid tumors. Anticancer Drug Des. 14:169–177. 1999.PubMed/NCBI
|
|
55
|
Huang L, Ao Q, Zhang Q, Yang X, Xing H, Li
F, Chen G, Zhou J, Wang S, Xu G, et al: Hypoxia induced paclitaxel
resistance in human ovarian cancers via hypoxia-inducible factor
1alpha. J Cancer Res Clin Oncol. 136:447–456. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Warfel NA and El-Deiry WS: p21WAF1 and
tumourigenesis: 20 years after. Curr Opin Oncol. 25:52–58. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ding Y, Wang Y, Chen J, Hu Y, Cao Z, Ren P
and Zhang Y: p21 overexpression sensitizes osteosarcoma U2OS cells
to cisplatin via evoking caspase-3 and Bax/Bcl-2 cascade. Tumour
Biol. 35:3119–3123. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Qin LF and Ng IO: Exogenous expression of
p21 (WAF1/CIP1) exerts cell growth inhibition and enhances
sensitivity to cisplatin in hepatoma cells. Cancer Lett. 172:7–15.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Abukhdeir AM, Vitolo MI, Argani P, De
Marzo AM, Karakas B, Konishi H, Gustin JP, Lauring J, Garay JP,
Pendleton C, et al: Tamoxifen-stimulated growth of breast cancer
due to p21 loss. Proc Natl Acad Sci USA. 105:288–293. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Ziller C, Lincet H, Muller CD, Staedel C,
Behr JP and Poulain L: The cyclin-dependent kinase inhibitor p21
(cip1/waf1) enhances the cytotoxicity of ganciclovir in HSV-tk
transfected ovarian carcinoma cells. Cancer Lett. 212:43–52. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ivanovska I, Ball AS, Diaz RL, Magnus JF,
Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson
AL, et al: MicroRNAs in the miR-106b family regulate p21/CDKN1A and
promote cell cycle progression. Mol Cell Biol. 28:2167–2174. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li Z, Yang CS, Nakashima K and Rana TM:
Small RNA-mediated regulation of iPS cell generation. EMBO J.
30:823–834. 2011. View Article : Google Scholar : PubMed/NCBI
|