Introduction
Lung cancer (LC) accounts for 13% (1.6 million) and
18% (1.4 million) of the global cancer incidence and
cancer-associated mortality, respectively, particularly in men
(1). In females, these rates have
increased in North America, representing the first cause of
cancer-associated mortality and the second most prevalent type of
cancer (1,2). Non-small cell LC (NSCLC) comprises 85%
of all LC cases (3). At the time of
diagnosis, ~60% of patients present with an advanced stage of the
disease (3). In Mexico, <1% of
NSCLC cases are diagnosed during early stages (3,4). The
1-year overall survival (OS) rate continues to be poor despite
treatment (5). A total of 30–35% of
patients respond to platinum-based chemotherapy, which improves the
quality of life compared with the best supportive care (6). Other strategies have been evaluated to
improve the survival rates of patients with advanced disease,
including combining molecular targeted therapies and chemotherapy,
but have produced contradictory results (7).
Certain cell surface proteins, such as the epidermal
growth factor receptor (EGFR), are used as prognostic biomarkers
and therapeutic targets that increase the response and OS rate of
patients with NSCLC (8). EGFR is
overexpressed in >60% of NSCLC cases (9). In combination with monoclonal antibodies
against EGFR, including cetuximab and necitumumab, first-line
chemotherapy has resulted in improved survival rates in patients
with advanced-stage disease (10–12).
Immunohistochemistry (IHC) is a standard method used
to identify the presence of EGFR. Currently, scoring systems assist
in determining the EGFR expression levels in tumor samples using
internationally validated antibodies (12–14). The
FLEX study assessed EGFR expression using an IHC scoring system
according to the intensity of cell membrane staining (scale of 0–3)
(15). In a subanalysis, the EGFR
expression levels determined during IHC were tested as a biomarker
to evaluate the efficacy of cisplatin and vinorelbine plus
cetuximab (12). The EGFR expression
data were used to generate IHC scores on a continuous scale of
0–300, and subsequently, the response data were used to select an
outcome-based, discriminatory threshold for an IHC score for EGFR
expression of 200 (10,13). The present study aimed to evaluate the
interobserver agreement of the results of the H-score method for
patients with advanced NSCLC among three senior pathologists using
the same system as the FLEX study (12).
Materials and methods
Samples
Tumor tissue samples from 85 patients with NSCLC,
who were treated at the Thoracic Oncology Unit, National Cancer
Institute of Mexico (INCan) (Mexico City, Mexico) were reviewed.
The samples were obtained via biopsy from patients treated between
January 2008 and December 2012. All samples were histologically
characterized. Three additional samples were used as internal
controls for EGFR expression and one negative control was included.
The general and clinical characteristics of each patient were
retrieved from clinical records. The variables selected for
analysis included age, gender, smoking history, wood-smoke and
asbestos exposure, Eastern Cooperative Oncology Group (ECOG) and
Karnofsky performance statuses, and therapy [platinum-based or
tyrosine kinase inhibitors (TKIs)].
IHC protocols
All 85 tumor samples were fixed in 10% formalin for
10 h at room temperature and embedded in paraffin. Sections (3 µm
thick) were prepared and mounted onto positively-charged glass
slides. Immunostaining was performed with an automated
immunostainer using the EGFR pharmDx™ kit (Dako, Glostrup,
Denmark), which was performed as previously described by the FLEX
study (15). EGFR expression was
evaluated by three senior pathologists (Department of Pathology,
INCan, Mexico City, Mexico), who were blinded to the clinical
outcomes. EGFR expression was assessed by IHC using the DAKO EGFR
pharmDx kit (Dako, Glostrup, Denmark), which was performed as
previously described by the FLEX study (15). The tumor samples were scored according
to the fraction of stained cells at each intensity. The staining
intensity of the cell membrane was scored within a scale ranging
from 0–3 and divided into 4 categories as follows: No staining, 0;
weak staining, 1+ (light brown membrane staining); intermediate
staining, 2+; and strong staining, 3+ (dark brown linear membrane
staining). For more reliable scoring definitions, strong staining
(3+) was clearly visible using a ×4 objective lens, moderate
staining (2+) required a ×10 or ×20 objective lens for clear
observation, and weak staining (1+) required a ×40 objective lens.
The EGFR H-score was defined as a continuous variable with a scale
ranging from 0–300 and was calculated using the following formula:
1 × (percentage of weakly stained cells, 1+) + 2 × (percentage of
moderately stained cells staining, 2+) + 3 × (percentage of
strongly stained cells, 3+). High and low scores of EGFR expression
were defined using 200 as the threshold.
Statistical analysis
For the descriptive analysis, the general variables
were summarized as the means and standard deviations or frequencies
and proportions, according to the nature of the variable
(continuous or categorical, respectively). The bivariate
correlation coefficient was calculated between and within each
pathologist's observations. The interobserver variations in the
EGFR total score were established using the mean Pearson
correlation test, whereas the interobserver variations in EGFR
scores ≥200 were established using the mean Spearman correlation
test. P<0.05 was considered to indicate a statistically
significant difference. χ2 or Fisher's exact tests were
used to assess the significance among the clinical factors and the
H-scores evaluated by each pathologist. OS time was analyzed by the
Kaplan-Meier method, and comparisons among subgroups were analyzed
by the log-rank test. For the survival curve analysis, all
variables were dichotomized. The adjustment for potential
confounders was performed using a multivariate Cox regression
model, and hazard ratios (HRs) were calculated along with their
corresponding 95% confidence intervals (CIs) as a measure of
association. All statistical analyses were performed using SPSS
v.20 (IBM SPSS, Armonk, NY, USA).
Results
Study population
A total of 85 patients diagnosed with NSCLC were
included in the present study; 49 (57.6%) of them were female and
36 (42.4%) were male. The mean age at diagnosis was 61±12.97 years
(range, 32–86 years). The majority of patients were smokers
(60.0%), with 27.57 as the mean tobacco index, and 57% of patients
were exposed to wood-smoke and 16.5% to asbestos. The most common
histological type was adenocarcinoma (77.6%). A total of 82% of
patients had stage IV cancer at diagnosis, 82.4% had a good ECOG
performance status (0–1) and 61.2% had pleural effusion at
diagnosis (Table I).
 | Table I.Baseline general characteristics
among all patients and by mean EGFR score index punctuation. |
Table I.
Baseline general characteristics
among all patients and by mean EGFR score index punctuation.
|
|
|
|
| Mean EGFR-score
index punctuation |
| Mean EGFR-score
index punctuation |
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| Characteristic | Overall (n=85) | Mean (± SD; mean
EGFR) score=125.41±96.36 | P-value | <100 (n=39) | ≥100 (n=46) | P-value | <200 (n=61) | ≥200 (n=24) | P-value |
|---|
| Gender |
|
| 0.096 |
|
| 0.048a |
|
| 0.123a |
|
Male | 42.4 (36/85) | 105.09
(±88.18) |
| 53.8 (21/39) | 32.6 (15/46) |
| 47.5 (29/61) | 29.2 (7/24) |
|
Female | 57.6 (49/85) | 140.34
(±100.21) |
| 46.2 (18/39) | 67.4 (31/46) |
| 52.5 (32/61) | 70.8 (17/24) |
|
| Mean age (±
SD) | 61.76 (12.97) | – | – | 62.3 (13.88) | 61.28 (12.28) | 0.712b | 62.54 (13.67) | 59.79 (11.02) | 0.382a |
| Median age,
years |
|
| 0.231 |
|
| 0.849a |
|
| 0.113a |
|
<60 | 44.7 (38/85) | 138.38
(±97.82) |
| 43.6 (17/39) | 45.7 (21/46) |
| 39.3 (24/61) | 58.3 (14/24) |
|
|
≥60 | 55.3 (47/85) | 114.11
(±94.69) |
| 56.4 (22/39) | 54.3 (25/46) |
| 60.7 (37/61) | 41.7 (10/24) |
|
| Smoking
exposure |
|
| 0.252 |
|
| 0.288a |
|
| 0.106a |
|
Non-smoker | 40.0 (34/85) | 141.47
(±108.33) |
| 33.3 (13/39) | 45.7 (21/46) |
| 34.4 (21/61) | 54.2 (13/24) |
|
|
Smoker | 60.0 (51/85) | 116.86
(±86.44) |
| 66.6 (26/39) | 54.3 (25/46) |
| 65.6 (40/61) | 45.8 (11/24) |
|
| Tobacco index |
|
|
|
|
| 0.384b |
|
| 0.696b |
| Mean (±
SD) | 27.57 (36.67) | – | – | 22.99 (18.44) | 32.16 (48.78) |
| 26.48 (30.70) | 31.45 (55.04) |
|
| Wood-smoke
exposure |
|
| 0.488 |
|
| 0.514a |
|
| 0.936a |
|
Absent | 57.6 (49/85) | 131.66
(±88.74) |
| 53.8 (21/39) | 60.9 (28/46) |
| 57.4 (35/61) | 58.3 (14/24) |
|
|
Present | 42.4 (36/85) | 116.89
(±106.55) |
| 46.2 (18/39) | 39.1 (18/46) |
| 42.6 (26/61) | 41.7 (10/24) |
|
| Wood-smoke
index |
|
|
|
|
| 0.193b |
|
| 0.696b |
| Mean (±
SD) | 90.41 (100.68) | – | – | 68.38 (72.94) | 112.44
(120.50) |
| 73.18 (81.17) | 135.20
(134.28) |
|
| Asbestos
exposure |
|
| 0.131 |
|
| 0.155a |
|
| 0.976a |
|
Absent | 83.5 (71/85) | 118.38
(±97.45) |
| 89.7 (35/39) | 78.3 (36/46) |
| 83.6 (51/61) | 83.3 (20/24) |
|
|
Present | 16.5 (14/85) | 161.07
(±84.99) |
| 10.3 (4/39) | 21.7 (10/46) |
| 16.4 (10/61) | 16.7 (4/24) |
|
| Histology |
|
| 0.974 |
|
| 0.883a |
|
| 0.344a |
|
Adenocarcinoma | 77.6 (66/85) | 125.22
(±97.14) |
| 76.9 (30/39) | 78.3 (36/46) |
| 80.3 (49/61) | 70.8 (17/24) |
|
|
Other | 22.4 (19/85) | 126.05
(±96.18) |
| 23.1 (9/39) | 21.7 (10/46) |
| 19.7 (12/61) | 29.2 (7/24) |
|
| Disease stage |
|
| 0.776 |
|
| 0.614a |
|
| 0.882a |
|
II–III | 17.6 (15/85) | 131.73
(±93.90) |
| 15.4 (6/39) | 19.6 (9/46) |
| 18.0 (11/61) | 16.7 (4/24) |
|
| IV | 82.4 (70/85) | 124.02
(±97.48) |
| 84.6 (33/39) | 80.4 (37/46) |
| 82.0 (50/61) | 83.3 (20/24) |
|
| ECOG PS |
|
| 0.131 |
|
| 0.227a |
|
| 0.158a |
|
0–1 | 82.4 (70/85) | 132.73
(±95.96) |
| 76.9 (30/39) | 87.0 (40/46 |
| 78.7 (48/61) | 91.7 (22/24) |
|
|
2–3 | 17.6 (15/85) | 91.22 (±93.80) |
| 23.1 (9/39) | 13.0 (6/46) |
| 21.3 (13/61) | 8.3 (2/24) |
|
| Pleural
effusion |
|
| 0.396 |
|
| 0.406a |
|
| 0.515a |
|
Yes | 61.2 (52/33) | 132.53
(±93.26) |
| 56.4 (22/39) | 65.2 (30/46) |
| 59.0 (36/61) | 66.7 (16/24) |
|
| No | 38.8 (33/85) | 114.19
(±101.48) |
| 43.6 (17/39) | 38.4 (16/46) |
| 41.0 (25/61) | 33.3 (8/24) |
|
| PB-CT |
|
| 0.955 |
|
| 0.748a |
|
| 0.969a |
| No | 24.7 (21/85) | 124.36
(±106.61) |
| 23.1 (9/39) | 26.1 (12/46) |
| 24.6 (16/61) | 25.0 (6/24) |
|
|
Yes | 75.3 (64/85) | 125.75
(±95.40) |
| 76.9 (30/39) | 73.9 (34/46) |
| 75.4 (46/61) | 75.0 (18/24) |
|
| DCR with PB-CT |
|
| 0.102 |
|
| 0.265a |
|
| 0.697a |
|
Yes | 30.6 (26/64) | 149.35
(±95.09) |
| 33.3 (10/30) | 47.1 (16/34) |
| 39.1 (18/46) | 44.4 (8/18) |
|
| No | 44.7 (38/64) | 109.60
(±93.43) |
| 66.7 (20/30) | 52.9 (18/34) |
| 60.9 (28/46) | 55.6 (10/18) |
|
| TKI |
|
| 0.287 |
|
| 0.227a |
|
| 0.158a |
| No | 82.4 (70/85) | 130.59
(±99.63) |
| 76.9 (30/39) | 87.0 (40/46) |
| 78.7 (48/61) | 91.7 (22/24) |
|
|
Yes | 17.6 (15/85) | 101.22
(±77.61) |
| 23.1 (9/39) | 13.0 (6/46) |
| 21.3 (13/61) | 8.3 (2/24) |
|
| DCR with TKI |
|
| 0.818 |
|
| 1.000a |
|
| 0.283a |
|
Yes | 33.3 (5/15) | 94.33 (±79.00) |
| 33.3 (3/9) | 33.3 (2/6) |
| 38.5 (5/13) | 0.0 (0/2) |
|
| No | 66.7 (10/15) | 104.66
(±80.98) |
| 66.7 (6/9) | 66.7 (4/6) |
| 61.5 (8/13) | 100.0 (2/2) |
|
EGFR score evaluation
A total of 85 tumor tissue samples were analyzed
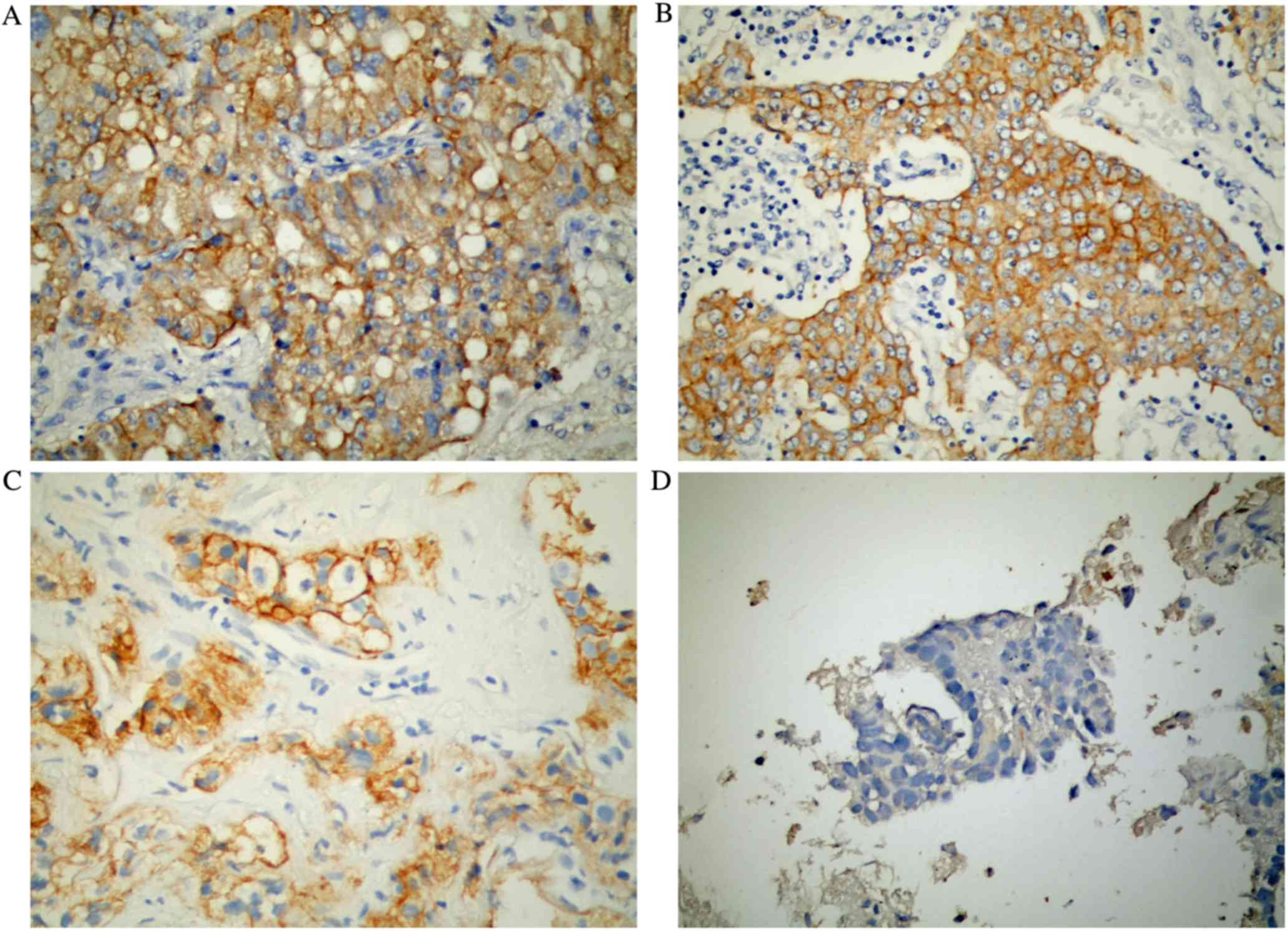
according to the FLEX study methodology (12). Fig. 1
presents the differences in EGFR staining among the tissues
histologically classified as adenocarcinoma. Each pathologist
assessed all tumor samples; the means and standard deviations of
the EGFR scores from pathologists A, B and C were 111±102, 127±103
and 128.53±104.03, respectively. When assessing the average EGFR
scores from the three pathologists, EGFR scores ≥1, ≥100, ≥200 and
between 250–300 were observed in 85.9, 54.1, 28.2 and 12.9% of the
tumor samples, respectively (Table
II).
 | Table II.EGFR immunohistochemistry scores
interobserver agreement. |
Table II.
EGFR immunohistochemistry scores
interobserver agreement.
| Score | Overall | Pathologist A | Pathologist B | Pathologist C |
|---|
| EGFR score |
|
|
|
|
| Mean (±
SD) | 125.41 (96.36) | 111 (102.08) | 126.65
(103.05) | 128.53
(104.037) |
|
Contingency coefficient |
| 0.982 | 0.980 | 0.988 |
| EGFR score 1 |
|
|
|
|
|
<1 | 14.1 (12/85) | 31.8 (27/85) | 30.6 (26/85) | 14.1 (12/85) |
| ≥1 | 85.9 (73/85) | 68.2 (27/85) | 69.4 (59/85) | 85.9 (73/85) |
|
Spearman's rho |
| 0.487 | 0.495 | 0.604 |
| κ
measurement agreement |
| 0.101 | 0.102 | 0.123 |
| EGFR score 100 |
|
|
|
|
|
<100 | 45.9 (39/85) | 47.1 (40/85) | 41.2 (35/85) | 42.4 (36/85) |
|
≥100 | 54.1 (46/85) | 52.9 (45/85) | 58.8 (50/85) | 57.6 (49/85) |
|
Spearman's rho |
| 0.834 | 0.857 | 0.734 |
| κ
measurement agreement |
| 0.201 | 0.193 | 0.082 |
| EGFR score 200 |
|
|
|
|
|
<200 | 71.8 (61/85) | 74.1 (63/85) | 62.4 (53/85) | 58.8 (50/85) |
|
≥200 | 28.2 (24/85) | 25.9 (22/85) | 37.6 (32/85) | 41.2 (35/85) |
|
Spearman's rho |
| 0.773 | 0.71 | 0.675 |
| κ
measurement agreement |
| 0.116 | 0.111 | 0.044 |
| EGFR score
250–300 |
|
|
|
|
|
<250 | 87.1 (74/85) | 84.7 (72/85) | 88.2 (75/85) | 82.4 (70/85) |
|
250–300 | 12.9 (11/85) | 15.3 (13/85) | 11.8 (10/85) | 17.6 (15/85) |
|
Spearman's rho |
| 0.635 | 0.545 | 0.541 |
| κ
measurement agreement |
| 0.057 | 0.056 | 0.021 |
Clinical characteristics associated
with EGFR scores >100 and >200
The mean overall EGFR score was 125.45 (±96.36). No
different was observed in the mean EGFR score in terms of any
clinical characteristic. Furthermore, no differences were observed
in the patients' age, tobacco smoking, exposure to wood-smoke or
asbestos, histological type, disease stage, ECOG performance status
or pleural effusion at diagnosis when patients with an EGFR score
<200 were compared with patients with an EGFR score ≥200, or
patients with an EGFR score <100 were compared with patients
with an EGFR score ≥100. Female gender was the only clinical
characteristic that was associated with a higher frequency of
patients with a mean EGFR score >100 (64.7 vs. 46.2%; P=0.048;
Table I).
EGFR score correlation
Regarding EGFR score evaluation, the agreement
contingency coefficient was 98%; the interobserver agreement
between the high IHC EGFR scores (≥1, 100, 200 and 250–300) ranged
from 0.487–0.604, 0.7.4–0.834, 0.675–0.773 and 0.541–0.635,
respectively (Table II).
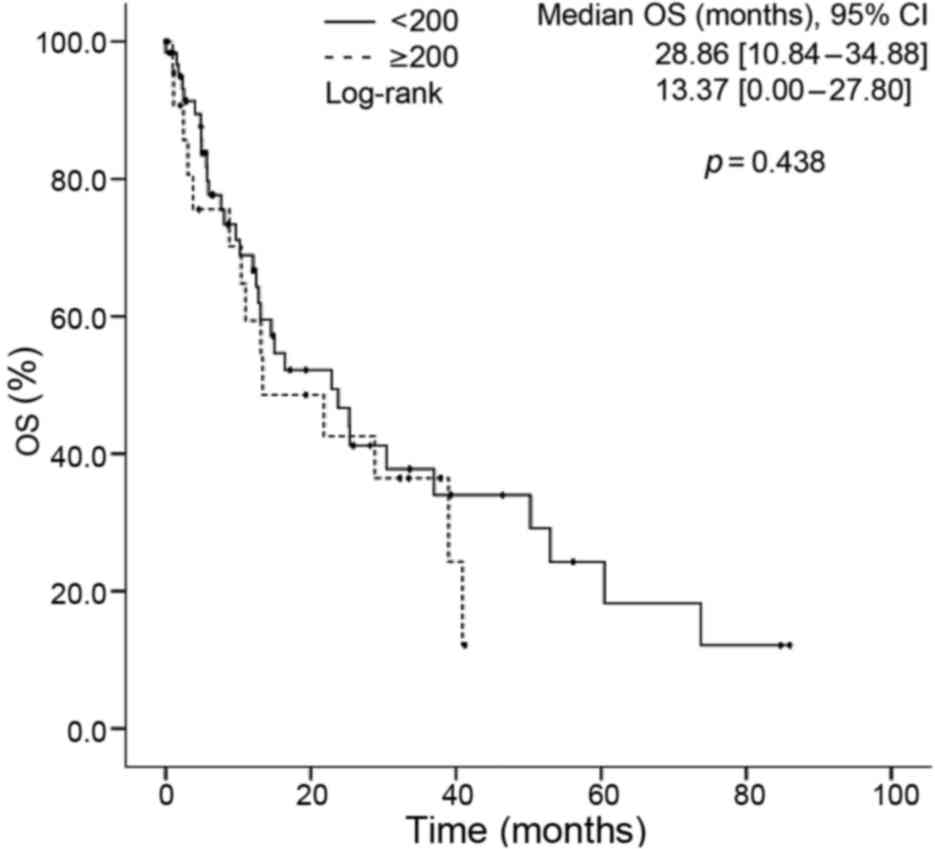
EGFR score and clinical outcomes
The median OS time among the study population was
21.81 months. No differences were observed in the OS time of
patients with regard to their gender, age, tobacco smoking,
wood-smoke or asbestos exposure, histological type, ECOG
performance status, disease stage, pleural effusion at baseline or
EGFR score (<200 vs. ≥200) (Fig.
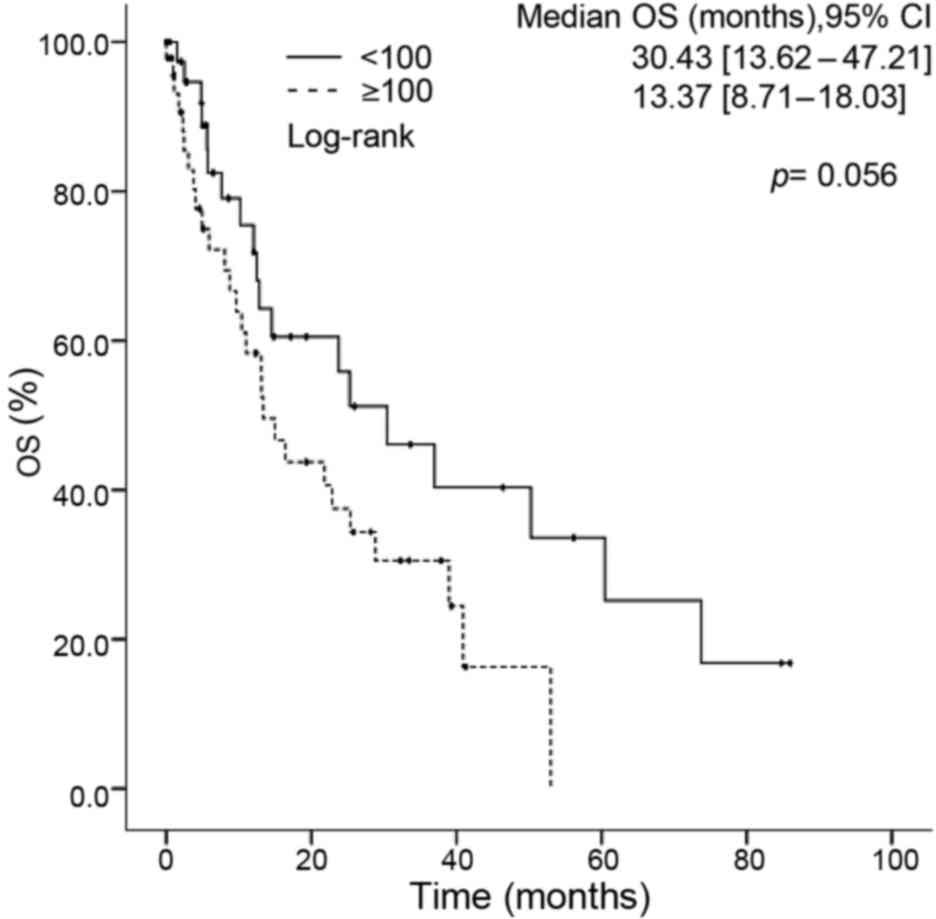
2) in the univariate analysis. An EGFR score >100 was the
only clinical characteristic associated with a shorter OS time
(13.37 vs. 30.43 months for patients with an EGFR score <100;
P=0.05) (Fig. 3). In the multivariate
analysis, EGFR score (<100 vs. ≥100) was the only factor that
was independently associated with OS time [HR, 2.56 (1.24–5.44);
P=0.015] (Table III).
 | Table III.Univariate and multivariate
analysis. |
Table III.
Univariate and multivariate
analysis.
|
| Overall
survival |
|---|
|
|
|
|---|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Characteristic | Median | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Overall | 21.81 | 10.32–33.39 |
|
|
|
|
| Gender |
|
|
|
|
|
|
|
Female | 25.29 | 7.63–18.58 |
|
|
|
|
|
Male | 13.1 | 11.19–39.40 | 0.195 | 0.49 | 0.21–1.12 | 0.091 |
| Median age,
years |
|
|
|
|
|
|
|
<60 | 28.84 | 16.78–40.90 |
|
|
|
|
|
≥60 | 14.94 | 10.08–19.81 | 0.214 | 1.46 | 0.75–2.84 | 0.260 |
| Smoking
exposure |
|
|
|
|
|
|
|
Non-smoker | 22.86 | 7.95–37.77 |
|
|
|
|
|
Smoker | 21.81 | 5.66–37.96 | 0.582 | 0.61 | 0.26–1.41 | 0.255 |
| Wood-smoke
exposure |
|
|
|
|
|
|
|
Absent | 14.52 | 11.90–17.14 |
|
|
|
|
|
Present | 25.29 | 8.19–41.36 | 0.386 | 0.66 | 0.30–1.45 | 0.306 |
| Asbestos
exposure |
|
|
|
|
|
|
|
Absent | 23.75 | 11.08–36.41 |
|
|
|
|
|
Present | 8.08 | 0.808–15.35 | 0.065 | 1.88 | 0.67–4.24 | 0.265 |
| Histology |
|
|
|
|
|
|
|
Adenocarcinoma | 22.86 | 11.5–34.23 |
|
|
|
|
Other | 13.1 | 2.87–23.37 | 0.634 | 1.55 | 0.65–3.67 | 0.313 |
| Disease stage |
|
|
|
|
|
|
|
II–III | 25.36 | 0.54–50.17 |
|
|
|
|
| IV | 21.81 | 11.07–32.55 | 0.589 | 1.82 | 0.78–4.23 | 0.164 |
| ECOG PS |
|
|
|
|
|
|
|
0–1 | 21.81 | 10.86–32.77 |
|
|
|
|
|
2–3 | 30.42 | 0.00–70.76 | 0.740 | 1.4 | 0.53–3.68 | 0.485 |
| Pleural
effusion |
|
|
|
|
|
|
|
Yes | 16.42 | 3.72–29.12 |
|
|
|
|
| No | 25.36 | 4.06–46.44 | 0.778 | 1.05 | 0.53 −2.08 | 0.884 |
| Mean EGFR
score |
|
|
|
|
|
|
|
<100 | 30.43 | 13.62 −47.21 |
|
|
|
|
|
≥100 | 13.37 | 8.701–18.03 | 0.056a | 2.56 | 1.20–5.44 | 0.015a |
| Mean EGFR
score |
|
|
|
|
|
|
|
<200 | 22.86 | 10.48–34.88 |
|
|
|
|
|
≥200 | 13.37 | 0.00–27.80 | 0.438 |
|
|
|
Discussion
EGFR, a gene that is frequently overexpressed in
40–80% of NSCLC cases, serves an important role in tumor cell
survival and proliferation (16,17).
Recent clinical trials with EGFR inhibitors have demonstrated
positive results in patients with NSCLC, particularly by increasing
the progression-free survival (PFS) time among patients harboring
EGFR mutations compared with patients with wild-type EGFR (7–12).
Meta-analyses regarding the use of TKIs in a population with mixed
EGFR-activating mutations have only reported PFS and relative risk
(RR) benefits, and no OS benefit (13–15).
Previous studies have demonstrated the benefits of afatinib on
patient OS using pooled data from the LUX-lung 3 and 6 clinical
trials (11,12,18).
Afatinib is hypothetically more effective at inhibiting EGFR
signaling than reversible TKIs, due it forming stable covalent
bonds and irreversibly inhibiting ATP from binding to the tyrosine
kinase domain of EGFR (19); however,
no randomized clinical trial or meta-analysis has demonstrated that
afatinib is superior to erlotinib or gefitinib regarding OS.
Irreversible TKIs have been reported to significantly improve the
OS time of patients with exon 19 deletions (20,21).
Despite such findings, the association between the
expression of EGFR mRNA and protein and treatment response is
currently unclear, as is the optimal method for determining EGFR
levels in tumors. A study of 183 tissue samples from patients with
NSCLC assessing the correlation between protein expression and gene
copy number by IHC observed that increased EGFR protein levels were
correlated with a high gene copy number (13). The same study reported that a high
copy number correlated with a poor prognosis and that this
phenomenon was more frequently observed in patients with squamous
cell carcinoma than in patients with other types of carcinoma
(13). In addition, the correlation
between the expression of ErbB receptor family proteins and
different clinical outcomes and therapeutic responses to monoclonal
antibodies has also been widely studied, although the results are
more heterogeneous (22). EGFR
expression is frequently observed in NSCLC patients with brain
metastases (23). A previous study
that performed IHC analysis of EGFR, human epidermal growth factor
receptor (HER) 2 and HER3 expression in tissue microarrays of 131
NSCLC brain metastases identified that ErbB receptor family members
were frequently overexpressed (23).
However, no significant correlations between the overexpression of
ErbB receptor family members and clinical pathological parameters,
including OS time, were observed (24). By contrast, a prospective study
assessing the development of brain metastasis in 293 patients with
advanced NSCLC reported that EGFR expression (RR, 1.6; 95% CI,
1.4–1.9; P=0.012) was independently associated with a shorter OS
time (25). Similarly, a report from
a retrospective cohort of patients with NSCLC that assessed HER2
levels demonstrated a higher objective response rate among patients
that overexpressed HER2 and were treated with trastuzumab (26). The median OS time of patients with a
H-score ≥200 may be improved with the addition of cetuximab to
their chemoradiation regimen (42 vs. 21 months), but cetuximab may
be detrimental for patients with a H-score <200 (27). EGFR expression level is a predictive
value for the response of patients with advanced NSCLC to
chemotherapy plus cetuximab treatment (14). In addition to gemcitabine and
cisplatin chemotherapy, a second generation of recombinant, human
immunoglobulin G1 EGFR monoclonal antibodies, including
necitumumab, improves OS time (28).
Tissues were evaluated by IHC to determine the level of EGFR
protein expression (28). EGFR
expression was high (≥200) in 38% of the tissues and low (<200)
in 62% of the tissues. The HR for OS for treatment with
necitumumab/gemcitabine/cisplatin vs. gemcitabine/cisplatin alone
was more favorable in patients bearing tumors with high EGFR
expression (29). However, there was
no difference observed between the low and high H-score groups when
assessing the PFS and OS time; in addition, an EGFR H-score ≥200
did not predict treatment efficacy in patients with NSCLC who
received necitumumab plus cisplatin and pemetrexed (29).
Pirker et al (15) analyzed EGFR IHC data to investigate
whether tumor EGFR expression level was predictive of the efficacy
of chemotherapy plus cetuximab. EGFR expression data were used to
generate IHC scores on a continuous scale of 0–300, and the
response data was subsequently employed to select an outcome-based,
discriminatory threshold IHC score for EGFR expression of 200
(10). Likewise, the phase III FLEX
study demonstrated that the addition of cetuximab, an EGFR
antibody, to cisplatin and vinorelbine significantly longer OS time
compared with chemotherapy alone as a first-line treatment for
patients with advanced NSCLC that overexpress EGFR (10). In this analysis, a total of 982 (90%)
patients were evaluated by IHC, and EGFR expression was high
(H-score ≥200) in 374 patients (38%) and low (H-score <200) in
608 patients (62%) (10). The HR for
OS time for necitumumab plus gemcitabine/cisplatin vs.
gemcitabine/cisplatin alone was more favorable in patients bearing
tumors with high EGFR expression (HR, 0.75 [95% CI, 0.60–0.94])
than in those with low EGFR expression (HR, 0.90 [95% CI,
0.75–1.07]). This previous study allowed the current study to
differentiate a patient subgroup that would derive a survival
benefit from the addition of cetuximab to chemotherapy, which was
associated with a score ≥200, compared with other subgroups that
would receive little or no benefit and whose score was <200
(15).
Regarding the reproducibility of the H-score,
Rüschoff et al (28) evaluated
the interobserver reproducibility of this EGFR IHC scoring system.
A high agreement was observed amongst the scores with an overall
concordance rate of 90.9% and a mean coefficient of 0.812 (29). Specimens with reference scores <200
and ≥200 exhibited mean concordance rates of 94.7 and 85.6%,
respectively (15). According to
these studies, EGFR expression measured by IHC is a potential
predictive biomarker for the response of patients with NSCLC to
cetuximab, with the advantage that IHC is a well-established,
widely used and low cost technique (15,29). The
reproducibility and validation of these results in other
populations has not been widely studied. According to Hirsch et
al (24), a lower cut-off score
for EGFR expression is able to better resolve positive and negative
EGFR IHC results. When higher cut-off points were used to define
positive staining, they did not improve the test's discrimination
(24).
In the present study, a good interobserver agreement
of 80–90% was observed with a mean coefficient of 0.983 among three
pathologists, and the positivity in the samples was 70%, which is
consistent with other studies (28,30). In
the present study, a better concordance for the H-scores was
observed when using a cut-off of 100 (73.4–83.4%); meanwhile, the
concordance of the cut-off of 200 ranged from 67.5–77.3%. Samples
with a reference EGFR H-score <200 and ≥200 demonstrated mean
concordance rates of 94.7 and 85.6%, respectively (15). An important hallmark of the study was
that the population was not selected based on IHC expression levels
or treatment regimen.
In other forms of cancer, including breast and
gastric cancer, IHC determination of molecular markers, such as
HER2 overexpression, is important for the treatment strategy
(31,32). Patients with high HER2-expressing
tumors derive the greatest benefit from trastuzumab therapy
(31). Additionally, it was
previously determined that the interlaboratory reproducibility of
HER2 expression in gastric cancer using two different antibodies
was 48.3 vs. 75.9%, while the interobserver reproducibility was
~90% (32). Testing and scoring is
important to ensure the accurate identification of patients who are
eligible for treatment.
Finally, in the present study, an EGFR H-score
>100 was frequently observed in women. Currently, the overall
incidence of LC is increasing in females and Hispanic women present
with a higher prevalence of EGFR mutations than men (36.9 vs.
18.5%, respectively) (4,33). Studies have demonstrated that EGFR
expression is closely associated with poor survival in females who
are undergoing conventional chemotherapy, with a higher mortality
than breast and colorectal cancer combined (33). An EGFR expression rate of 50% has been
reported in women with NSCLC, highlighting that EGFR may be used as
an indicator of the increasing incidence, poor prognosis and
disease progression in female patients with NSCLC (33,34).
Conversely, alterations in the EGFR gene represent a better
response to TKI treatment and OS time for women, who otherwise
would have a poor prognosis in response to chemotherapy (35).
In conclusion, EGFR expression is a hallmark of
several neoplasms, particularly LC, where it is a determinant for
targeted treatment. The present study demonstrated that
determination of EGFR expression levels by IHC is highly
reproducible between pathologists. According to this data, high
EGFR expression levels are associated with a poorer prognosis for
patients with NSCLC; however, these levels may be associated with a
better OS time in patients with EGFR mutations who undergo EGFR TKI
treatment.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arrieta O, Guzmán-de Alba E, Alba-López
LF, Acosta-Espinoza A, Alatorre-Alexander J, Alexander-Meza JF,
Allende-Pérez SR, Alvarado-Aguilar S, Araujo-Navarrete ME,
Argote-Greene LM, et al: National consensus of diagnosis and
treatment of non-small cell lung cancer. Rev Invest Clin. 65:(Suppl
1). S5–S84. 2013.PubMed/NCBI
|
|
4
|
Arrieta O, Ramírez-Tirado LA, Báez-Saldana
R, Peña-Curiel O, Soca-Chafre G and Macedo-Perez EO: Different
mutation profiles and clinical characteristics among Hispanic
patients with non-small cell lung cancer could explain the
‘Hispanic paradox’. Lung Cancer. 90:161–166. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
National Cancer Institute, . SEER Stat
Fact Sheets: Lung and Bronchus Cancer. Available from. http://seer.cancer.gov/statfacts/html/lungb.htmlAccessed
on April 1, 2016.
|
|
6
|
Azzoli CG, Giaccone G and Temin S:
American society of clinical oncology clinical practice guideline
update on chemotherapy for stage IV non-small-cell lung cancer. J
Oncol Pract. 6:39–43. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun S, Schiller JH, Spinola M and Minna
JD: New molecularly targeted therapies for lung cancer. J Clin
Invest. 117:2740–2750. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Siegelin MD and Borczuk AC: Epidermal
growth factor receptor mutations in lung adenocarcinoma. Lab
Invest. 94:129–137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gazdar AF: Epidermal growth factor
receptor inhibition in lung cancer: The evolving role of
individualized therapy. Cancer Metastasis Rev. 29:37–48. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pirker R, Pereira JR, Szczesna A, von
Pawel J, Krzakowski M, Ramlau R, Vynnychenko I, Park K, Yu CT,
Ganul V, et al: Cetuximab plus chemotherapy in patients with
advanced non-small-cell lung cancer (FLEX): An open-label
randomised phase III trial. Lancet. 373:1525–1531. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Paz-Ares L, Mezger J, Ciuleanu TE, Fischer
JR, von Pawel J, Provencio M, Kazarnowicz A, Losonczy G, de Castro
G Jr, Szczesna A, et al: Necitumumab plus pemetrexed and cisplatin
as first-line therapy in patients with stage IV non-squamous
non-small-cell lung cancer (INSPIRE): An open-label, randomised,
controlled phase 3 study. Lancet Oncol. 16:328–337. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Douillard JY, Pirker R, O'Byrne KJ, Kerr
KM, Störkel S, von Heydebreck A, Grote HJ, Celik I and Shepherd FA:
Relationship between EGFR expression, EGFR mutation status, and the
efficacy of chemotherapy plus cetuximab in FLEX study patients with
advanced non-small-cell lung cancer. J Thorac Oncol. 9:717–724.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hirsch FR, Dziadziuszko R, Thatcher N,
Mann H, Watkins C, Parums DV, Speake G, Holloway B, Bunn PA Jr and
Franklin WA: Epidermal growth factor receptor immunohistochemistry:
Comparison of antibodies and cutoff points to predict benefit from
gefitinib in a phase 3 placebo-controlled study in advanced
nonsmall-cell lung cancer. Cancer. 112:1114–1121. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
O'Byrne KJ, Gatzemeier U, Bondarenko I,
Barrios C, Eschbach C, Martens UM, Hotko Y, Kortsik C, PazAres L,
Pereira JR, et al: Molecular biomarkers in non-small-cell lung
cancer: A retrospective analysis of data from the phase 3 FLEX
study. Lancet Oncol. 12:795–805. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pirker R, Pereira JR, von Pawel J,
Krzakowski M, Ramlau R, Park K, de Marinis F, Eberhardt WE,
Paz-Ares L, Störkel S, et al: EGFR expression as a predictor of
survival for first-line chemotherapy plus cetuximab in patients
with advanced non-small-cell lung cancer: Analysis of data from the
phase 3 FLEX study. Lancet Oncol. 13:33–42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Herbst RS and Shin DM: Monoclonal
antibodies to target epidermal growth factor receptor-positive
tumors: A new paradigm for cancer therapy. Cancer. 94:1593–1611.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Toyooka S, Mitsudomi T, Soh J, Aokage K,
Yamane M, Oto T, Kiura K and Miyoshi S: Molecular oncology of lung
cancer. Gen Thorac Cardiovasc Surg. 59:527–537. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brugger W, Triller N, BlasinskaMorawiec M,
Curescu S, Sakalauskas R, Manikhas GM, Mazieres J, Whittom R, Ward
C, Mayne K, et al: Prospective molecular marker analyses of EGFR
and KRAS from a randomized, placebo-controlled study of erlotinib
maintenance therapy in advanced non-small-cell lung cancer. J Clin
Oncol. 29:4113–4120. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kosaka T, Yamaki E, Mogi A and Kuwano H:
Mechanisms of resistance to EGFR TKIs and development of a new
generation of drugs in non-small-cell lung cancer. J Biomed
Biotechnol. 2011:1652142011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Johnson ML, Sima CS, Chaft J, Paik PK, Pao
W, Kris MG, Ladanyi M and Riely GJ: Association of KRAS and EGFR
mutations with survival in patients with advanced lung
adenocarcinomas. Cancer. 119:356–362. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kuan FC, Kuo LT, Chen MC, Yang CT, Shi CS,
Teng D and Lee KD: Overall survival benefits of first-line EGFR
tyrosine kinase inhibitors in EGFR-mutated non-small-cell lung
cancers: A systematic review and meta-analysis. Br J Cancer.
113:1519–1528. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Merrick DT, Kittelson J, Winterhalder R,
Kotantoulas G, Ingeberg S, Keith RL, Kennedy TC, Miller YE,
Franklin WA and Hirsch FR: Analysis of c-ErbB1/epidermal growth
factor receptor and c-ErbB2/HER-2 expression in bronchial
dysplasia: Evaluation of potential targets for chemoprevention of
lung cancer. Clin Cancer Res. 12:2281–2288. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Berghoff AS, Magerle M, IlhanMutlu A,
Dinhof C, Widhalm G, Dieckman K, Marosi C, Wohrer A, Hackl M,
Zöchbauer-Müller S, et al: Frequent overexpression of ErbB-receptor
family members in brain metastases of non-small cell lung cancer
patients. APMIS. 121:1144–1152. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hirsch FR, VarellaGarcia M, Cappuzzo F,
McCoy J, Bemis L, Xavier AC, Dziadziuszko R, Gumerlock P, Chansky
K, West H, et al: Combination of EGFR gene copy number and protein
expression predicts outcome for advanced non-small-cell lung cancer
patients treated with gefitinib. Ann Oncol. 18:752–760. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Arrieta O, SaavedraPerez D, Kuri R,
AvilesSalas A, Martinez L, MendozaPosada D, Castillo P, Astorga A,
Guzman E and De la Garza J: Brain metastasis development and poor
survival associated with carcinoembryonic antigen (CEA) level in
advanced non-small cell lung cancer: A prospective analysis. BMC
Cancer. 9:1192009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Langer CJ, Stephenson P, Thor A, Vangel M
and Johnson DH: Eastern Cooperative Oncology Group Study 2598:
Trastuzumab in the treatment of advanced non-small-cell lung
cancer: Is there a role? Focus on Eastern Cooperative Oncology
Group study 2598. J Clin Oncol. 22:1180–1187. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bradley JD, Paulus R, Komaki R, Masters G,
Blumenschein G, Schild S, Bogart J, Hu C, Forster K, Magliocco A,
et al: Standard-dose versus high-dose conformal radiotherapy with
concurrent and consolidation carboplatin plus paclitaxel with or
without cetuximab for patients with stage IIIA or IIIB
non-small-cell lung cancer (RTOG 0617): A randomised, two-by-two
factorial phase 3 study. Lancet Oncol. 16:187–199. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rüschoff J, Kerr KM, Grote HJ, Middel P,
von Heydebreck A, Alves VA, Baldus SE, Buttner R, Carvalho L, Fink
L, et al: Reproducibility of immunohistochemical scoring for
epidermal growth factor receptor expression in non-small cell lung
cancer: Round robin test. Arch Pathol Lab Med. 137:1255–1261. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thatcher N, Hirsch FR, Luft AV, Szczesna
A, Ciuleanu TE, Dediu M, Ramlau R, Galiulin RK, Bálint B, Losonczy
G, et al: Necitumumab plus gemcitabine and cisplatin versus
gemcitabine and cisplatin alone as first-line therapy in patients
with stage IV squamous non-small-cell lung cancer (SQUIRE): An
open-label, randomised, controlled phase 3 trial. Lancet Oncol.
16:763–774. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mazières J, Peters S, Lepage B, Cortot AB,
Barlesi F, BeauFaller M, Besse B, Blons H, MansuetLupo A, Urban T,
et al: Lung cancer that harbors an HER2 mutation: Epidemiologic
characteristics and therapeutic perspectives. J Clin Oncol.
31:1997–2003. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rüschoff J, Hanna W, Bilous M, Hofmann M,
Osamura RY, PenaultLlorca F, van de Vijver M and Viale G: HER2
testing in gastric cancer: A practical approach. Mod Pathol.
25:637–650. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Prenzel N, Fischer OM, Streit S, Hart S
and Ullrich A: The epidermal growth factor receptor family as a
central element for cellular signal transduction and
diversification. Endocr Relat Cancer. 8:11–31. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun G, Liu B, He J, Zhao X and Li B:
Expression of EGFR is closely related to reduced 3-year survival
rate in Chinese female NSCLC. Med Sci Monit. 21:2225–2231. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hirsch FR, VarellaGarcia M, McCoy J, West
H, Xavier AC, Gumerlock P, Bunn PA Jr, Franklin WA, Crowley J,
Gandara DR; Southwest Oncology Group, ; et al: Increased epidermal
growth factor receptor gene copy number detected by fluorescence in
situ hybridization associates with increased sensitivity to
gefitinib in patients with bronchioloalveolar carcinoma subtypes: A
Southwest Oncology Group Study. J Clin Oncol. 23:6838–6845. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rotella V, Fornaro L, Vasile E, Tibaldi C,
Boldrini L, Chella A, D'Incecco A, Cirigliano G, Chioni A, Lupi C,
et al: EGFR and K-Ras mutations in women with lung adenocarcinoma:
Implications for treatment strategy definition. J Exp Clin Cancer
Res. 33:772014. View Article : Google Scholar : PubMed/NCBI
|