Introduction
Epithelioid hemangioendothelioma (EHE) is a rare
disease that was initially described by Weiss and Enzinger in 1982
(1). EHE develops from a malignant
transformation of the vascular endothelium and is generally located
in soft tissues and internal organs (1). Hepatic epithelioid hemangioendothelioma
(HEH) is the most common type of EHE; forms of EHE in other organs,
including the lungs, brain, spleen, bones, breast, heart and
stomach have been reported only rarely (2–6). HEH is a
rare sarcoma of the liver, which typically presents as a number of
nodules and may be misdiagnosed as a metastatic carcinoma (7,8).
Additionally, HEH may recur locally and metastasize to distant
organs (8); HEH has previously been
considered an intermediate malignancy (9). However, the World Health Organization
has previously categorized EHE as having full malignant potential
(10). Therefore, it is crucial to
identify the grade of malignancy and to distinguish HEH from other
types of epithelioid vascular tumors, including epithelioid
hemangioma and epithelioid angiosarcoma. However, due to the
morphological similarities, it is challenging to differentiate
these tumors solely using histological features (7). The current study focused on microRNAs
(miRNAs) and performed miRNA profiling, in order to identify novel
specific biomarkers for the accurate diagnosis of HEH.
Case report
A 72-year-old female was referred to Kagawa
University Hospital in 2014 (Kagawa, Japan) for further evaluation
of epigastralgia. The patient had no significant medical history
and no familial history of genetic disorders or cancers. Abdominal
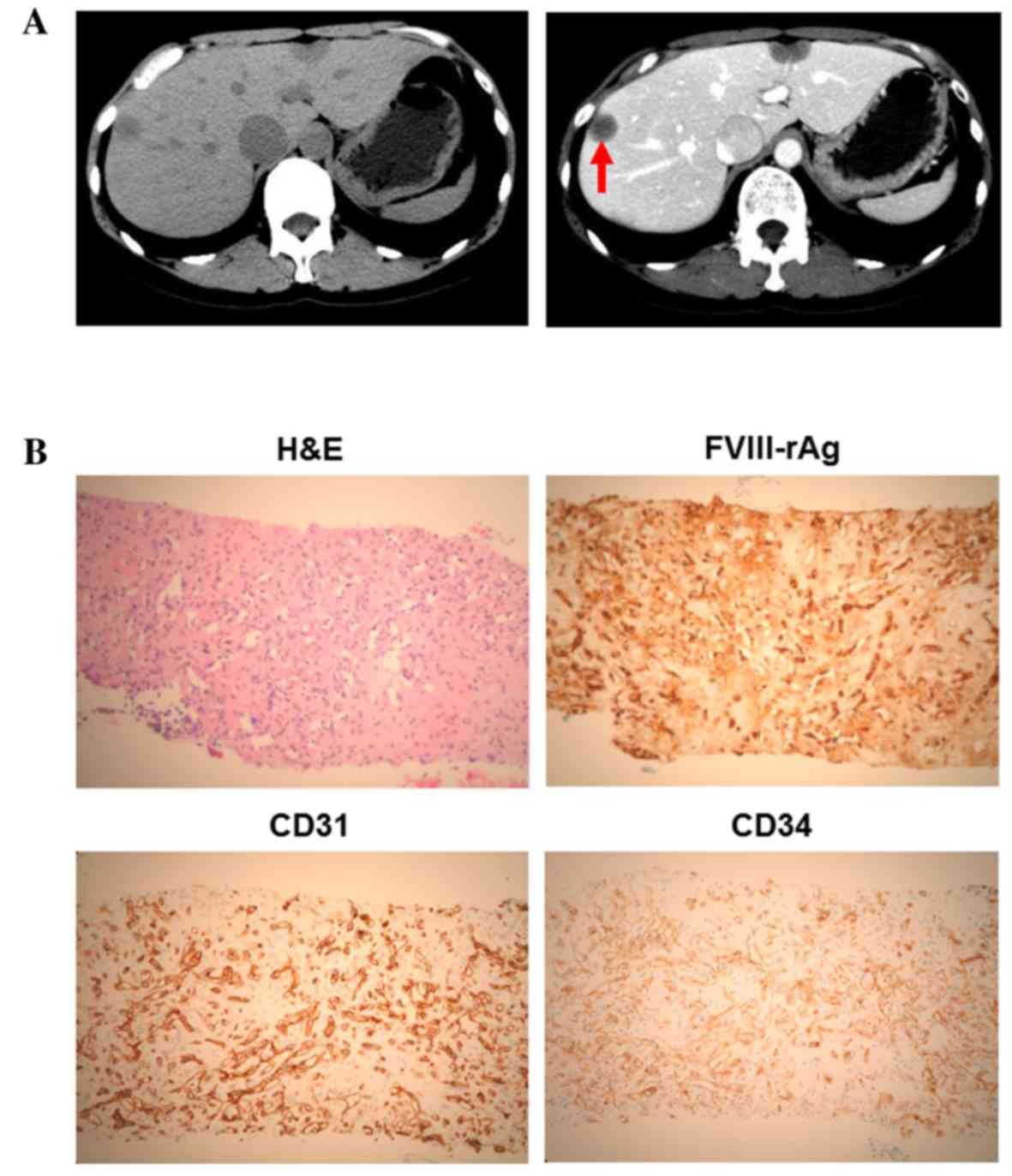
contrast computed tomography (CT) revealed a number of liver tumors
(Fig. 1A); multiple nodules with
capsules located peripherally in the liver were apparent, with
coarse calcification and peripheral enhancement following the
administration of contrast medium. A liver biopsy and
immunohistochemical staining revealed that the tumor cells were
positive for factor VIII-associated antigen (FVIII-rAg), platelet
endothelial cell adhesion molecule 1 (CD31) and human hematopoietic
progenitor cell antigen (CD34), consistent with a diagnosis of HEH
(Fig. 1B). Primary antibodies against
human FVIII-rAg (Dako, Tokyo, Japan), CD31 (Dako, JC70A, Tokyo,
Japan), and CD34 (Leica Microsystems, Tokyo, Japan) were added to
paraffin-embedded liver sections. Secondary antibodies, including
ImmPRESS™ REAGENT Anti-Rat Ig and ImmPRESS™ REAGENT Anti-Rabbit Ig,
were also used (Vector Laboratories, Inc., Burlingame, CA, USA) and
incubated with a DAB Substrate kit (Vector Laboratories, Inc.).
The patient was treated with a partial
ultrasonography-guided hepatectomy for the multiple HEHs. Currently
the patient is still alive and has been relapse-free for 2 years
post-surgery.
To elucidate the etiology of HEH, particularly the
miRNA profiles, tissue samples obtained from three separate
sections of normal and tumor tissues were analyzed. Total RNA was
extracted from the human liver samples using the miRNeasy Mini kit
(Qiagen, Inc., Venlo, Netherlands), according to the manufacturer's
instructions. Following RNA measurement with an RNA 6000 Nano kit
(Agilent Technologies, Santa Clara, CA, USA), the samples were
labeled using a miRCURY Hy3 Power Labeling kit (Exiqon, Vedbaek,
Denmark) and then hybridized onto a mouse miRNA Oligo chip (version
19; Toray Industries, Inc., Tokyo, Japan). Scanning was conducted
with the 3D-Gene Scanner 3000 (Toray Industries, Inc.). The 3D-Gene
extraction version 1.2 software (Toray Industries, Inc.) was used
to read the raw intensities of the image. To determine changes in
miRNA expression between normal tissue and tumor tissue samples,
the raw data were analyzed via GeneSpringGX version 10.0 (Agilent
Technologies, Inc., Tokyo, Japan).
A total of 14 miRNAs were significantly upregulated
and 93 miRNAs were significantly downregulated in the tumor
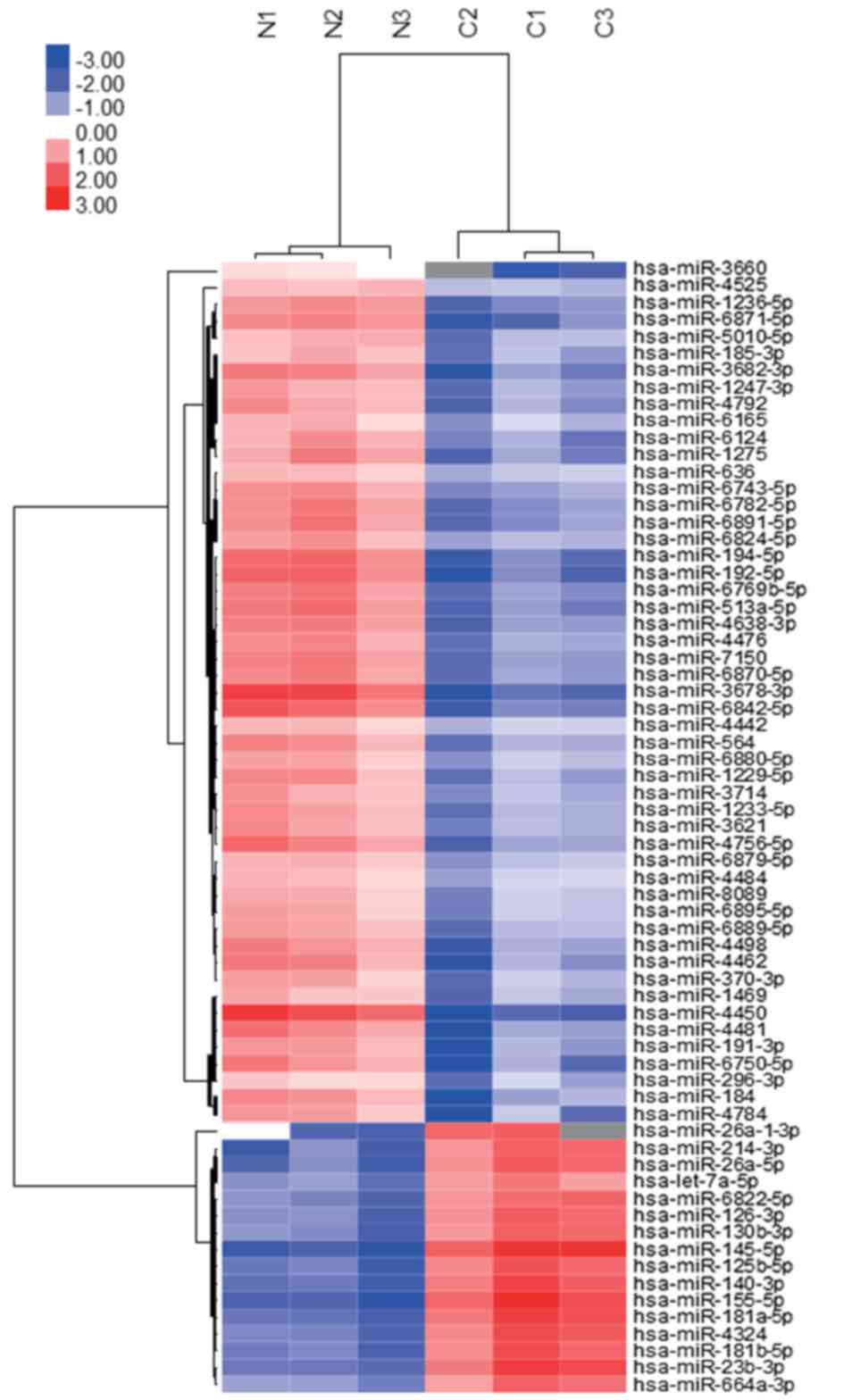
tissues, as compared with the normal tissues (Table I). Additionally, these data were
organized using unsupervised hierarchical clustering and analysis
of variance (ANOVA) functions in the GeneSpring software.
Unsupervised hierarchical clustering was performed by using the
clustering function (condition tree) with Pearson's correlation.
Two-way ANOVA and asymptotic P-value computation without any error
correction on the samples were performed to identify the miRNAs
that varied most prominently across the different groups. The
P-value cutoff was set as 0.05. In the present study, the miRNAs in
tumor tissues clustered separately from those in normal tissues
(Fig. 2). The microarray data used in
the present study were submitted as a complete data set to the NCBI
Gene Expression Omnibus (no. GSE66794). Each expression of 107
miRNAs, which were statistically up-and down-regulated in HEH
tissues, was analyzed as compared to that profile in angiosarcoma
which exhibits a similar histology to HEH by using
‘Sarcoma-microRNA Database’. The sarcoma miRNA expression database
was accessed through http://www.oncomir.umn.edu/.
 | Table I.Comparison of miRNAs between normal
and HEH tumor tissues (P<0.01). |
Table I.
Comparison of miRNAs between normal
and HEH tumor tissues (P<0.01).
| A, Upregulated
miRNAs |
|---|
|
|---|
| miRNA | P-value | Fold change (T/N),
mean ± SD |
|---|
| hsa-miR-145-5p | 0.0059 | 26.4833±12.9514 |
| hsa-miR-155-5p | 0.0099 | 20.5192±8.659 |
| hsa-miR-181a-5p | 0.0071 | 13.1044±4.978 |
| hsa-miR-23b-3p | 0.0096 | 12.362±4.993 |
| hsa-miR-140-3p | 0.0076 | 12.7157±5.4437 |
| hsa-miR-125b-5p | 0.0056 | 11.0117±4.9514 |
| hsa-miR-4324 | 0.0096 | 10.1675±4.1997 |
| hsa-miR-214-3p | 0.0056 | 12.0701±7.5642 |
| hsa-miR-26a-5p | 0.0058 | 10.9722±5.6222 |
| hsa-miR-126-3p | 0.0064 | 8.2668±3.5601 |
| hsa-miR-130b-3p | 0.0074 | 8.3378±4.1174 |
| hsa-miR-6822-5p | 0.0055 | 7.8268±3.3700 |
| hsa-let-7a-5p | 0.0058 | 5.2697±1.8241 |
| hsa-miR-34c-3p | 0.0043 | 1.6188±0.1349 |
|
| B, Downregulated
miRNAs |
|
| miRNA | P-value | Fold change (T/N),
mean ± SD |
|
| hsa-miR-4463 | 0.0076 | 0.5185±0.1658 |
| hsa-miR-4713-5p | 0.0049 | 0.4745±0.1267 |
| hsa-miR-3191-5p | 0.0090 | 0.4616±0.1886 |
| hsa-miR-296-3p | 0.0059 | 0.441±0.1595 |
|
hsa-miR-1290a | 0.0089 | 0.4589±0.1978 |
| hsa-miR-6716-5p | 0.0066 | 0.4225±0.1967 |
| hsa-miR-4442 | 0.0016 | 0.4199±0.1121 |
| hsa-miR-4534 | 0.0056 | 0.4224±0.1531 |
| hsa-miR-4484 | 0.0019 | 0.4176±0.1387 |
| hsa-miR-615-3p | 0.0065 | 0.3907±0.114 |
| hsa-miR-6791-5p | 0.0067 | 0.4044±0.1801 |
| hsa-miR-636 | 0.0006 | 0.3943±0.0982 |
| hsa-miR-6808-5p | 0.0076 | 0.4024±0.143 |
| hsa-miR-4800-3p | 0.0039 | 0.3808±0.1669 |
| hsa-miR-6855-3p | 0.0022 | 0.3805±0.1372 |
| hsa-miR-4525 | 0.0014 | 0.3655±0.0679 |
| hsa-miR-6165 | 0.0053 | 0.3614±0.1487 |
| hsa-miR-7114-5p | 0.0065 | 0.3395±0.128 |
| hsa-miR-6879-5p | 0.0012 | 0.3438±0.1193 |
| hsa-miR-3622a-5p | 0.0078 | 0.3342±0.1694 |
| hsa-miR-4497 | 0.0081 | 0.3358±0.1514 |
| hsa-miR-8089 | 0.0024 | 0.3367±0.1433 |
| hsa-miR-6880-5p | 0.0025 | 0.3261±0.1262 |
| hsa-miR-7854-3p | 0.0068 | 0.3168±0.1401 |
| hsa-miR-6895-5p | 0.0027 | 0.316±0.1378 |
| hsa-miR-885-5p | 0.0080 | 0.3281±0.1762 |
|
hsa-miR-7110-5p | 0.0078 | 0.3198±0.1633 |
|
hsa-miR-6877-5p | 0.0062 | 0.3072±0.1433 |
|
hsa-miR-3194-5p | 0.0050 | 0.3039±0.1679 |
| hsa-miR-1469 | 0.0008 | 0.2858±0.1107 |
|
hsa-miR-5010-5p | 0.0048 | 0.2964±0.1442 |
| hsa-miR-370-3p | 0.0043 | 0.2966±0.1479 |
|
hsa-miR-4749-5p | 0.0099 | 0.3068±0.1709 |
| hsa-miR-6076 | 0.0084 | 0.2912±0.1304 |
|
hsa-miR-664b-5p | 0.0081 | 0.2968±0.1903 |
|
hsa-miR-4745-5p | 0.0043 | 0.276±0.0822 |
| hsa-miR-3714 | 0.0009 | 0.2668±0.0619 |
|
hsa-miR-6824-5p | 0.0053 | 0.2791±0.0895 |
|
hsa-miR-6825-5p | 0.0070 | 0.2823±0.1436 |
|
hsa-miR-6889-5p | 0.0007 | 0.2673±0.1206 |
|
hsa-miR-5585-3p | 0.0071 | 0.2779±0.1291 |
| hsa-miR-4428 | 0.0023 | 0.2574±0.1187 |
| hsa-miR-184 | 0.0069 | 0.2667±0.0888 |
|
hsa-miR-642b-3p | 0.0068 | 0.2536±0.1318 |
| hsa-miR-3621 | 0.0014 | 0.2451±0.0755 |
|
hsa-miR-6769a-5p | 0.0045 | 0.2436±0.1112 |
|
hsa-miR-1247-3p | 0.0002 | 0.2335±0.0803 |
|
hsa-miR-6757-5p | 0.0074 | 0.2381±0.1004 |
|
hsa-miR-1233-5p | 0.0009 | 0.2357±0.0937 |
| hsa-miR-8060 | 0.0031 | 0.2276±0.0573 |
|
hsa-miR-6825-3p | 0.0041 | 0.2303±0.0685 |
| hsa-miR-1254 | 0.0050 | 0.2409±0.1457 |
|
hsa-miR-3925-5p | 0.0096 | 0.248±0.0242 |
|
hsa-miR-6851-5p | 0.0047 | 0.2196±0.1064 |
|
hsa-miR-6748-5p | 0.0081 | 0.2358±0.1399 |
| hsa-miR-4257 | 0.0071 | 0.2184±0.090 |
|
hsa-miR-6743-5p | 0.0017 | 0.2173±0.0827 |
| hsa-miR-564 | 0.0020 | 0.2139±0.0959 |
| hsa-miR-8063 | 0.0060 | 0.2046±0.1137 |
|
hsa-miR-1229-5p | 0.0044 | 0.2149±0.0950 |
| hsa-miR-6124 | 0.0072 | 0.2171±0.1188 |
| hsa-miR-4792 | 0.0006 | 0.1942±0.072 |
| hsa-miR-4476 | 0.0019 | 0.2045±0.0877 |
| hsa-miR-4481 | 0.0094 | 0.1983±0.0423 |
| hsa-miR-4435 | 0.0025 | 0.1843±0.0804 |
|
hsa-miR-3150a-3p | 0.0043 | 0.1788±0.0622 |
|
hsa-miR-122-5pa | 0.0094 | 0.1912±0.0978 |
|
hsa-miR-6750-5p | 0.0097 | 0.1705±0.0376 |
| hsa-miR-4498 | 0.0011 | 0.1771±0.0938 |
|
hsa-miR-6775-3p | 0.0038 | 0.167±0.0633 |
|
hsa-miR-6870-5p | 0.0032 | 0.1774±0.0767 |
|
hsa-miR-3682-3p | 0.0044 | 0.1664±0.0194 |
| hsa-miR-765 | 0.0033 | 0.1515±0.081 |
| hsa-miR-8071 | 0.0045 | 0.1602±0.0773 |
|
hsa-miR-6782-5p | 0.0042 | 0.1646±0.0752 |
|
hsa-miR-1236-5p | 0.0006 | 0.157±0.0564 |
| hsa-miR-7150 | 0.0022 | 0.161±0.0669 |
|
hsa-miR-4756-5p | 0.0024 | 0.1585±0.081 |
|
hsa-miR-6891-5p | 0.0073 | 0.1647±0.0828 |
|
hsa-miR-4638-3p | 0.0009 | 0.1561±0.0716 |
|
hsa-miR-6769b-5p | 0.0031 | 0.1561±0.0639 |
|
hsa-miR-6871-5p | 0.0022 | 0.1439±0.0416 |
| hsa-miR-4443 | 0.0071 | 0.1427±0.0639 |
|
hsa-miR-513a-5p | 0.0038 | 0.1287±0.0552 |
| hsa-miR-8078 | 0.0061 | 0.1079±0.0482 |
|
hsa-miR-6842-5p | 0.0012 | 0.0943±0.0427 |
| hsa-miR-194-5p | 0.0015 | 0.0948±0.0434 |
|
hsa-miR-6893-5p | 0.0045 | 0.0858±0.0367 |
| hsa-miR-192-5p | 0.0016 | 0.0816±0.0414 |
|
hsa-miR-3162-5p | 0.0063 | 0.0799±0.0308 |
| hsa-miR-4450 | 0.0052 | 0.0558±0.0073 |
|
hsa-miR-3678-3p | 0.0026 | 0.0546±0.0283 |
| hsa-miR-3649 | 0.0045 | 0.0417±0.0192 |
Of a total of 107 significantly upregulated and
downregulated miRNAs identified in HEH, only miR-122-5p and
miR-1290 demonstrated a differential expression pattern in
angiosarcoma (Table I).
Discussion
HEH is a rare hepatic, vascular, soft-tissue tumor
with malignant potential; the incidence of HEH is <1/100,000
(11). The majority of patients
present with nonspecific symptoms. HEH was first identified
incidentally by imaging, including ultrasonography and computed
tomography (12). The etiological
factors of HEH require further study; however, various risk factors
have been identified, including oral contraceptives, alcohol,
trauma, viral infections and chronic liver diseases (11,12).
Thorotrast (thorium dioxide) (13)
and vinyl chloride (14) have also
been reported as risk factors.
Laboratory findings, including biochemical
examination of blood, are typically non-diagnostic in cases of EHE.
Tumor markers, including α-fetoprotein, carcinoembryonic antigen
and carbohydrate antigen 19–9 have a potential role in excluding
the diagnosis of other hepatic neoplasms (15,16).
Examination of blood samples has revealed that an increase in
alkaline phosphatase is the most frequently observed abnormality of
HEH (12). Radiological studies,
including CT, magnetic resonance imaging and angiography indicate
that there are two subtypes of HEH: The nodular subtype manifests
as multifocal nodules in the early stages of HEH, and these nodules
grow and eventually coalesce, whereas the diffuse subtype manifests
as large confluent masses preferentially involving the peripheral
liver (17). In the present case,
multiple nodules with capsules were observed, located peripherally
in the liver, appearing with coarse calcification and peripheral
enhancement when imaged using a contrast medium (Fig. 1A).
A definitive diagnosis of HEH requires
histopathological examination; the predominant histological
features include nests and cords of epithelioid endothelial cells
and the presence of intracytoplasmic lumina. CD31, CD34 and
FVIII-rAg are established endothelial cell markers that are
commonly used for the diagnosis of EHE (18). As shown in Fig. 1B, the immunohistochemistry of HEH
tissue samples revealed the presence of these endothelial
markers.
EHE has previously been considered as an
intermediate malignancy; however, the World Health Organization has
now classified EHE as having full malignant potential (9,10). The
metastatic rate of EHE is ~25% and mortality occurs in ~15% of all
cases (1,9). By contrast, angiosarcoma of deep soft
tissues metastasizes in ~50% of epithelioid angiosarcomas, and
mortality occurs in over half of patients within a year of the
diagnosis (19). Prognostic factors
to distinguish the two biological subsets of tumors require further
study to be identified.
In the present study, miRNA profiles were examined
using tumor tissue samples and adjacent normal tissue samples to
identify the distinguishing features of EHE and angiosarcoma.
Unsupervised hierarchical clustering analysis using Pearson's
correlation demonstrated that the tumor tissues clustered
separately from the normal tissues (Fig.
2). Of the 107 significantly upregulated and downregulated
miRNAs identified in HEH, only miR-122-5p and miR-1290 demonstrated
a differential expression pattern between HEH and angiosarcoma
(Table I). miR-122-5p has previously
been identified to be downregulated in patients with hepatocellular
carcinoma, as compared with the healthy controls (20). The expression of miR-1290 has also
previously been demonstrated to be associated with breast cancer
prognosis (21). Interestingly, those
previous reports indicate that miR-122-5p and miR-1290 were
down-regulated in epithelial cancer cells. Therefore, the results
of the present study suggest that EHE has similar miRNA expression
patterns to epithelial cancer cells and that these expression
patterns in EHE may form a different phenotype from that in
angiosarcoma. In conclusion, miRNA profiling has potential as a
novel biological diagnostic tool for HEH. Furthermore, miR-122-5p
and miR-1290 are potential biomarkers for the accurate diagnosis of
HEH in primary mesenchymal tumors of the liver.
References
|
1
|
Weiss SW and Enzinger FM: Epithelioid
hemangioendothelioma: A vascular tumor often mistaken for a
carcinoma. Cancer. 50:970–981. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tiu CM, Chou YH, Wang HT and Chang T:
Epithelioid hemangioendothelioma of spleen with intrasplenic
metastasis: Ultrasound and computed-tomography appearance. Comput
Med Imaging Graph. 16:287–290. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee KC, Ng WF and Chan JK: Epithelioid
haemangioendothelioma presenting as a gastric polyp.
Histopathology. 12:335–337. 1988.PubMed/NCBI
|
|
4
|
Ellis GL and Kratochvil FJ III:
Epithelioid hemangioendothelioma of the head and neck: A
clinicopathologic report of twelve cases. Oral Surg Oral Med Oral
Pathol. 61:61–68. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marchiano D, Fisher F and Hofstetter S:
Epithelioid hemangioendothelioma of the heart with distant
metastases. A case report and literature review. J Cardiovasc Surg
(Torino). 34:529–533. 1993.PubMed/NCBI
|
|
6
|
Kamath SM, Nagaraj HK and Mysorekar VV:
Hepatic and adrenal hemangioendothelioma-a case report. J Clin
Diagn Res. 7:2583–2584. 2013.PubMed/NCBI
|
|
7
|
Lee SE, Park HY, Kim S, Bang H, Min JH and
Choi YL: Epithelioid hemangioendothelioma with extensive cystic
change and CAMTA1 rearrangement. Pathol Int. 63:502–505. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pokharna RK, Garg PK, Gupta SD, Dutta U
and Tandon RK: Primary epithelioid haemangioendothelioma of the
liver: Case report and review of the literature. J Clin Pathol.
50:1029–1031. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mentzel T, Beham A, Calonje E, Katenkamp D
and Fletcher CD: Epithelioid hemangioendothelioma of skin and soft
tissues: Clinicopathologic and immunohistochemical study of 30
cases. Am J Surg Pathol. 21:363–374. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Moulai N, Chavanon O, Guillou L, Noirclerc
M, Blin D, Brambilla E and Lantuejoul S: Atypical primary
epithelioid hemangioendothelioma of the heart. J Thorac Oncol.
1:188–189. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Neofytou K, Chrysochos A, Charalambous N,
Dietis M, Petridis C, Andreou C and Petrou A: Hepatic epithelioid
hemangioendothelioma and the danger of misdiagnosis: Report of a
case. Case Rep Oncol Med. 2013:2439392013.PubMed/NCBI
|
|
12
|
Mehrabi A, Kashfi A, Fonouni H, Schemmer
P, Schmied BM, Hallscheidt P, Schirmacher P, Weitz J, Friess H,
Buchler MW and Schmidt J: Primary malignant hepatic epithelioid
hemangioendothelioma: A comprehensive review of the literature with
emphasis on the surgical therapy. Cancer. 107:2108–2121. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Soslow RA, Yin P, Steinberg CR and Yang
GC: Cytopathologic features of hepatic epithelioid
hemangioendothelioma. Diagn Cytopathol. 17:50–53. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Darras T, Moisse R and Colette JM:
Epithelioid hemangioendothelioma of the liver. J Belge Radiol.
71:722–723. 1988.PubMed/NCBI
|
|
15
|
Okuda K, Kotoda K, Obata H, Hayashi N and
Hisamitsu T: Clinical observations during a relatively early stage
of hepatocellular carcinoma, with special reference to serum
alpha-fetoprotein levels. Gastroenterology. 69:226–234.
1975.PubMed/NCBI
|
|
16
|
Haglund C, Lindgren J, Roberts PJ and
Nordling S: Difference in tissue expression of tumour markers CA
19–9 and CA 50 in hepatocellular carcinoma and cholangiocarcinoma.
Br J Cancer. 63:386–389. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miller WJ, Dodd GD III, Federle MP and
Baron RL: Epithelioid hemangioendothelioma of the liver: Imaging
findings with pathologic correlation. AJR Am J Roentgenol.
159:53–57. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Makhlouf HR, Ishak KG and Goodman ZD:
Epithelioid hemangioendothelioma of the liver: A clinicopathologic
study of 137 cases. Cancer. 85:562–582. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Meis-Kindblom JM and Kindblom LG:
Angiosarcoma of soft tissue: A study of 80 cases. Am J Surg Pathol.
22:683–697. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tan Y, Ge G, Pan T, Wen D and Gan J: A
pilot study of serum microRNAs panel as potential biomarkers for
diagnosis of nonalcoholic fatty liver disease. PLoS One.
9:e1051922014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Endo Y, Yamashita H, Takahashi S, Sato S,
Yoshimoto N, Asano T, Hato Y, Dong Y, Fujii Y and Toyama T:
Immunohistochemical determination of the miR-1290 target arylamine
N-acetyltransferase 1 (NAT1) as a prognostic biomarker in breast
cancer. BMC Cancer. 14:9902014. View Article : Google Scholar : PubMed/NCBI
|