Introduction
Prostate cancer is one of the most common types of
cancer in men, with >40,000 new cases diagnosed every year in
the UK (1). Prostate cancer is
typically slow-growing and the majority of men do not notice the
symptoms until the cancer has become large enough to press against
the urethra and interfere with urination. The standard treatment
approach for prostate cancer is surgical removal of the gland or
prostatectomy, coupled with chemotherapy and radiotherapy (1). These approaches may be followed by
hormone therapy, which interrupts the cancer cells' supply of
testosterone, and therefore inhibits their ability to grow. The
high incidence of prostate cancer and the lack of treatment
efficacy of traditional treatment seriously affects the normal life
of men (1).
Tumors are caused by abnormal cell proliferation,
differentiation and apoptosis due to the activation of certain
proto-oncogenes, inactivation of tumor suppressors and changes to
apoptosis-associated genes (2,3). p53
upregulated modulator of apoptosis (PUMA), also known as
Bcl-2-binding component 3 (BBC3), is a pro-apoptotic protein and is
a member of the B-cell lymphoma 2 (Bcl-2) protein family (4,5). The
underlying mechanism of PUMA-mediated apoptosis has been
extensively evaluated (6–9). The tumor suppressor protein p53 has a
dual role associated with PUMA gene expression and function. The
efficiency of PUMA as an apoptosis-inducing protein and its
association with p53 depends on the cell lineage, the status of p53
(deficiency vs. mutation) and the type of stimulus. Thus, PUMA
activates apoptosis through p53-dependent and -independent
signaling pathways, induced by a range of signals (10–13). In
the majority of cell types, p53 expression results in increased
PUMA gene expression, and subsequent PUMA-mediated apoptosis
requires functional p53. The majority of PUMA-induced apoptosis
occurs via activation of p53 (10–12). p53
is activated by survival signals, including glucose deprivation
(14), which leads to an increase in
the expression levels of PUMA. PUMA expression leads to apoptosis
by displacement of p53 from Bcl-extra large (xL), allowing p53 to
increase mitochondrial permeability (15). The subsequent increase in PUMA levels
is able to induce apoptosis via mitochondrial dysfunction. p53, as
well as PUMA, is activated as a result of DNA damage caused by a
range of genotoxic agents. Alternative agents that are able to
induce p53-dependent apoptosis are neurotoxins (16,17),
proteasome inhibitors (18),
microtubule poisons (19) and
transcription inhibitors (20).
However, in certain cell types, PUMA apoptosis may additionally be
induced independently of p53 activation by alternative stimuli,
including oncogenic stress (21,22),
growth factor and/or cytokine withdrawal and kinase inhibition, ER
stress, altered redox status, ischemia, immune modulation, and
infection (23,24). Regardless of the method of signaling
pathway activation, PUMA interacts with the anti-apoptotic proteins
Bcl-2, Bcl-xL, Bcl-W and myeloid cell leukemia 1, via its BH3
structural domain, to induce cell apoptosis (5,25) and
obstructs the interaction of these proteins with the pro-apoptotic
molecules Bcl-2-like protein 4 (Bax) and Bcl-2 homologous
antagonist/killer (Bak), therefore freeing Bax and/or Bak, which
are subsequently able to signal apoptosis to the mitochondria
(5,23,25).
Following mitochondrial dysfunction, the caspase cascade is
activated, which leads to cell death (23).
A number of studies have demonstrated that PUMA
functioning is altered or absent in cancer cells (26,27). In
addition, the loss of p53-mediated apoptosis has been implicated as
a significant occurrence during tumor progression (26,28). The
majority of cancer types exhibit p53 gene deficiency or mutations,
meaning that the application of gene therapies that target this
gene is not possible (28). However,
an alternative potential method may involve focusing on PUMA as a
therapeutic target, leading to the induction of apoptosis in cancer
cells. In the present study, the influence of exogenous PUMA on
proliferation and apoptosis was investigated in prostate cancer
PC-3 cells, and it was determined whether PUMA requires functional
p53 in these cells.
Materials and methods
Cell culture and treatments
PC-3 human prostate adenocarcinoma cancer cells were
purchased from American Type Culture Collection (Manassas, VA,
USA). Cells were cultured with RPMI-1640 supplemented with 10%
heat-inactivated fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Cells were maintained in
culture at 37°C in a humidified atmosphere of 5% CO2 and
95% humidity.
Scrambled RNA and p53 small interfering RNA (siRNA)
were obtained from Dharmacon (GE Healthcare Life Sciences,
Chalfont, UK). pCEP4-[hemagglutinin (HA)] 2-PUMA recombinant
plasmid and non-carrier plasmid pCEP4-(HA) 2-C1 were kindly
provided by Dr B. Vogelstein (Johns Hopkins Oncology Center,
Baltimore, MD, USA). A total of 2×106 cells were
transfected by scrambled RNA or p53 siRNA using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) for 72 h according to
the manufacturer's protocol. Subsequently, the cells were cultured
in 100-mm dishes and transfected by PUMA or pCEP4 plasmid and left
to grow for 24 h, prior to detection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA was isolated from PC-3 cells with the Total RNA
Isolation kit (A&A Biotechnology, Gdynia, Poland) according to
the manufacturer's protocol. First strand complementary DNA (cDNA)
was obtained by reverse transcription of 1 µg of total RNA with the
RevertAid First Strand cDNA synthesis kit (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Amplification of the cDNA was performed using the
TaqMan® Gene Expression Assay (Applied Biosystems;
Thermo Fisher Scientific, Inc.), with fluorogenic fluorescein
amidite-labeled probes, sequence-specific primers of gene coding
p53 and the internal control glyceraldehyde-3-phosphate
dehydrogenase (GAPDH), according to the manufacturer's protocol.
Samples with no cDNA or no reverse transcription were used as
controls. Genes were amplified by a first step of 120 sec at 95°C,
followed by 45 cycles of 30 sec at 95°C, 30 sec at 60°C and 30 sec
at 72°C. The real-time fluorescence detection was performed with
the ABI PRISM 7700 Sequence Detector (Perkin-Elmer Applied
Biosystem; Thermo Fisher Scientific, Inc.). Fold differences in p53
expression, normalized to the level of GAPDH, were calculated with
the formula 2ΔΔCq (29).
Relative quantities of messenger RNA (mRNA) in siRNA-treated cells
were indicated as a percentage of the amount of mRNA in the
untreated cells.
Western blotting
Cells were lysed in cell lysis buffer (50 mM HEPES,
pH 8.0; 150 mM NaCl; 1% Triton X-100; 10% glycerol; 1 mM
MgCl2; 1.5 mM ethylenediaminetetraacetic acid; 20 mM
β-glycerophosphate; 50 mM NaF; 1 mM Na3VO4;
10 µg/ml aprotonin; 1 µM pepstatin A and 1 mM phenylmethylsulfonyl
fluoride; catalogue no. P0013; Beyotime Institute of Biotechnology,
Haimen, China) containing a protease inhibitor cocktail (Roche
Applied Science, Madison, WI, USA). Proteins were isolated from the
total cells using 14,000 × g centrifugation for 10 min at
4°C and the supernatant was collected. Protein samples (50 µg) were
separated by 12% sodium dodecyl sulfate polyacrylamide gel
electrophoresis and transferred to Immobilon-P polyvinyl difluoride
membranes (EMD Millipore, Billerica, MA, USA). The membrane was
blocked with 5% non-fat dried milk in Tris-buffered saline with
Tween 20 for 1 h at room temperature and then incubated overnight
at 4°C with mouse monoclonal anti-p53 (1:1,000 dilution; catalog
no. sc-126), anti-HA tag (1:2,000 dilution; catalog no. sc-7392),
rabbit polyclonal anti-p21 (1:2,000 dilution; catalog no. sc-397),
anti-Bcl-2 (1:2,000 dilution; catalog no. sc-783), anti-Bax
(1:2,000 dilution; catalog no. sc-526) and anti-GAPDH (1:2,000
dilution; catalog no. sc-25778) antibodies (Santa Cruz
Biotechnology Inc., Dallas, TX, USA). Goat anti-mouse (1:2,000
dilution; catalog no. sc-2005) and goat anti-rabbit immunoglobulin
G (1:2,000 dilution; catalog no. sc-2004) were used for incubation
for 2 h at room temporature. Enhanced chemiluminescence-detecting
reagent (GE Healthcare Life Sciences) was used to visualize the
membranes. The protein blots were quantified by densitometry using
QuantityOne software (Bio-Rad Laboratories, Inc., Hercules, CA,
USA), and the amounts were expressed relative to the internal
reference GAPDH.
Cell proliferation assay
Cell proliferation was assessed by
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. Briefly, cells were seeded (1×104 cells per well)
into a 96-well flat-bottom plate following transfection. Cells were
cultured at 37°C in an atmosphere of 5% CO2 for 48 h,
followed by an additional 4 h of incubation subsequent to the
addition of 20 µl 5 mg/ml MTT (Sigma-Aldrich; Merck Millipore,
Darmstadt, Germany) to each well. Cells were lysed by addition of
200 µl dimethylsulfoxide (Sigma-Aldrich; Merck Millipore).
Absorbance was measured at 490 nm with an enzyme-linked
immunosorbent assay (ELISA) reader (Tecan Austria GmbH, Grödig,
Austria).
Cell apoptosis assay
Apoptosis was assessed using a Cell Death Detection
ELISAPLUS kit (Sigma-Aldrich; Merck Millipore). PC-3
cells (4×103) were seeded into each well of a 96-well
plate following transfection. A total of 9 h later, samples were
collected and ELISA was performed in accordance with the
manufacturer's protocol. The results are presented as the fold
induction compared with the control. To confirm the role of
apoptosis, caspase-3 activation was additionally determined in
transfected cells using the Caspase-3 (Active) Human ELISA kit
(Thermo Fisher Scientific, Inc.). PUMA or non-carrier plasmids were
transfected into p53 siRNA-transfected or scrambled
siRNA-transfected cells. The cells were subsequently cultured at
4.5×105 cells per well in 6-well plates. A total of 8 h
later, the cells were lysed and assayed for western blotting.
Statistical analysis
SPSS (version 11.0; SPSS, Inc., Chicago, IL, USA)
was used to analyze the experimental data. Data are presented as
the mean ± standard error. Statistical significance was analyzed by
Student's t-test using SPSS 11.0 software (SPSS, Inc., Chicago, IL,
USA). All experiments were repeated at least 3 times. P<0.05 was
considered to indicate a statistically significant difference.
Results
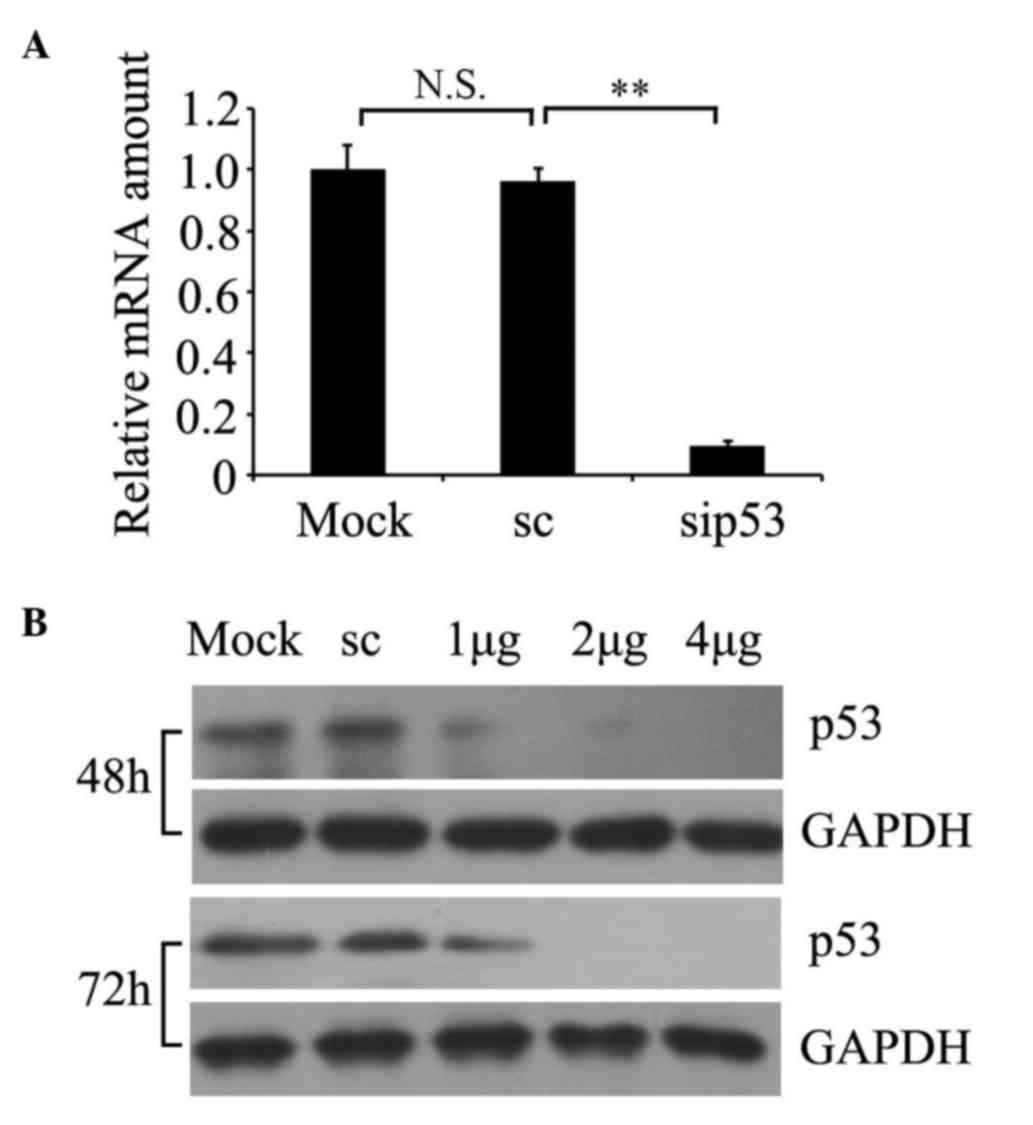
Silencing of p53 protein in prostate
cancer PC-3 cells
To determine whether siRNA was able to knockdown p53
expression in cultured prostate cancer PC-3 cells, p53 RNA
expression was examined by RT-qPCR (Fig.
1A) and protein expression was detected by western blotting
(Fig. 1B). A representative time
course and dose response is presented in Fig. 1B. The results demonstrated that the
relative p53 mRNA amount was significantly decreased in p53
siRNA-treated cells relative to non-silencing scrambled
siRNA-transfected cells (P=0.002; Fig.
1A), and the residual expression of p53 protein in the cells
with 2 µg of siRNA transfected for 72 h was markedly suppressed.
For the subsequent experiments, 2 µg of siRNA transfected for 72 h
was selected as the optimum condition.
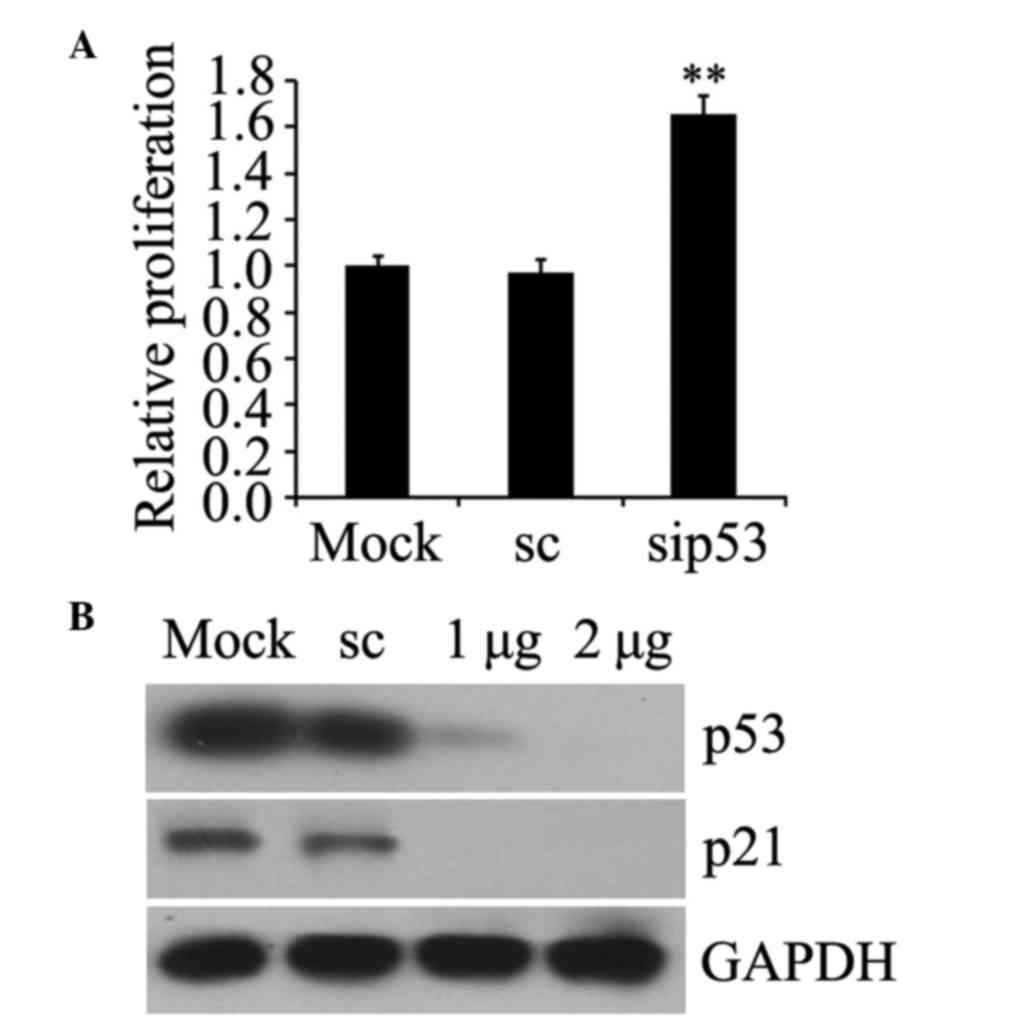
Effect of p53 deficiency on cellular
function
The functional effects of p53 knockdown were
assessed by detection of cell proliferation and p21 protein
expression, which is normally induced by p53 (30). The results revealed that the growth of
prostate cancer PC-3 cells was increased following p53 siRNA
transfection compared with the mock-transfected cells (P=0.007;
Fig. 2A) and the expression of p21
protein was blocked in p53-silenced cells (Fig. 2B). These data confirmed that p53
deficiency with siRNA contributed to relevant functional
alterations, which were consistent with p53 interference.
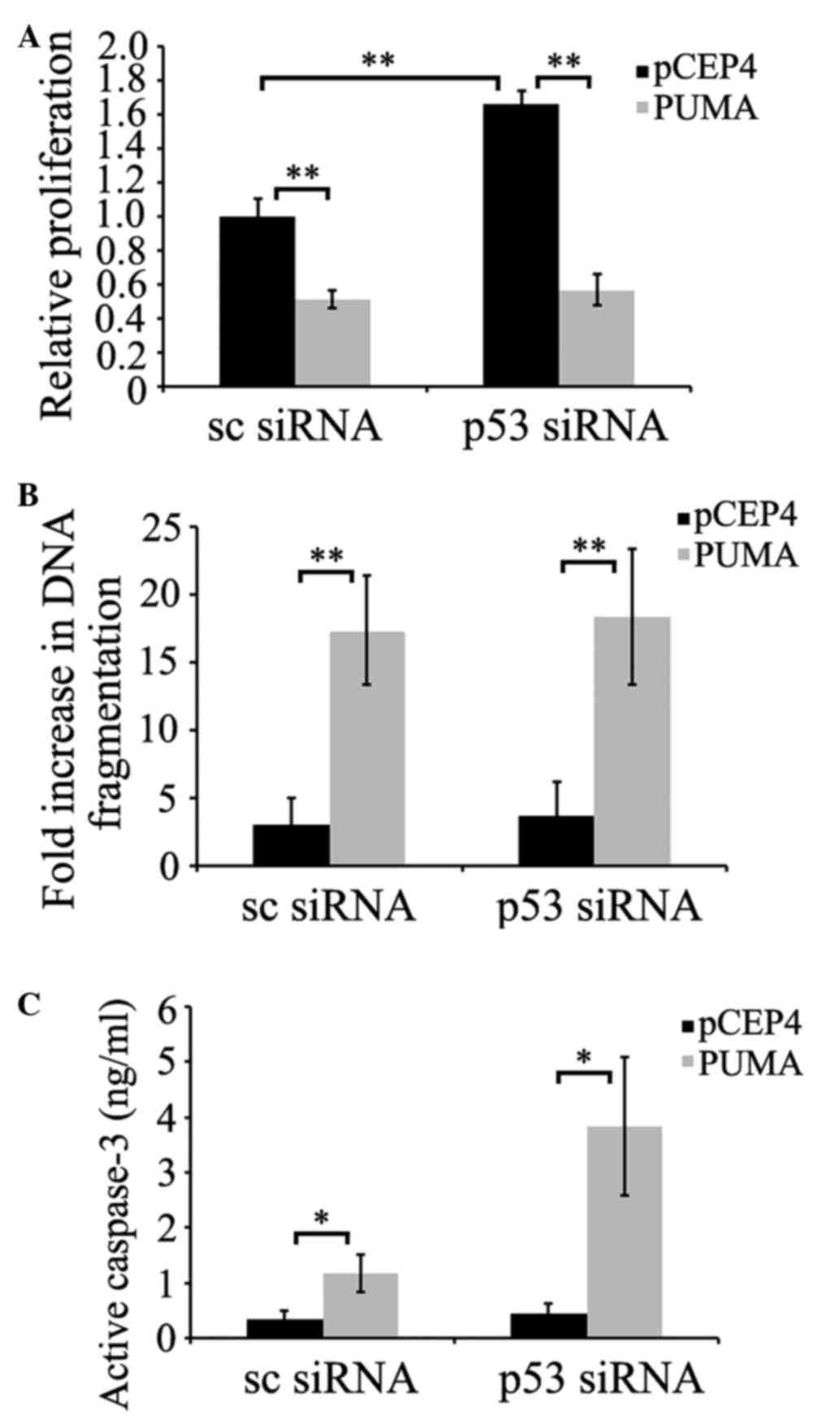
PUMA induces apoptosis independently
of p53
To confirm whether p53 was required for PUMA-induced
apoptosis, prostate cancer PC-3 cells were transfected with p53
siRNA. Once p53 reached its minimum level of expression 72 h
subsequent to transfection, cells were transfected with
pCEP4-(HA)-PUMA, or empty pCEP4 plasmids. As demonstrated in
Fig. 3A and B, PUMA-induced apoptosis
was assessed by measuring cell viability or histone release. The
results indicated that p53-silenced cells did not show significant
differences in PUMA-induced apoptosis levels compared with
scrambled siRNA-transfected cells (P=0.095 and P=0.126; Fig. 3A and B, respectively). PUMA-induced
apoptosis in scrambled siRNA-transfected cells was similar to that
in p53 siRNA-transfected cells. As additional evidence of
PUMA-mediated apoptosis, caspase-3 activation was evaluated in
p53-deficient PC-3 cells. The data demonstrated that PUMA induced
higher caspase-3 activation in p53-deficient PC-3 cells compared
with scrambled siRNA-transfected cells (P=0.045 and P=0.021;
Fig. 3C), suggesting that PUMA did
not require the participation of p53 to induce this apoptosis
signaling pathway.
Influence of exogenous PUMA on Bcl-2
and Bax protein expression in human prostate cancer PC-3 cells
The western blot analysis results revealed HA-PUMA
protein expression in scrambled siRNA-transfected and p53
siRNA-transfected cells, indicating successful transfection
(Fig. 4A). Compared with the control
transfected with empty pCEP4 plasmid, the PUMA group demonstrated
significantly increased expression of the pro-apoptotic protein Bax
and significantly reduced expression of the anti-apoptotic protein
Bcl-2, regardless of p53 deficiency (Fig.
4A). In addition, the Bax/Bcl-2 ratios were significantly
increased compared with the control with empty plasmid pCEP4
(P=0.003 and P=0.002; Fig. 4B). These
results verified that PUMA was able to engage the apoptotic
signaling pathway by modulating Bcl-2 and Bax protein expression
independently of p53.
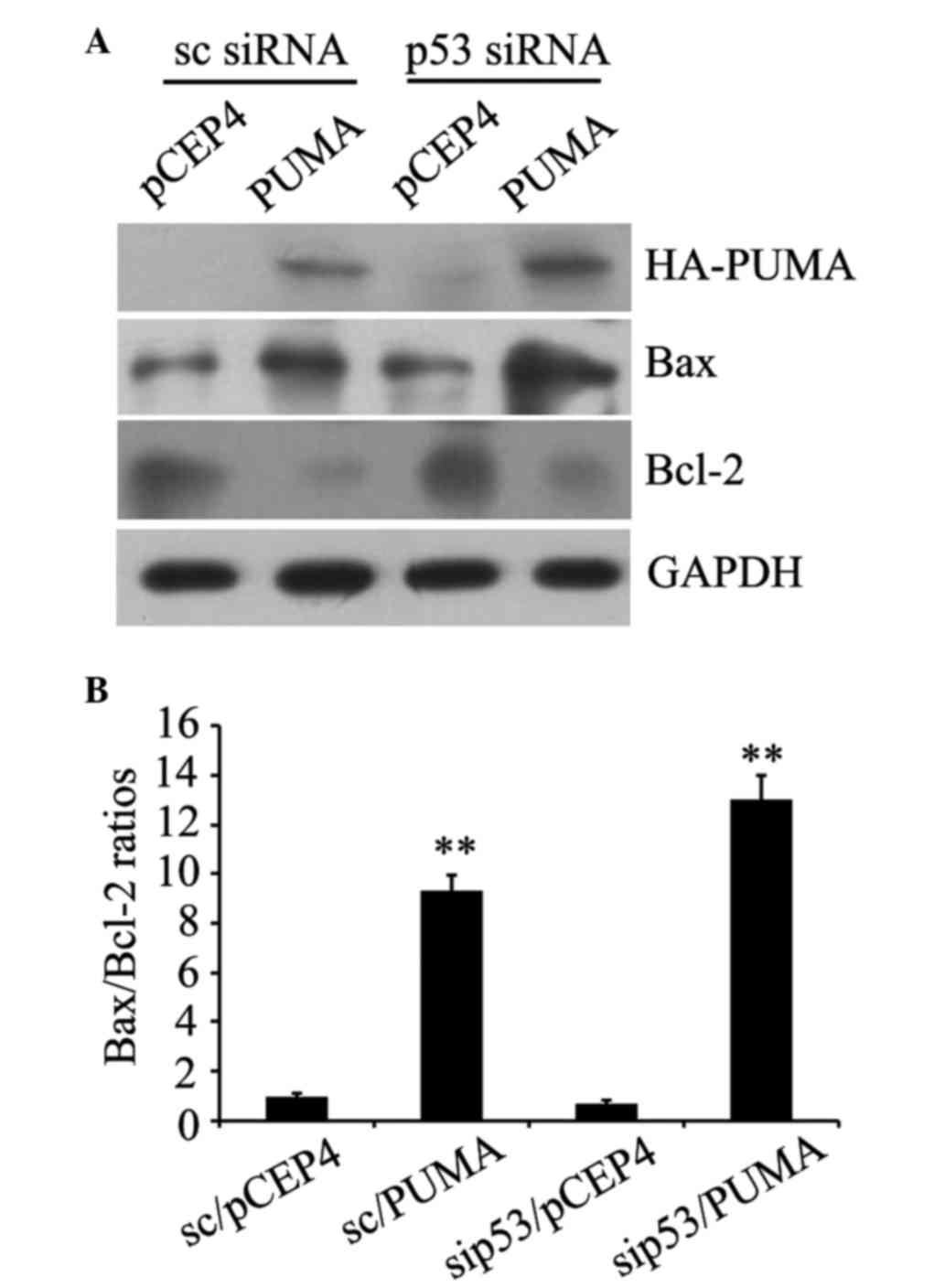 | Figure 4.Influence of exogenous PUMA on Bcl-2
and Bax protein expression in human prostate cancer PC-3 cells. (A)
Exogenous PUMA expression, and alterations in Bax or Bcl-2 protein
expression, in control and p53-deficient PC-3 cells were assessed
by western blot analysis, respectively, with anti-HA, anti-Bax or
anti-Bcl-2 antibodies. (B) Bax/Bcl-2 ratios were counted by the
quantitation of Bax protein blots divided by the quantitation of
Bcl-2 protein blots. **P<0.01 vs. pCET4 empty vector-transfected
control cells. Data are presented as the mean ± standard error.
PUMA, p53 upregulated modulator of apoptosis; Bcl-2, B-cell
lymphoma 2; Bax, bcl-2-like protein 4; HA, hemagglutinin; sc,
control; siRNA, small interfering RNA; GAPDH,
glyceraldehyde-3-phosphate dehydrogenase; sip53; siRNA
duplexes. |
Discussion
Prostate cancer is one of the most common malignant
tumors in men (1). Chemotherapy kills
tumor cells by activating apoptosis, while apoptotic signaling
pathway defects are associated with tumor resistance to
chemotherapy. Due to side effects and tumor cell insensitivities,
the clinical application of chemotherapeutic drugs may be greatly
restricted (31). A number of studies
concerning gene therapy have attracted widespread attention
(32–34). Therefore, searching for ideal target
genes has become a significant issue to address.
The introduction of apoptotic genes has the
potential to increase sensitivity to chemotherapy drugs. The loss
of p53-mediated apoptosis has been implicated as a significant
event in tumor progression. p53 is able to induce or potentiate
apoptosis via a number of mechanisms, including by regulating the
expression of genes that are able to participate in the apoptotic
response and via transcriptionally independent means. p53 is a
notable potential therapeutic gene as it is able to induce
apoptosis in a range of cell types. Defects in p53 structure and
function in specific unhealthy cells have been described,
suggesting that forced expression of this tumor suppressor protein
may be beneficial (35–38). However, enhancing p53 gene expression
has been observed to have only modest efficacy (39). PUMA has a significant role in
apoptotic signaling pathways, is rapidly induced by p53 and has
powerful apoptosis-promoting effects. Therefore, a possible
explanation for the limited efficacy of p53 gene enhancement may be
that p53 did not readily induce apoptosis, potentially because PUMA
expression was not increased (40).
Thus, directly targeting PUMA may be an effective treatment
approach to cause rapid cell death. In vitro studies have
demonstrated that PUMA affects tumor cell proliferation and induces
cellular apoptosis independently of p53, which has led to hopes of
using PUMA for tumor therapy (41,42). Yu
et al (43) reported that PUMA
induced lung cancer cell apoptosis and inhibited cell proliferation
via caspase activation and cytochrome c release in
non-chemoradiotherapy sensitive lung cancer cells, providing
evidence that PUMA is able to increase sensitivity to radiotherapy
and chemotherapy drugs.
In the present study, using siRNA to decrease p53
expression demonstrated that prostate cancer PC-3 cells are
sensitive to PUMA-induced death. Western blot analysis revealed
that, following PUMA gene transfection, pro-apoptotic protein Bax
significantly increased and anti-apoptotic protein Bcl-2
significantly decreased. In the Bcl-2 family, the ratio of
pro-apoptotic to anti-apoptotic proteins is a significant factor in
determining the occurrence and level of apoptosis (44). The proportion of pro-apoptotic
proteins in cells additionally determines the cellular response to
death signals and cell fate. It has been reported that decreased
Bax protein expression is associated with the sensitivity of tumor
cells to chemotherapy and the length of patient survival times
(45,46). Therefore, the results of the present
study suggested that PUMA was an effective mediator of apoptosis,
regardless of the p53 status in PC-3 cells.
In conclusion, the results of the present study
imply that PUMA is able to efficiently decrease the growth of
prostate cancer PC-3 cells by promoting apoptosis independently of
p53. This supports the potential use of PUMA as a novel and
promising target for prostate cancer therapy. A number of cancer
types exhibit deletions or mutations in p53, so PUMA shows great
significance in cancer treatment. In future studies, we will verify
whether the phenomenon exists in other cell lines of prostate
cancer. Furthermore, the potential functions of PUMA in the
chemosensitivity and treatment of protate cancer remain to be
investigated.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81302234), the Shandong
Provincial Natural Science Foundation, China (Grant no.
ZR2016HP38), Projects of Medical and Health Technology Development
program in Shandong province (Grant no. 2016WS0714), Science and
Technology Project of Yantai (grant nos. 2016ZH084 and 2016WS018)
and the Scientific research startup Project of Binzhou Medical
University (grant no. BY2014KYQD24).
References
|
1
|
Mandal A: Prostate Cancer, . NEWS MEDICAL.
The Latest Developments in Life Sciences & Medicine. http://www.news-medical.net/health/Prostate-Cancer.aspxAccessed.
February 27–2015.
|
|
2
|
Tsai SC, Lu CC, Lee CY, Lin YC, Chung JG,
Kuo SC, Amagaya S, Chen FN, Chen MY, Chan SF and Yang JS: AKT
serine/threonine protein kinase modulates bufalin-triggered
intrinsic pathway of apoptosis in CAL 27 human oral cancer cells.
Int J Oncol. 41:1683–1692. 2012.PubMed/NCBI
|
|
3
|
Umit UM, Berna T, Handan K, Ipek E, Berrak
Y, Can E and Bahadir GM: Role of melatonin and luzindole in rat
mammary cancer. J Invest Surg. 25:345–353. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakano K and Vousden KH: PUMA, a novel
proapoptotic gene, is induced by p53. Mol Cell. 7:683–694. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Han J, Flemington C, Houghton AB, Gu Z,
Zambetti GP, Lutz RJ, Zhu L and Chittenden T: Expression of bbc3, a
pro-apoptotic BH3-only gene, is regulated by diverse cell death and
survival signals. Proc Natl Acad Sci USA. 98:11318–11323. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu H, Li W, Yu X, Gao F, Duan Z, Ma X,
Tan S, Yuan Y, Liu L, Wang J, et al: EZH2-mediated Puma gene
repression regulates non-small cell lung cancer cell proliferation
and cisplatin-induced apoptosis. Oncotarget. Jul 26–2016.(Epub
ahead of print).
|
|
7
|
Jang Y, Kim J, Ko JW and Kwon YH:
Homocysteine induces PUMA-mediated mitochondrial apoptosis in
SH-SY5Y cells. Amino Acids. 48:2559–2569. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma J, Feng Y, Liu Y and Li X: PUMA and
survivin are involved in the apoptosis of HepG2 cells induced by
microcystin-LR via mitochondria-mediated pathway. Chemosphere.
157:241–249. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun Y, Xia P, Zhang H, Liu B and Shi Y:
P53 is required for Doxorubicin-induced apoptosis via the TGF-beta
signaling pathway in osteosarcoma-derived cells. Am J Cancer Res.
6:114–125. 2015.PubMed/NCBI
|
|
10
|
Wang P, Yu J and Zhang L: The nuclear
function of p53 is required for PUMA-mediated apoptosis induced by
DNA damage. Proc Natl Acad Sci USA. 104:4054–4059. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jeffers JR, Parganas E, Lee Y, Yang C,
Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, et al:
Puma is an essential mediator of p53-dependent and -independent
apoptotic pathways. Cancer Cell. 4:321–328. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Avila JL, Grundmann O, Burd R and Limesand
KH: Radiation-induced salivary gland dysfunction results from
p53-dependent apoptosis. Int J Radiat Oncol Biol Phys. 73:523–529.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Niizuma K, Endo H, Nito C, Myer DJ and
Chan PH: Potential role of PUMA in delayed death of hippocampal CA1
neurons after transient global cerebral ischemia. Stroke.
40:618–625. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao Y, Coloff JL, Ferguson EC, Jacobs SR,
Cui K and Rathmell JC: Glucose metabolism attenuates p53 and
Puma-dependent cell death upon growth factor deprivation. J Biol
Che. 283:36344–36353. 2008. View Article : Google Scholar
|
|
15
|
Chipuk JE, Bouchier-Hayes L, Kuwana T,
Newmeyer DD and Green DR: PUMA couples the nuclear and cytoplasmic
proapoptotic function of p53. Science. 309:1732–1735. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gomez-Lazaro M, Galindo MF,
Fernandez-Gomez FJ, Prehn JH and Jordán J: Activation of p53 and
the pro-apoptotic p53 target gene PUMA during
depolarization-induced apoptosis of chromaffin cells. Exp. Neurol.
196:96–103. 2005.
|
|
17
|
Wong HK, Fricker M, Wyttenbach A,
Villunger A, Michalak EM, Strasser A and Tolkovsky AM: Mutually
exclusive subsets of BH3-only proteins are activated by the p53 and
c-Jun N-terminal kinase/c-Jun signaling pathways during cortical
neuron apoptosis induced by arsenite. Mol Cell Biol. 25:8732–8747.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu J, Wang P, Ming L, Wood MA and Zhang L:
SMAC/Diablo mediates the proapoptotic function of PUMA by
regulating PUMA-induced mitochondrial events. Oncogene.
26:4189–4198. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Giannakakou P, Nakano M, Nicolaou KC,
O'Brate A, Yu J, Blagosklonny MV, Greber UF and Fojo T: Enhanced
microtubule-dependent trafficking and p53 nuclear accumulation by
suppression of microtubule dynamics. Proc Natl Acad Sci USA.
99:10855–10860. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kalousek I, Brodska B, Otevrelova P and
Röselova P: Actinomycin D upregulates proapoptotic protein Puma and
downregulates Bcl-2 mRNA in normal peripheral blood lymphocytes.
Anticancer Drugs. 18:763–772. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fernandez PC, Frank SR, Wang L, Schroeder
M, Liu S, Greene J, Cocito A and Amati B: Genomic targets of the
human c-Myc protein. Genes Dev. 17:1115–1129. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Maclean KH, Keller UB, Rodriguez-Galindo
C, Nilsson JA and Cleveland JL: c-Myc augments gamma
irradiation-induced apoptosis by suppressing Bcl-XL. Mol Cell Biol.
23:7256–7270. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu J and Zhang L: PUMA, a potent killer
with or without p53. Oncogene. 27:(Suppl 1). S71–S83. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Castedo M, Perfettini JL, Piacentini M and
Kroemer G: p53 - A pro-apoptotic signal transducer involved in
AIDS. Biochem Biophys Res Commun. 331:701–706. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yee KS and Vousden KH: Contribution of
membrane localization to the apoptotic activity of PUMA. Apoptosis.
13:87–95. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vogelstein B and Kinzler KW: Cancer genes
and the pathways they control. Nat Med. 10:789–799. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu J and Zhang L: The transcriptional
targets of p53 in apoptosis control. Biochem Biophys Res Commun.
331:851–858. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hao and Cho WC: Battle against cancer: An
everlasting saga of p53. Int J Mol Sci. 15:22109–22127. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak and Schmittgen, . Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pap T, Aupperle KR, Gay S, Firestein GS
and Gay RE: Invasiveness of synovial fibroblasts is regulated by
p53 in the SCID mouse in vivo model of cartilage invasion.
Arthritis Rheum. 44:676–681. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pommier Y, Sordet O, Antony S, Hayward RL
and Kohn KW: Apoptosis defects and chemotherapy resistance:
Molecular interaction maps and networks. Oncogene. 23:2934–2949.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hirscheler B: Europe gives green light to
first gene therapy for children. http://www.arabtimesonline.com/wp-content/uploads/pdf/2016/apr/03/29.pdfAccessed.
April 13–2016.
|
|
33
|
Coghlan A: Gene Therapy Approved. New
Scientist. 230:8–9. 2016. View Article : Google Scholar
|
|
34
|
Cyranoski D: Chinese scientists to pioneer
first human CRISPR trial. Nature. 535:476–477. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yamanishi Y, Boyle DL, Rosengren S, Green
DR, Zvaifler NJ and Firestein GS: Regional analysis of p53
mutations in rheumatoid arthritis synovium. Proc Natl Acad Sci USA.
99:10025–10030. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Firestein GS, Echeverri F, Yeo M, Zvaifler
NJ and Green DR: Somatic mutations in the p53 tumor suppressor gene
in rheumatoid arthritis synovium. Proc Natl Acad Sci USA.
94:10895–10900. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rème T, Travaglio A, Gueydon E, Adla L,
Jorgensen C and Sany J: Mutations of the p53 tumour suppressor gene
in erosive rheumatoid synovial tissue. Clin Exp Immunol.
111:353–358. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kullmann F, Judex M, Neudecker I, Lechner
S, Jüsten HP, Green DR, Wessinghage D, Firestein GS, Gay S,
Schölmerich J and Müller-Ladner U: Analysis of the p53 tumor
suppressor gene in rheumatoid arthritis synovial fibroblasts.
Arthritis Rheum. 42:1594–1600. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yao Q, Wang S, Glorioso JC, Evans CH,
Robbins PD, Ghivizzani SC and Oligino TJ: Gene transfer of p53 to
arthritic joints stimulates synovial apoptosis and inhibits
inflammation. Mol Ther. 3:901–910. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cha HS, Rosengren S, Boyle DL and
Firestein GS: PUMA regulation and proapoptotic effects in
fibroblast-like synoviocytes. Arthritis Rheum. 54:587–592. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Thin TH, Li L, Chung TK, Sun H and Taneja
R: Stra13 is induced by genotoxic stress and regulates
ionizing-radiation-induced apoptosis. EMBO Rep. 8:401–407. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dudgeon C, Peng R, Wang P, Sebastiani A,
Yu J and Zhang L: Inhibiting oncogenic signaling by sorafenib
activates PUMA via GSK3β and NF-κB to suppress tumor cell growth.
Oncogene. 31:4848–4858. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu J, Yue W, Wu B and Zhang L: PUMA
sensitizes lung cancer cells to chemotherapeutic agents and
irradiation. Clin Cancer Res. 12:2928–2936. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gupta S, Afaqa F and Mukhtar H:
Involvement of nuclear factor-kappa B, Bax and Bcl-2 in induction
of cell cycle arrest and apoptosis by apigenin in human prostate
carcinoma cells. Oncogene. 21:3727–3738. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xiao D, Vogel V and Singh SV: Benzyl
isothiocyanate-induced apoptosis in human breast cancer cells is
initiated by reactive oxygen species and regulated by Bax and Bak.
Mol Cancer Ther. 5:2931–2945. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xiao D, Lew KL, Kim YA, Zeng Y, Hahm ER,
Dhir R and Singh SV: Diallyl trisulfide suppresses growth of PC-3
human prostate cancer xenograft in vivo in association with Bax and
Bak induction. Clin Cancer Res. 12:6836–6843. 2006. View Article : Google Scholar : PubMed/NCBI
|